1. Introduction
Viruses are intracellular parasites that rely on their hosts for replication and transmission. While viral infections in humans are rarely fatal, mortality often occurs when viruses cross species barriers or when the immune system is weakened. Defenses against viruses typically involve various immune components, and the effectiveness of these mechanisms varies based on how specific viruses enter, replicate, and spread within the host [
1,
2].
COVID-19 is a disease caused by the SARS-CoV-2 virus, first reported in December 2019 in Wuhan, China. Coronaviruses are a large family of viruses that affect humans and a wide range of animals. SARS-CoV-2 evolves through mutations during genome replication, leading to the emergence of new variants [
2]. The pandemic reached Venezuela on March 13, 2020, with a significant rise in cases by mid-May. By February 19, 2021, health authorities had launched a national vaccination campaign using the Sputnik V and Sinopharm BBIBP-CorV vaccines [
3,
4].
Upon SARS-CoV-2 infection, the innate immune response is rapidly activated in host cells through pathogen recognition receptors (PRRs) that recognize viral components. This activation triggers the expression of virus-stimulated genes (VSGs) through transcription factors such as IRF3 and NF-κB, leading to the production of type I and III interferons (IFNs). These IFNs are secreted by infected cells, initiating a broader antiviral state by inducing interferon-stimulated genes (ISGs) in neighboring cells. Type I and III IFNs signal through the Jak-STAT pathway and differ in their receptor distribution. Dysregulation of IFN responses, influenced by factors such as the viral protein ORF9b and age, may enhance susceptibility to SARS-CoV-2, particularly in elderly individuals [
5,
6,
7,
8,
9]
Investigating adaptive immunity within the context of SARS-CoV-2 infection and vaccination is critical for achieving a comprehensive understanding of COVID-19 as a disease [
5,
6,
7,
8,
9]. Multiple studies have explored the interaction between innate and adaptive immunity, highlighting the significant roles of CD4+ and CD8+ T lymphocytes, B cells, and neutralizing antibodies [
10,
11,
12]. However, the roles of other cell populations, including monocytes, natural killer (NK) cells, natural killer T (NKT) cells, mucosal T cells (MAIT), follicular cells, and T gamma delta (Tγδ) cells, have been documented in infection and, rarely, upon vaccination [
13]. These cells may have important implications for COVID-19 vaccination strategies and the development of immune memory with reinfection and trained immunity [14}.
Recent advancements in quantifying antigen-specific T-cell responses have increasingly utilized MHC class I and II tetramers to visualize the responses of CD8+ and CD4+ T lymphocytes, respectively [
15,
16]. Research using MHC class I tetramers has demonstrated that a significant proportion of activated CD8+ T lymphocytes during viral infections are virus-specific [
15,
16]. CD8 cytotoxic T lymphocytes (CTLs) perform the function of recognizing and eliminating virus-infected cells through the mechanisms of perforin and granzymes or by engaging with Fas ligand on target lymphocytes, thereby inducing apoptosis [
16]. CD8+ T lymphocytes secrete cytokines such as IFN-γ and TNF-α, and their activation is regulated [
16,
17]. Normal T cells express and secrete RANTES, which exerts antiviral effects without necessarily leading to the death of the infected cells [
17].
CD8+ T lymphocyte responses have been recognized against SARS-CoV-2 S protein, M protein, N protein, nsp6, and ORF3a [
11,
17,
18]. Memory CD8+ T lymphocytes circulate by 20-50 days post-symptom onset and have a half-life of 225 days [
19]. The preponderance of these circulating CD8+ T lymphocytes is CD45RA (TEMRA), with lesser amounts of effector memory (TEM) and CD8 naïve central memory (TCM) [
20]. TEMRA plays a role in protection against severe disease, as shown in other viral infections [
21]. In the context of acute SARS-CoV-2 infection, virus-specific CD8+ T lymphocytes with cytolytic capacity against infected cells can be detected as early as day 1 post-symptom onset (PSO) [
21]. Their rapid induction commonly occurred within the first 7 days and peaked at 2 weeks PSO. This virus-specific CD8+ T lymphocyte dynamic has been associated with better COVID-19 outcomes (as has their capacity to produce high levels of cytotoxic effector molecules, such as IFN-γ, granzyme B, and perforin [
5,
6,
19,
21].
On the other hand, a group of CD8 cells expresses the CD314 (NKG2D) receptor, which is involved in the antiviral response [
22]. However, its activation is independent of antigens and is dependent on the expression of stress receptors, which serve as ligands for NKG2D, including MICA/B and ULBP. These bystander cells can aid in the elimination of infected cells that express those markers. The expression of CD314 is also essential for NK anti-viral and cytotoxic responses [
23].
This study aimed to determine the effectiveness of the Sinopharm vaccine/BIBP in inducing long-term immune memory in a group of Venezuelan adult volunteers. Upon stimulation with viral peptides and inactivated virus, CD4 cells (CD4/CD154), memory CD8 cells (CD8/CD107a), bystander CD8 cells (CD8/CD314), memory B cells (CD19/CD86), NK cells (CD56/CD314) and the secretion of IFNγ and granzyme B upon stimulation.
2. Materials and Methods
2.1. Characteristics of the Volunteers
The research study was conducted between February 2023 and June 2023. On February 15, 2023, approval for Study 001/2023 was granted by the Ethical Committee of the Institute of Immunology at the Faculty of Medicine of the Central University of Venezuela. Before undergoing heparin-anticoagulated venous sampling, the recruited volunteers were required to read and sign an informed consent form, thereby entrusting the Ethical Committee with oversight of the generated data. Additionally, the Ethical Committee authorized the submission of the manuscript associated with this study.
The following inclusion criteria were established for the study: 1) completion of the basic vaccination regimen of two doses of BBIBP-CorV vaccine [
24], 2) absence of any recent infectious diseases at the time of sample collection, including a negative SARS-CoV-2 antigen test, 3) no history of autoimmune diseases as determined through laboratory screening, 4) no administration of immunosuppressive treatment, 5) participation from individuals of a genetically admixed Venezuelan population, as verified in our laboratory, and 6) adult individuals. Pregnant women, individuals who tested positive for the SARS-CoV-2 antigen, those from diverse genetic backgrounds, and those who had received alternative vaccines were excluded from participation. A total of 52 individuals, comprising 30 females and 22 males, were successfully recruited for the study. Most individuals (58%) had a documented moderate SARS-CoV-2 infection, and 42% had a mild infection. All of them were highly exposed to the virus and were either healthcare or service personnel.
2.2. Screening of Anti-RGD S Protein
The antibodies against the receptor binding domain (RBD) of the S protein were analyzed using the commercial LEGEND MAX™ Spike SARS-CoV-2 (RBD) kit from BioLegend, following the manufacturer's instructions. The kit was previously validated at our Institution [
25].
2.3. Stimulation of the Samples
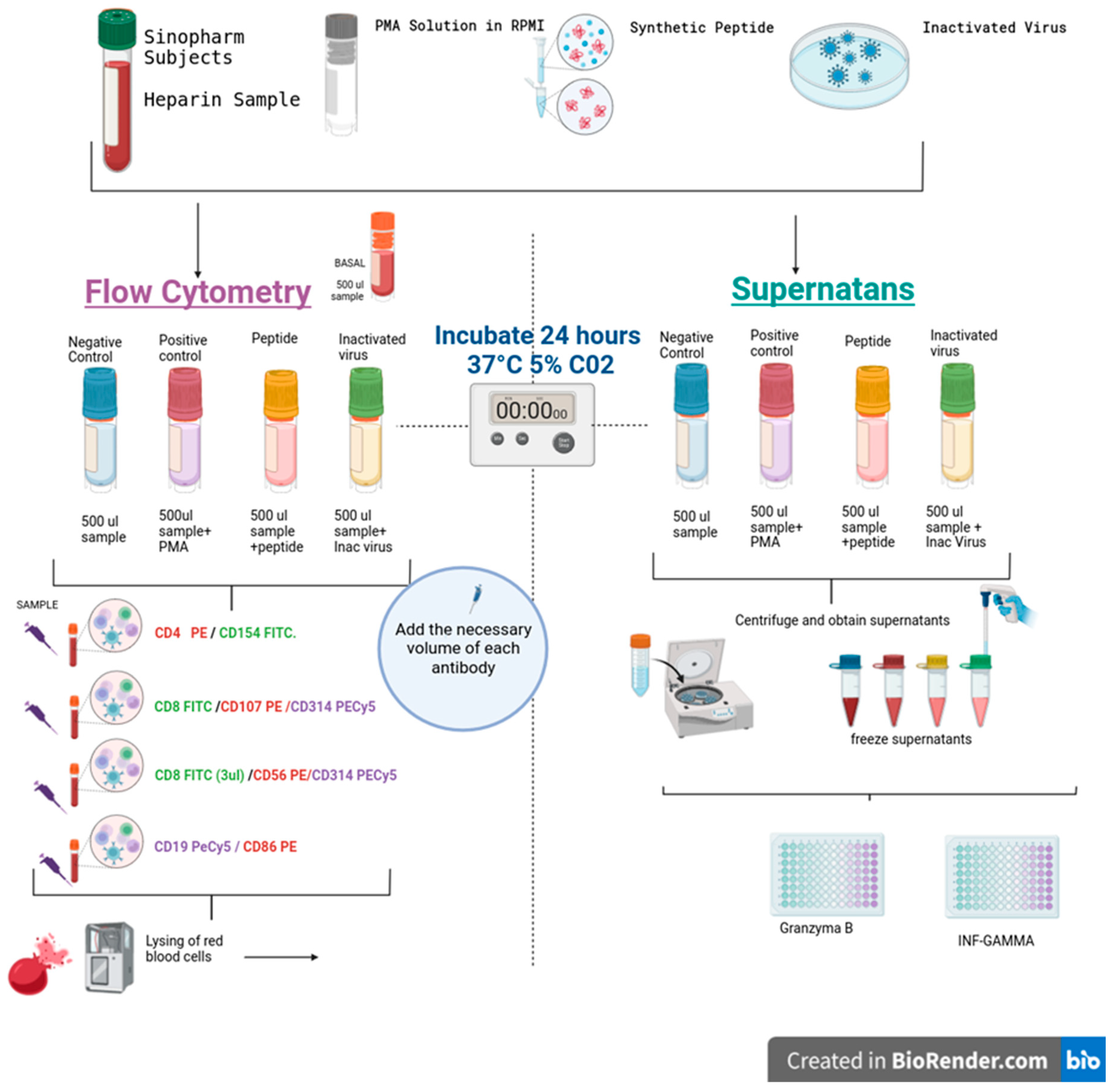
Cell stimulation specific to SARS-CoV-2 was conducted to illustrate alterations in the expression of various activation markers on B cells, T cells (CD4 and CD8), and natural killer (NK) cells. Additionally, the analysis included the production of granzyme B and interferon-gamma (IFN-γ), as depicted in
Figure 1. Four tubes per individual were used, as illustrated in
Figure 1. A volume of 0.5 mL of blood was seeded in 24-well plates (Corning). One well was a control; the second was stimulated with SARS-CoV-2-specific synthetic peptides (PepPool: SARS-CoV-2 (SNMO), human code: 3622-1) from MABTECH; the third was stimulated with 10 plaque-forming units of heat-inactivated virus (Wuhan Strain) obtained from the supernatants of Vero cell cultures and its genetic integrity verified by PCR. As a positive control of the assay, cells were stimulated with Cell Activation Cocktail (without brefeldin A) from BioLegend (cat. no. 423302). Plates were assembled in duplicate and incubated at 37°C in a 5% CO2 atmosphere for 24 hours. Lymphocytes without stimulation were used as the negative control.
2.4. Expression of Activation Markers in Different Lymphocyte Populations and Subpopulations
After incubation, as shown in
Figure 1, the cells were transferred to 12 × 75 mm tubes, and the samples were washed and labeled with the specific antibodies. Then, the erythrocytes were lysed with the automatic lyser (Beckman Coulter), and the samples were analyzed by flow cytometry using the Epics XL equipment from Beckman Coulter. The assay was performed in duplicate. The following panels of antibodies were used: 1) T-helper T-lymphocytes (CD4PE/CD154 FITC), 2) cytotoxic T-lymphocytes degranulation (CD8 FITC/CD107aPE), 3) bystander CD8 lymphocytes and NK cells (CD8 FITC/CD56 PE/CD314 PECY5), and 4) B lymphocytes (CD19 PECY5/CD86 PE). All antibodies used were obtained from BioLegend, and gating was performed on the lymphocyte population, with a minimum of 5,000 events per sample.
It is important to note that all the samples responded to the positive stimulus (PMA/ionomycin) as expected.
2.5. IFNγ and Granzyme B ELISA
The supernatants of the stimulated cells were stored at -20 C until use. Measurement of IFN-γ and granzyme B was performed in triplicate using a sandwich ELISA from BioLegend, following the manufacturer's recommended methodology. For the LEGEND MAX™ Human IFN-γ ELISA kit, the minimum detectable concentration was 5.6 pg/mL. In contrast, for the LEGEND MAX™ Human granzyme B ELISA kit, the minimum detectable concentration was 2.4 ± 1.2 pg/mL. The concentration of the cytokines was determined using a standard curve, and all diluted samples fell within the limits of the kit. As shown in
Figure 2, the positive control for the cellular assays also served as the positive control for the ELISA.
2.6. Statistical Analysis
The GraphPad Prism version 6 program was used for the statistical analysis. The comparison among groups was performed using ANOVA with Bonferroni corrections for the different groups. In specific cases, paired and unpaired Student’s t-tests were used. Pearson correlations were performed with different parameters, and significance was assessed in each case. Significance was considered when p<0.05.
3. Results
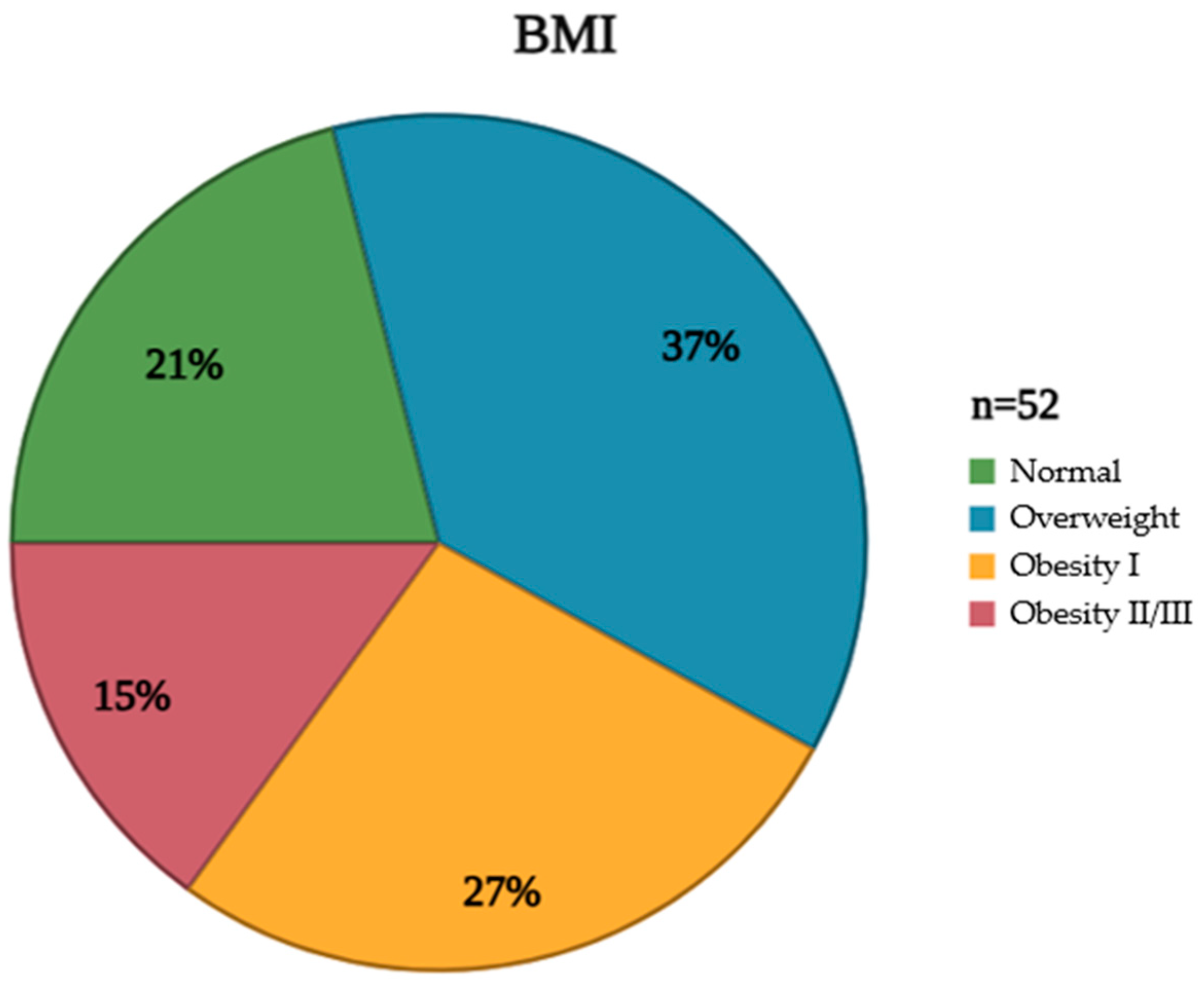
The studied population exhibited a diverse range of characteristics, meticulously documented to provide a comprehensive understanding of the cohort's composition. These attributes, encompassing demographic, clinical, and anthropometric data, are crucial for contextualizing the subsequent analyses and interpreting the study's findings. Furthermore, an assessment of clinical characteristics revealed the prevalence of various comorbidities, including hypertension and diabetes mellitus, providing insight into the overall health status of the population (Table 1). In addition to these clinical parameters, BMI distribution within the studied group was carefully evaluated and visually represented. This distribution, illustrating the frequency of individuals falling within different BMI categories (underweight, normal weight, overweight, and obese), provides a valuable perspective on the population's weight status and its potential impact on the vaccine response (
Figure 2).
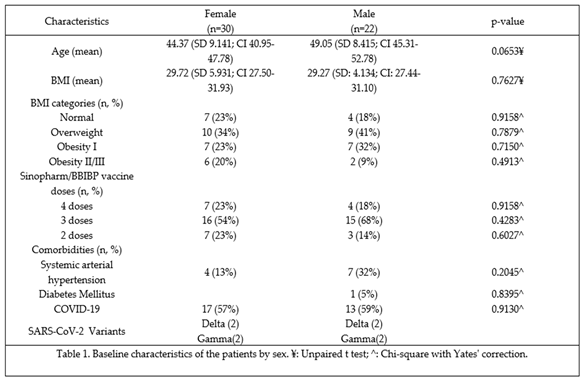
Figure 2.
Distribution of patients by BMI categories according to the World Health Organization. BMI: Body mass index. Created on Biorender.com.
Figure 2.
Distribution of patients by BMI categories according to the World Health Organization. BMI: Body mass index. Created on Biorender.com.
Most individuals from the cohort had antibodies against the RBD of the Spike protein (70.5%) with values ≥ 40 IU/ml, while 29.5% had values lower than 40 IU/ml. From the entire cohort, 10.5% had values lower than 10 IU/ml but higher than 2 IU/ml, which is the cutoff value of the kit. These individuals with low titers received two doses of the vaccine.
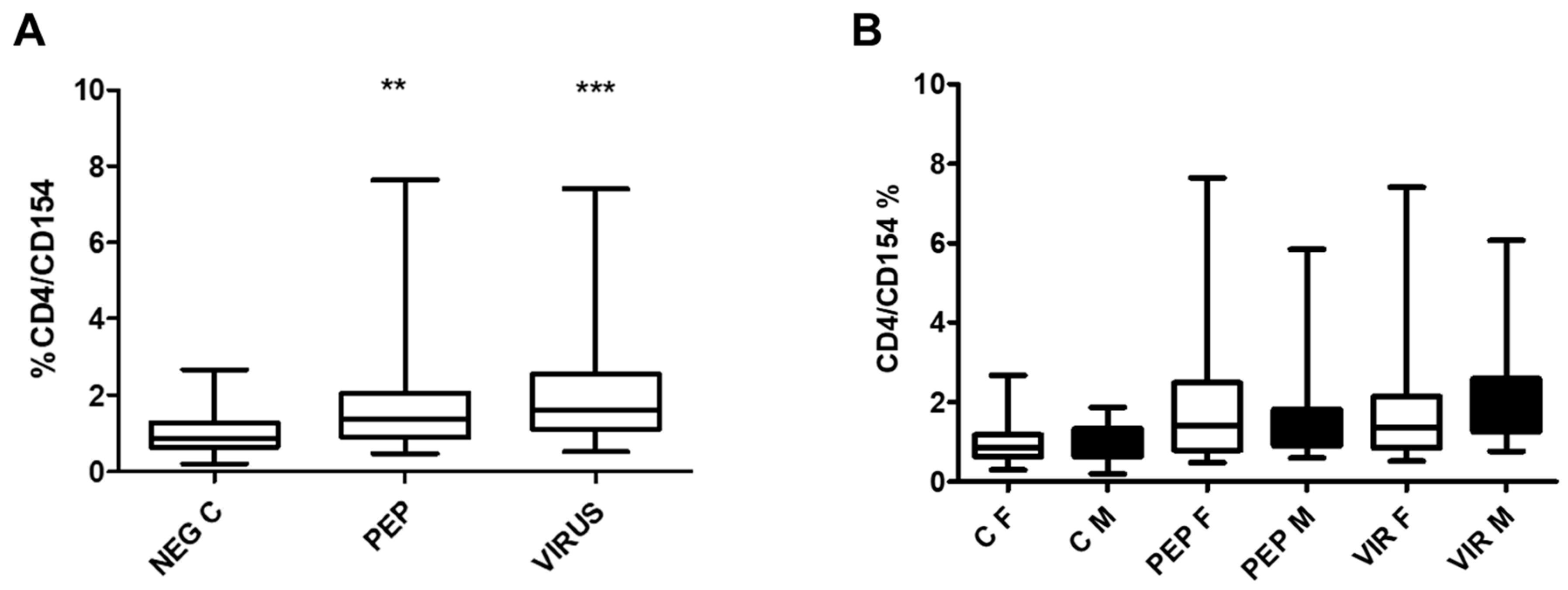
Figure 3 illustrates the effect of peptides and inactivated virus on the expression of CD154 in CD4 cells in the whole group (part A) and divided by gender (part B). Significant differences were obtained in stimulated cells as compared to the control. However, when the results were analyzed by gender (Part B), no significant difference was found in the expression of the antigen CD154 in females vs males. Significant differences were maintained when the stimulated groups were compared to the negative controls.
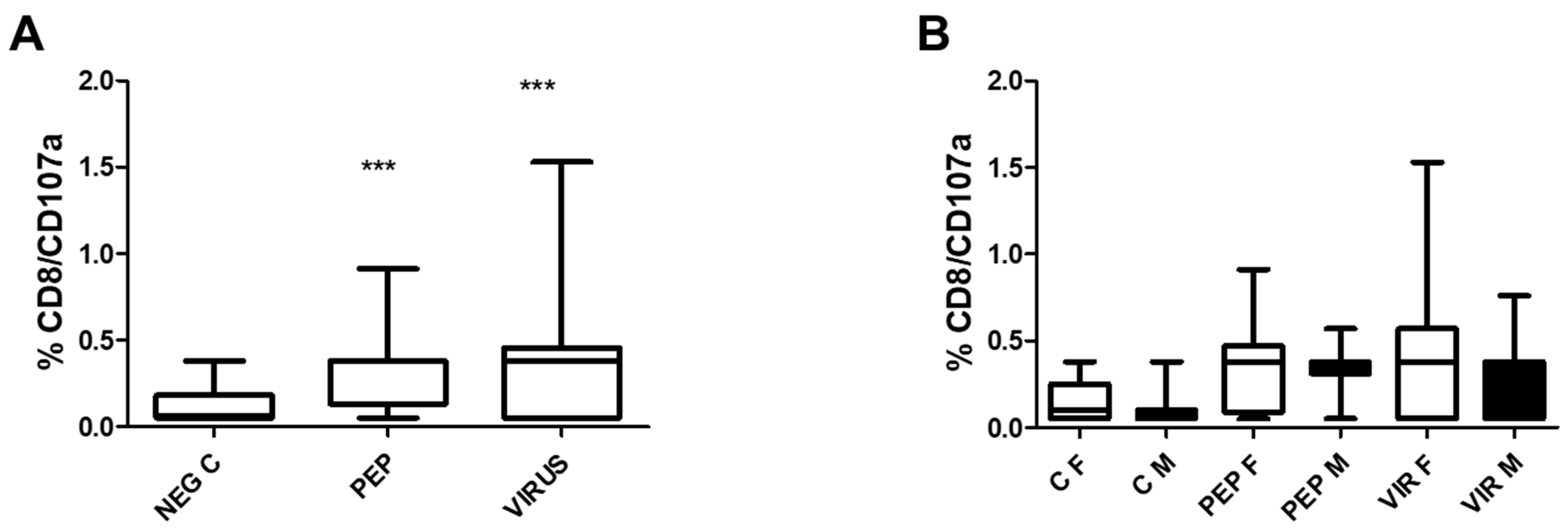
The induction of degranulation in response to stimulation by CD8 T cells is illustrated in
Figure 4. Part A represents the effect of the whole group in which significant differences were observed when the stimuli were compared to the control. Despite the significance recorded in part A of the Figure, there were no significant differences between genders. Nevertheless, as recorded in the previous Figure, significant differences were observed between the stimuli and the negative control.
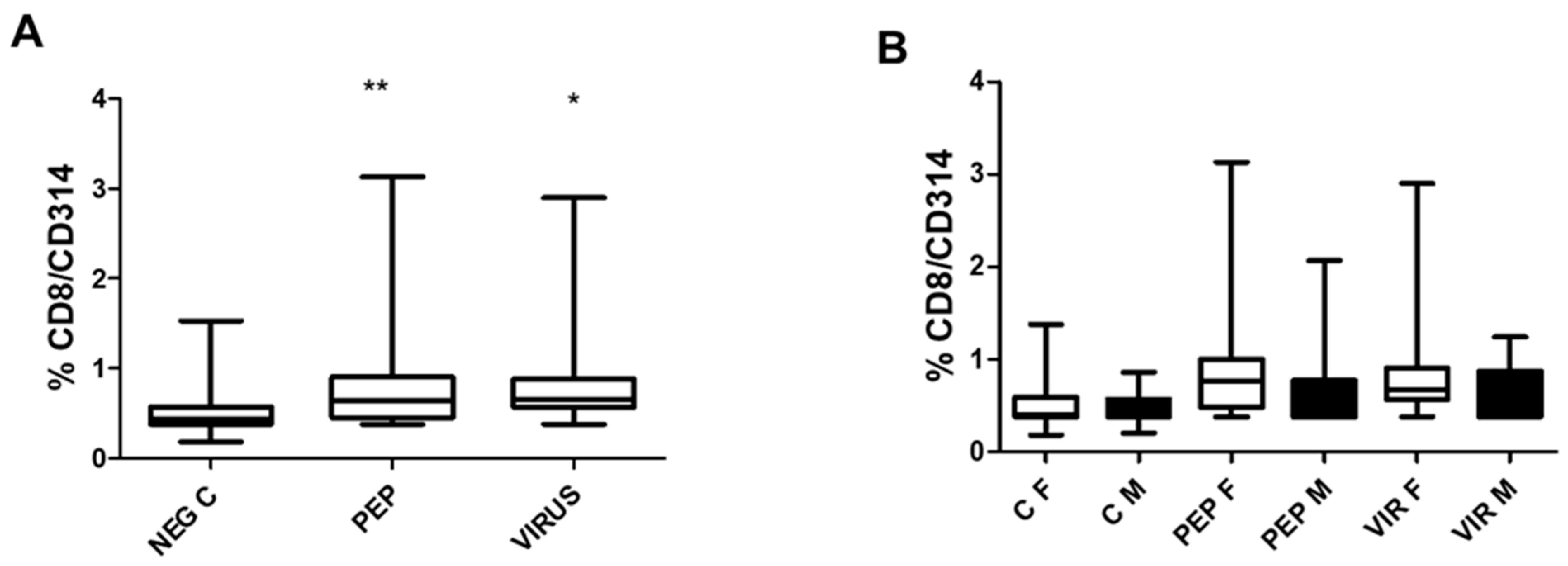
The effect of peptide and inactivated virus on the induction of CD314 (NKG2D) expression, representing T CD8 bystander cells, is depicted in
Figure 5. Both stimuli induced a significant increase in CD314 expression, as shown in part A of the Figure. In part B of the Figure, the effect of gender is represented, which is similar to that observed for the other markers CD4/CD154 and CD8/CD107a. The specific response to each situation is identical in both genders.
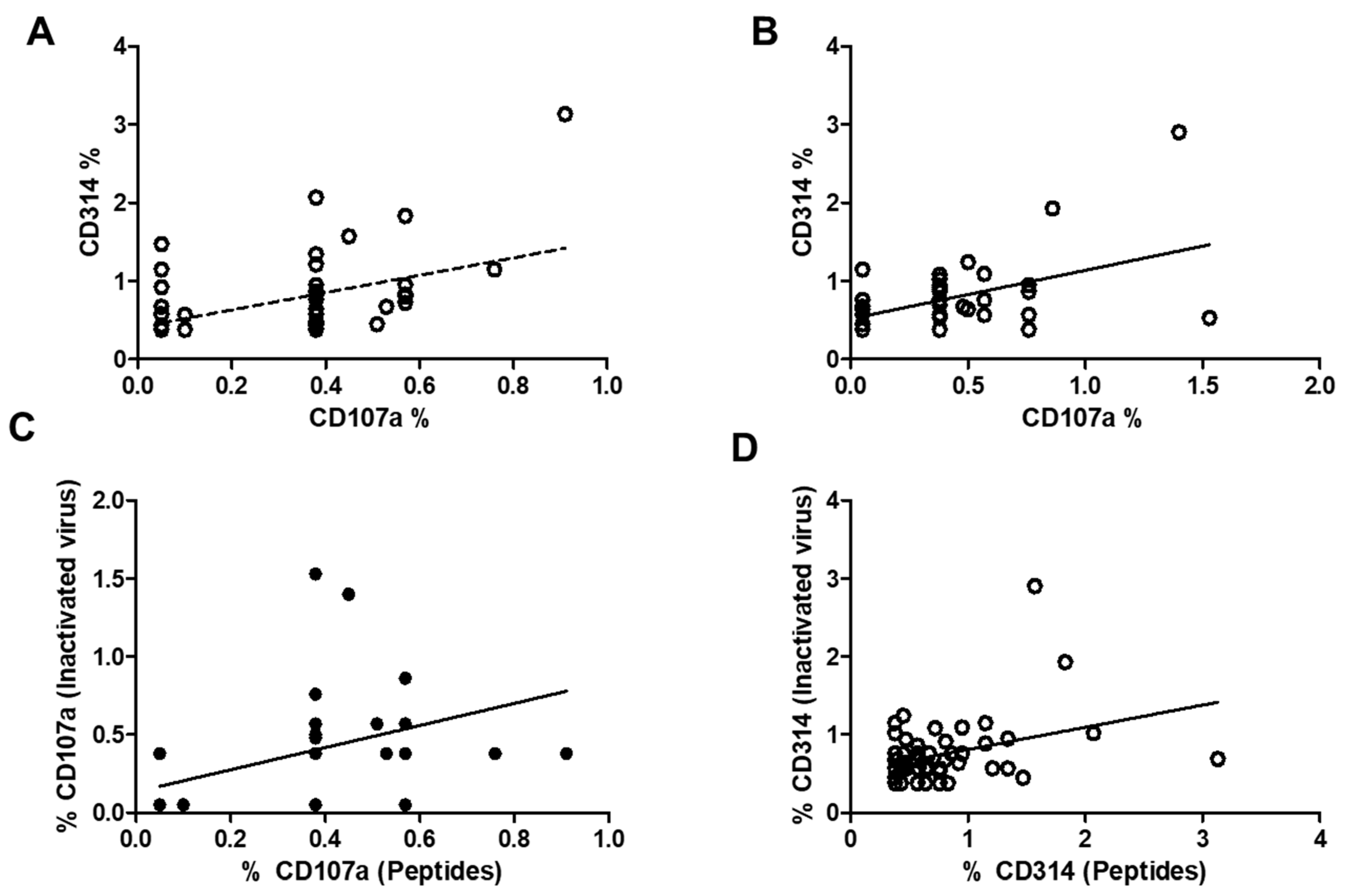
Figure 6 illustrates the correlation between the expression of CD107a and CD314 in stimulated CD8 cells, memory CD8 cells, and bystander CD8 cells using viral peptides (part A) or inactivated virus (part B), as well as the correlation between both markers in CD107a expression (part C) and CD314 expression. Correlations were observed in all the graphs and are statistically significant. To address the question regarding the differences in the increase in both markers depending on the stimulus, parts C and D of the figure represent both correlations. In part C of the Figure, representing CD107a, it is clear that some individuals did not respond to the inactivated virus, with two individuals having very low responses to both stimuli, while others had a higher response. For the CD314 expression, most values are concentrated in the lower part of the figure, suggesting similar expression post-stimulation; only four individuals differed from that response.
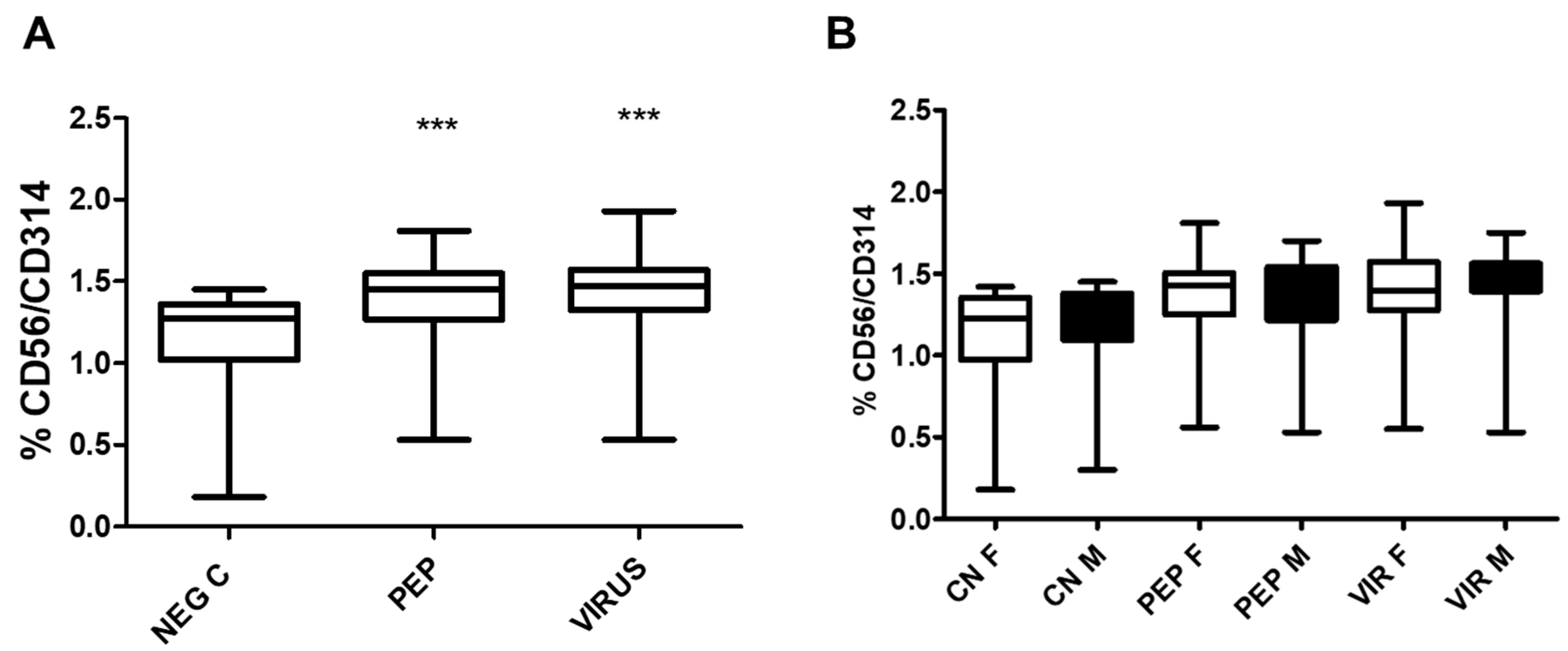
The expression of the killing receptor CD314 was also assessed in NK cells, as illustrated in
Figure 7, which shows an increase in CD314 expression upon activation by viral peptides and inactivated virus. As observed in CD8 cells for CD107a and CD314 cells, no differences were found when the groups were separated by gender. However, a positive increase was recorded with peptides and inactivated virus for each stimulus analyzed.
Even though the expression of CD314 in CD8+ T cells and NK cells increases upon stimulation with viral peptides or inactivated virus, there is no correlation between the percentage of positivity observed for both cell types (r = 0.1 for peptides and r = 0.01 for inactivated virus).
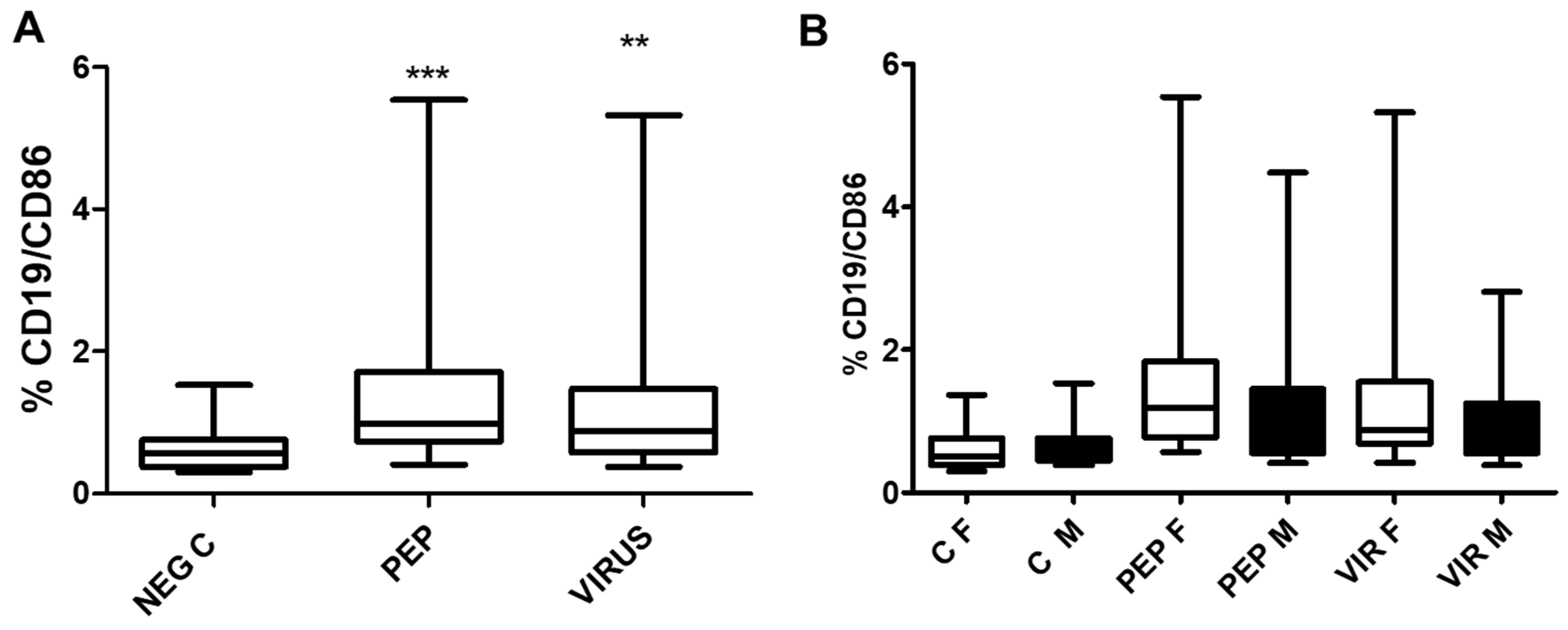
The effect of viral peptides and inactivated virus on the expression of the B lymphocyte activation marker C86 in B cells is illustrated in
Figure 8. The increase in expression is significant for both stimuli, and, as observed with other markers, there is no statistical difference between genders. However, the difference in stimulated cells versus control is significant (p<0.01) for both stimulators.
The secretion of IFN-γ and granzyme B in response to stimulation is illustrated in
Figure 9 (parts A and B), and the effect of gender is shown in parts C and D of the Figure. Significant differences were observed in the secretion of the entire cohort; however, only male volunteers produced significantly more granzyme B as compared to women in response to stimulation with an inactivated virus. Nonetheless, this difference in secretion of granzyme B was not associated with any other parameter.
There is no significant correlation among the values obtained for IFNγ and granzyme B (r = 0.1), between the expression of CD154 and IFNγ (r = 0.05), and between granzyme B and CD8/CD107a (r = 0.2), CD8/CD314 (r = 0.12) and CD56/CD314 (r = 0.08).
There is no correlation between the titers of RGD antibody and any of the cellular parameters analyzed, or the values of granzyme B and IFN-γ. Moreover, there was no significant difference when the group was separated by vaccine dose or COVID-19 complications in the different BMI groups (normal weight vs. overweight/obese); the statistical analysis revealed a p-value greater than 0.2 for each marker analyzed.
4. Discussion
The BBIBP-CorV vaccine is an inactivated vaccine that consists of virus particles grown in culture and then inactivated to lose their ability to cause disease while still stimulating an immune response [
24,
26]. This product is prepared by inoculating African green monkey kidney cells (Vero cells) with SARS-CoV-2 strain WIV04, which is representative of and identical to the sequences of the first outbreak. The Wuhan Institute of Virology isolated the Strain WIV04 from a clinical bronchoalveolar lavage fluid (BALF) sample collected at Wuhan Jinyintan Hospital in Hubei Province on December 30, 2019, from a symptomatic patient, a retailer working at the Huanan Seafood Wholesale Market [
2].
According to the entities in charge of genomic surveillance in Venezuela, Instituto Venezolano de Investigaciones Científicas (IVIC) and Instituto Nacional de Higiene (INH), Omicron was the variant circulating in Venezuela at the time of the study [
4]. In fact, during that period, Venezuela submitted 141 sequences to the GISAID database, and all of them belonged to the Omicron variant.
Various studies on COVID-19 vaccines have focused on the neutralizing antibody response, with little emphasis on cell-mediated immunity [
10,
11,
12]. However, current data suggest that the T-cell response plays a crucial role in protecting against severe forms of COVID-19 following vaccination, particularly against the various viral variants that partially evade recognition by antibodies [
11]. The development of COVID-19 vaccines has marked a turning point in the management of the SARS-CoV-2 pandemic [
10,
11,
12].
Lymphocyte T-cell activation studies confirm that the Sinopharm vaccine induces a memory T-cell response. This is a crucial aspect that confers protection beyond the initial antibody response [
27]. Ning J. and coworkers [
28] have shown that both CD4+ and CD8+ T cells are activated following vaccination with Sinopharm. These T cell subsets play distinct roles in immune defense [
28]. CD4+ T cells help coordinate the immune response, while CD8+ T cells can directly kill infected cells. However, very few reports have analyzed B cell and NK cell activation with this particular vaccine, as well as the possible role of CD8 bystander cells in this process. In addition, the response to the Sinopharm vaccine appears to differ by gender and obesity, as with other vaccines [
29], although no well-designed clinical trials have addressed this particular issue.
Variant-Specific Responses:
The variant-specific response is a significant area of ongoing research focusing on the effectiveness of Sinopharm-induced T lymphocyte responses against emerging SARS-CoV-2 variants. Some studies indicate that the T-cell response elicited by Sinopharm tends to be more strongly oriented toward the original Wuhan strain [
27,
28]. This has led to investigations into how well these T cells can recognize and respond to newer variants with mutations. Research indicates that a T lymphocyte response is present against newer variants; however, the reaction is stronger against the original Wuhan strain [
30,
31].
It's widely acknowledged that cellular immunity, particularly T-cell responses, plays a vital role in protecting against severe COVID-19 [
5,
6]. Even when antibody levels decline over time, T-cell memory can provide a lasting layer of defense. In essence, the BBIBP-CorV vaccine triggers a cellular immune response involving T-cell activation [
27,
28,
30,
31]. The protection afforded by vaccines prevents severe disease and reduces mortality caused by this virus. A study conducted by Ma J. et al. (2022) found that during infection or vaccination with BBIBP-CorV, plasma B lymphocytes produce antibodies against SARS-CoV-2, which decrease progressively over time [
32].
Tong et al. [
33] conducted a study with nine healthy individuals (five men and four women, aged 27-66) who had not received a vaccination in the past year. All tested negative for SARS-CoV-2 by RT-qPCR and for SARS-CoV-2-specific IgM and IgG antibodies by ELISA before vaccination and had no prior contact with COVID-19 patients [
33]. Their analyses revealed an enrichment of monocytes, central memory CD4+ T lymphocytes, type 2 helper T lymphocytes, and memory B lymphocytes after vaccination [
33]. TCR-seq and RNA-seq analyses of individual lymphocytes revealed a clonal expansion of CD4+ T lymphocytes following booster vaccination, which corresponded with a decrease in TCR diversity among central memory CD4+ T lymphocytes and type 2 helper T lymphocytes [
33]. However, their analysis of TCR repertoires revealed that CD4+ T lymphocytes (but not CD8+ T lymphocytes) showed expansion after vaccination with BBIBP-CorV. These data advise that inactivated vaccines primarily induce a CD4+ T-cell immune response [
33].
In a recent review, Mortari et al. [
34] compared different responses to vaccines in the European population, concluding that long-lasting memory B cell responses are observed in individuals vaccinated with heterologous vaccines. This conclusion, however, does include the possibility that infection with different variants of the virus after a homologous vaccination would produce similar effects as the heterolous vaccination. It is possible that in countries where the BBIBP-CorV vaccine or similar inactivated viral vaccines are used, herd immunity may play a crucial role in maintaining a memory response compared to vaccines based solely on the viral spike protein. Additionally, the presence of other viral proteins, such as the immunogenic N protein, can be more effective in maintaining a memory antiviral response. The roles of various cell types, including bystander CD8, NK, NKT, and Tγδ cells, in the protective antiviral response induced by vaccines are still not well-defined.
The analysis conducted in this manuscript utilizing commercial viral peptides and inactivated virus enhances our understanding of cellular responses upon activation. The observation that all markers were significantly upregulated across various cell populations and subpopulations underscores the extensive memory response that occurs following viral exposure. Most studies focusing on SARS-CoV-2 vaccines prioritize the induction of immune responses through the generation of antibodies, particularly neutralizing antibodies, and the activation of T cells (predominantly CD4+), often neglecting other critical components of the immune response [
1,
5,
6,
34]. Notably, the activation of CD8 bystander cells, which are independent of antigen recognition, illustrates the diversity of the immune response [
21,
22]. A significant correlation was identified between the expression of CD107a and CD314 in CD8 cells, specifically in the context of viral stimulation. These findings introduce new perspectives regarding the importance of comprehensively analyzing the immune response. Similarly, the activation of NK cells, as indicated by CD314, provides valuable insights into the role of cellular stress induced by viral contact, which serves as a crucial signal to activate the immune response.
Memory NK responses upon viral infection have been studied in several viral infections [
35,
36]; however, no clear consensus has been reached regarding SARS-CoV-2 infection [
35]. A similar event may be possible for CD8 bystander cells. There are still many unanswered questions regarding B cells and the long-term memory response following vaccination [
34]. Moreover, the effects of gender, obesity, and age remain an unresolved issue in terms of vaccine response [
29].
The differences observed in the secretion of IFNγ and granzyme B are interesting. The response is higher than expected, as observed in the analysis of subpopulations. The increase in response to viral peptides compared to inactivated virus raises the question of proper cell activation; however, there is no indication of any differences in cell response. Interestingly, there is no explanation for the difference in the granzyme B secretion between females and males with inactivated virus. The effect of the secreted cytokine and enzyme may be due to multiple cells, and this point should be further explored.
It is essential to note that the present report was conducted in an admixed population from Venezuela, which differs from other South American admixed populations. The response observed was lower than expected; conversely, it was similar to those previously reported [
27,
30,
31,
32]. Although herd immunity may play a crucial role in assessing the antiviral response in 2023, the effect observed here suggests that many questions remain about the memory response induced by the vaccine, which should be further studied.
The absence of differences in response between females and males in the cohort represents another key finding of this study, particularly in light of existing literature that reports gender disparities in responses to viral infections and vaccinations. Additionally, no variations in immune response were noted based on BMI, although the sample size of our patient cohort is limited. In summary, while the observed increases in various markers following stimulation may not be substantial, it is imperative to acknowledge the presence of a memory response, which may play a significant role in the overall immune response of vaccinated individuals. More research should be conducted
5. Conclusions
The Sinopharm/BBIBP vaccine is capable of inducing a memory response in various lymphocyte populations. The memory against the virus is maintained. The memory response is sustained and independent of gender.
6. Limitations of the Study
The major limitation of the study is the absence of a comparison group with other vaccines; however, the Sinopharm vaccine was the primary vaccine available to the Venezuelan population. Additionally, there was a limitation in identifying patients who were not vaccinated, as vaccination was considered mandatory. The lack of uniform BMI among the volunteers is another limitation; however, in our small group, we did not find any differences in response. Ththe conclusions of the study are based solely on the reaction of the vaccine analyzed, and no other findings could be reported.
Author Contributions
Conceptualization, AHG, and JBDS.; methodology, AHG, SJM, CM, and JBDS.; validation, AHG, and JBDS; formal analysis, AHG, CM, SJM, WYM, IB, FC.; investigation, AHG, CM, SJM, WYM, IB, FC.; resources, AHG, JBDS.; data curation, JBDS; writing—original draft preparation AHG, SJM, CM.; writing—review and editing, AHG, JBDS; supervision, JBDS.; project administration, AHG.; funding acquisition, AHG. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Fund for Science, Technology, and Innovation (FONACIT), an entity attached to the Ministry of Popular Power for Science and Technology of the Bolivarian Republic of Venezuela (MINCYT), Project number ACTA N° 023-2022. J.B.D.S. is partially financed by the National Institute of Virology and Bacteriology (Programme EXCELES, ID Project No. LX22NPO5103)—Funded by the European Union—Next Generation EU from the Ministry of Education, Youth and Sports of the Czech Republic (MEYS).
Institutional Review Board Statement
The study was conducted by the Declaration of Helsinki and was approved by the Institutional Review Board of the Institute of Immunology "Dr. Nicolas E. Bianco C.", Faculty of Medicine. Central University of Venezuela. Study number: 001/2023. Approved on February 15th, 2023.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study. The Ethical Committee authorized the publication of the data.
Data Availability Statement
The data of the study are available upon request.
Acknowledgments
The authors would like to thank all the volunteers who participated in the study and Drs: Mercedes Zabaleta, Leopoldo Deibis, and Félix Toro for their valuable comments and support.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Strauss J.H.; Strauss E.G.. Overview of Viruses and Virus Infection. Viruses and Human Disease. 2008:1–33. [CrossRef]
- Awwal, N.; Dweik, F.; Mahdi, S.; El-Dweik, M.; Anderson, S.H. A Review of SARS-CoV-2 Disease (COVID-19): Pandemic in Our Time. Pathogens. 2022 Mar 17;11(3):368. [CrossRef]
- Hernández, C.; Garcés, M.F.; Hernández, E. COVID-19 pandemic in Venezuela: The first quarantine. Acta Científ Sociedad Ven Bioanal Espe. 2020; Vol 23(1):101-117 . [CrossRef]
- Covid Pandemic data https://ourworldindata.org/coronavirus/country/venezuela. Accesed March 1, 2025.
- Sette, A.; Crotty, S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021 Feb 18;184(4):861-880. [CrossRef]
- De Sanctis, J.B.; García, A.H.; Moreno, D.; Hajduch, M. Coronavirus infection: An immunologists' perspective. Scand J Immunol. 2021 Jun;93(6):e13043. [CrossRef]
- Silva, M.J.A.; Ribeiro, L.R.; Lima K,V.B.; Lima, L.N.G.C. Adaptive immunity to SARS-CoV-2 infection: A systematic review. Front Immunol. 2022 Oct 10;13:1001198. [CrossRef]
- Maison, D.P.; Deng, Y.; Gerschenson, M. SARS-CoV-2 and the host-immune response. Front Immunol. 2023 Jun 19; 14:1195871. [CrossRef]
- Savan, R.; Gale, M Jr. Innate immunity and interferon in SARS-CoV-2 infection outcome. Immunity. 2023 Jul 11;56(7):1443-1450. [CrossRef]
- Najimi, N.; Kadi, C.; Elmtili, N.; Seghrouchni, F.; Bakri, Y. Unravelling humoral immunity in SARS-CoV-2: Insights from infection and vaccination. Hum Antibodies. 2024;32(3):85-106. [CrossRef]
- Karl, V.; Hofmann, M.; Thimme, R. Role of antiviral CD8+ T cell immunity to SARS-CoV-2 infection and vaccination. J Virol. 2025 Mar 3:e0135024. [CrossRef]
- Notarbartolo, S. T-Cell Immune Responses to SARS-CoV-2 Infection and Vaccination. Vaccines (Basel). 2024 Sep 30;12(10):1126. [CrossRef]
- Jordan, S.C. Innate and adaptive immune responses to SARS-CoV-2 in humans: relevance to acquired immunity and vaccine responses. Clin Exp Immunol. 2021 Jun;204(3):310-320. [CrossRef]
- Bahl, A.; Pandey, S.; Rakshit, R.; Kant, S.; Tripathi, D. Infection-induced trained immunity: a twist in paradigm of innate host defense and generation of immunological memory. Infect Immun. 2025 Jan 31;93(1):e0047224. [CrossRef]
- Kurtulus, S.; Hildeman, D. Assessment of CD4+ and CD8+ T cell responses using MHC class I and II tetramers. Methods Mol Biol. 2013; 979:71-9. [CrossRef]
- Poluektov, Y.; George, M.; Daftarian, P.; Delcommenne, M.C. Assessment of SARS-CoV-2-specific CD4+ and CD8+ T cell responses using MHC class I and II tetramers. Vaccine. 2021 Apr 8;39(15):2110-2116. [CrossRef]
- Koh, C. H.; Lee, S.; Kwak, M.; Kim, B. S.; Chung, Y. CD8 T-cell subsets: heterogeneity, functions, and therapeutic potential. Exper Mol Med, 2023; 55(11), 2287–2299. [CrossRef]
- Wu, J.; Shi, Y.; Pan, X.; Wu, S.; Hou, R.; Zhang, Y, et al. SARS-CoV-2 ORF9b inhibits RIG-I-MAVS antiviral signaling by interrupting K63-linked ubiquitination of NEMO. Cell Rep 2021 34(7):108761. [CrossRef]
- Dan, J. M.; Mateus, J.; Kato, Y.; Hastie, K. M.; Yu, E. D.; Faliti, C. E.; et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021; 371(6529), eabf4063. [CrossRef]
- Meraviglia, S.; Di Carlo, P.; Pampinella, D.; Guadagnino, G.; Presti, E. L.; Orlando, V.; et al. T-Cell Subsets (TCM, TEM, TEMRA) and Poly-Functional Immune Response in Patients with Human Immunodeficiency Virus (HIV) Infection and Different T-CD4 Cell Response. Ann Clin Lab Sci, 2019; 49(4), 519–528.
- Paniskaki, K.; Konik, M. J.; Anft, M.; Heidecke, H.; Meister, T. L.; Pfaender, S.; et al. Low avidity circulating SARS-CoV-2 reactive CD8+ T cells with proinflammatory TEMRA phenotype are associated with post-acute sequelae of COVID-19. Front Microbiol, 2023; 14, 1196721. [CrossRef]
- Paprckova, D.; Salyova, E.; Michalik, J.; Stepanek, O. Bystander activation in memory and antigen-inexperienced memory-like CD8 T cells. Curr Opin Immunol. 2023 Jun;82:102299. [CrossRef]
- Mariuzza, R.A.; Singh, P.; Karade, S.S.; Shahid, S.; Sharma, V.K. Recognition of Self and Viral Ligands by NK Cell Receptors. Immunol Rev. 2025;329(1):e13435. [CrossRef]
- https://covid-19pharmacovigilance.paho.org/sinopharmbibp. Assessed March 1, 2025.
- Mayora, S.; Martínez, W.; Guerrero, M.; Belisario, I.; De Sanctis, J.B.; García, A. Pilot study: Analysis and detection of specific IgM and IgG antibodies against the receptor binding domain of the spike protein of SARS-CoV-2. Gac Méd Caracas 2022;130(1): 85-94. [CrossRef]
- Khoshnood, S.; Arshadi, M.; Akrami, S.; Koupaei, M.; Ghahramanpour, H.; Shariati, A.; Sadeghifard, N.; Heidary, M. An overview on inactivated and live-attenuated SARS-CoV-2 vaccines. J Clin Lab Anal, 2022; 36(5), e24418. [CrossRef]
- Vályi-Nagy, I.; Matula, Z.; Gönczi, M.; Tasnády, S.; Bekő, G.; Réti, M.; Ajzner, É.; Uher, F. Comparison of antibody and T cell responses elicited by BBIBP-CorV (Sinopharm) and BNT162b2 (Pfizer-BioNTech) vaccines against SARS-CoV-2 in healthy adult humans. GeroScience, 2021; 43(5), 2321–2331. [CrossRef]
- Ning, J.; Wang, Q.; Chen, Y.; He, T.; Zhang, F.; Chen, X.; Shi, L.; Zhai, A.; Li, B.; Wu, C. Immunodominant SARS-CoV-2-specific CD4+ and CD8+ T-cell responses elicited by inactivated vaccines in healthy adults. J Med Virol. 2023 Apr;95(4):e28743. [CrossRef]
- De Sanctis, J.B.; Balda Noria, G.; García, A.H. Exploring How Adipose Tissue, Obesity, and Gender Influence the Immune Response to Vaccines: A Comprehensive Narrative Review. Int J Mol Sci. 2025 Jan 20;26(2):862. [CrossRef]
- Musa, S.; Merdrignac, L.; Skocibusic, S.; Nedic, R.; Cilovic-Lagarija, S.; Kissling, E. BBIBP-CorV vaccine effectiveness against COVID-19 in patients aged 60 years and older during the Delta-dominant period in the Federation of Bosnia and Herzegovina, a test-negative case-control study. Vaccine. 2024 May 31;42(15):3467-3473. [CrossRef]
- Albreiki, M.; Mousa, M.; Azman, S.K.; Vurivi, H.; Alhalwachi, Z.; Alshehhi, F.; et al. Risk of hospitalization and vaccine effectiveness among COVID-19 patients in the UAE during the Delta and Omicron outbreaks. Front Immunol. 2023 Feb 13;14:1049393. [CrossRef]
- Ma J, Cheng ZJ, Xue M, Huang H, Li S, Fang Y, Zeng Y, Lin R, Liang Z, Liang H, Deng Y, Cheng Y, Huang S, Wang Q, Niu X, Li S, Zheng P, Sun B. Investigation of Antibody Levels During Three Doses of Sinopharm/BBIBP Vaccine Inoculation. Front Immunol. 2022 Jun 22;13:913732. [CrossRef]
- Tong, R.; Luo, L.; Zhao, Y.; Sun, M.; Li, R.; Zhong, J.; et al. Characterizing the cellular and molecular variabilities of peripheral immune cells in healthy recipients of BBIBP-CorV inactivated SARS-CoV-2 vaccine by single-cell RNA sequencing. Emerg Microbes Infect. 2023 Dec;12(1):e2187245. [CrossRef]
- Piano Mortari, E.; Ferrucci, F.; Zografaki, I.; Carsetti, R.; Pacelli, L. T and B cell responses in different immunization scenarios for COVID-19: a narrative review. Front. Immunol. 2025; 16:1535014.
- De Sanctis, J.B.; Garmendia, J.V.; Hajdúch, M. Overview of Memory NK Cells in Viral Infections: Possible Role in SARS-CoV-2 Infection. Immuno 2022, 2, 52-67. [CrossRef]
- Herrera, L.; Martin-Inaraja, M.; Santos, S.; Inglés-Ferrándiz, M.; Azkarate, A.; Perez-Vaquero, M. A.; et al. Identifying SARS-CoV-2 'memory' NK cells from COVID-19 convalescent donors for adoptive cell therapy. Immunology, 2022; 165(2), 234–249. [CrossRef]
Figure 1.
The figure illustrates the procedures used for cell stimulation and analysis, as well as the determination of cytokines using commercial ELISA kits.
Figure 1.
The figure illustrates the procedures used for cell stimulation and analysis, as well as the determination of cytokines using commercial ELISA kits.
Figure 3.
Effect of Viral Peptides and Inactivated Virus on the Activation of CD4+ T Cells. Part A of the Figure depicts the impact on the whole group. The increased expression of CD154 was significant (**p < 0.01 and ***p<0.001, Bonferroni post-hoc test) using repeated measures ANOVA, with n = 52 individuals in paired analysis. In part B of the Figure, the group was separated by gender. There were no significant differences between genders (30 females and 22 males) in each condition; however, significant differences were observed within each gender between negative controls and viral peptides, as well as between negative controls and inactivated virus (p < 0.01 in both cases). C represents control, PEP viral peptides, VIR inactivated virus, F female, and M male.
Figure 3.
Effect of Viral Peptides and Inactivated Virus on the Activation of CD4+ T Cells. Part A of the Figure depicts the impact on the whole group. The increased expression of CD154 was significant (**p < 0.01 and ***p<0.001, Bonferroni post-hoc test) using repeated measures ANOVA, with n = 52 individuals in paired analysis. In part B of the Figure, the group was separated by gender. There were no significant differences between genders (30 females and 22 males) in each condition; however, significant differences were observed within each gender between negative controls and viral peptides, as well as between negative controls and inactivated virus (p < 0.01 in both cases). C represents control, PEP viral peptides, VIR inactivated virus, F female, and M male.
Figure 4.
Effect of Viral Peptides and Inactivated Virus on the Degranulation of CD8+ T Cells, as Measured by CD107a Expression. Part A of the Figure depicts the impact on the whole group. The increased expression of CD107a was significant (***p<0.001, Bonferroni post-hoc test) using repeated measures ANOVA, with n = 52 individuals in paired analysis. In part B of the Figure, the group was separated by gender. There were no significant differences between genders (30 females and 22 males) in each condition. However, significant differences were observed within each gender between negative controls with viral peptides and negative control with inactivated virus (p < 0.01 for both cases). C represents control, PEP viral peptides, VIR inactivated virus, F female, and M male.
Figure 4.
Effect of Viral Peptides and Inactivated Virus on the Degranulation of CD8+ T Cells, as Measured by CD107a Expression. Part A of the Figure depicts the impact on the whole group. The increased expression of CD107a was significant (***p<0.001, Bonferroni post-hoc test) using repeated measures ANOVA, with n = 52 individuals in paired analysis. In part B of the Figure, the group was separated by gender. There were no significant differences between genders (30 females and 22 males) in each condition. However, significant differences were observed within each gender between negative controls with viral peptides and negative control with inactivated virus (p < 0.01 for both cases). C represents control, PEP viral peptides, VIR inactivated virus, F female, and M male.
Figure 5.
Effect of Viral Peptides and Inactivated Virus on the Expression of CD314 (NKG2D). Part A of the Figure depicts the impact on the whole group. The increased expression of CD314 was significant (* < 0.05, **p < 0.001, Bonferroni post-hoc test) using repeated measures ANOVA, with n = 52 individuals in paired analysis. In part B of the Figure, the group was separated by gender. There were no significant differences between genders (30 females and 22 males) in each condition. However, significant differences were observed within each gender between negative controls and viral peptides, as well as between negative controls and inactivated virus (p < 0.01 in both cases). C represents control, PEP viral peptides, VIR inactivated virus, F female, and M male.
Figure 5.
Effect of Viral Peptides and Inactivated Virus on the Expression of CD314 (NKG2D). Part A of the Figure depicts the impact on the whole group. The increased expression of CD314 was significant (* < 0.05, **p < 0.001, Bonferroni post-hoc test) using repeated measures ANOVA, with n = 52 individuals in paired analysis. In part B of the Figure, the group was separated by gender. There were no significant differences between genders (30 females and 22 males) in each condition. However, significant differences were observed within each gender between negative controls and viral peptides, as well as between negative controls and inactivated virus (p < 0.01 in both cases). C represents control, PEP viral peptides, VIR inactivated virus, F female, and M male.
Figure 6.
represents the correlation between the expression of CD314 and CD107a in stimulated samples. Part A represents the correlation using viral peptides (r = 0.32, p < 0.01, n = 52). Part B represents the correlation using an inactivated virus (r = 0.47, p < 0.001). Part C of the Figure illustrates the correlation between the expression of CD107a and both stimuli, which was significant (r = 0.44, p < 0.005). Part D of the Figure illustrates the correlation between the expression of CD314 and both stimuli, which was barely significant (r = 0.35, p = 0.01).
Figure 6.
represents the correlation between the expression of CD314 and CD107a in stimulated samples. Part A represents the correlation using viral peptides (r = 0.32, p < 0.01, n = 52). Part B represents the correlation using an inactivated virus (r = 0.47, p < 0.001). Part C of the Figure illustrates the correlation between the expression of CD107a and both stimuli, which was significant (r = 0.44, p < 0.005). Part D of the Figure illustrates the correlation between the expression of CD314 and both stimuli, which was barely significant (r = 0.35, p = 0.01).
Figure 7.
The effect of Viral Peptides and Inactivated viruses on the expression of CD314 (NKG2D) is depicted in the figure. Part A of the Figure illustrates the impact on the whole group. The increased expression of CD314 was significant (***p < 0.0001, Bonferroni post-hoc test) using repeated measures ANOVA, with n = 52 individuals in paired analysis. In part B of the Figure, the group was separated by gender. There were no significant differences between genders (30 females and 22 males) in each condition; however, significant differences were observed within each gender between negative controls with viral peptides and negative control with inactivated virus (p < 0.01 in both cases). C represents control, PEP viral peptides, VIR inactivated virus, F female, and M male.
Figure 7.
The effect of Viral Peptides and Inactivated viruses on the expression of CD314 (NKG2D) is depicted in the figure. Part A of the Figure illustrates the impact on the whole group. The increased expression of CD314 was significant (***p < 0.0001, Bonferroni post-hoc test) using repeated measures ANOVA, with n = 52 individuals in paired analysis. In part B of the Figure, the group was separated by gender. There were no significant differences between genders (30 females and 22 males) in each condition; however, significant differences were observed within each gender between negative controls with viral peptides and negative control with inactivated virus (p < 0.01 in both cases). C represents control, PEP viral peptides, VIR inactivated virus, F female, and M male.
Figure 8.
The figure illustrates the effect of viral peptides and inactivated virus on the expression of CD86 in B lymphocytes. Part A of the Figure illustrates the impact on the whole group. The increased expression of CD86 was significant (**p < 0.001, ***p < 0.0001, Bonferroni post-hoc test) using repeated measures ANOVA, with n = 52 individuals in paired analysis. In part B of the Figure, the group was separated by gender. There were no significant differences between genders (30 females and 22 males) in each condition. However, significant differences were observed within each gender between negative controls and viral peptides, as well as between negative control and inactivated virus (p < 0.01 for both). C represents control, PEP viral peptides, VIR inactivated virus, F female, and M male.
Figure 8.
The figure illustrates the effect of viral peptides and inactivated virus on the expression of CD86 in B lymphocytes. Part A of the Figure illustrates the impact on the whole group. The increased expression of CD86 was significant (**p < 0.001, ***p < 0.0001, Bonferroni post-hoc test) using repeated measures ANOVA, with n = 52 individuals in paired analysis. In part B of the Figure, the group was separated by gender. There were no significant differences between genders (30 females and 22 males) in each condition. However, significant differences were observed within each gender between negative controls and viral peptides, as well as between negative control and inactivated virus (p < 0.01 for both). C represents control, PEP viral peptides, VIR inactivated virus, F female, and M male.
Figure 9.
The figure illustrates the levels of IFNγ (part A) and granzyme B (part B) following stimulation with peptides and inactivated virus. Significant differences were observed for both stimuli in IFN-γ (***p < 0.001) and granzyme B (***p < 0.001, and *p < 0.01). When the groups were separated by gender, there was no significant difference in IFN-γ secretion; however, a substantial increase in granzyme B was observed in male individuals stimulated with inactivated virus (**p < 0.005).
Figure 9.
The figure illustrates the levels of IFNγ (part A) and granzyme B (part B) following stimulation with peptides and inactivated virus. Significant differences were observed for both stimuli in IFN-γ (***p < 0.001) and granzyme B (***p < 0.001, and *p < 0.01). When the groups were separated by gender, there was no significant difference in IFN-γ secretion; however, a substantial increase in granzyme B was observed in male individuals stimulated with inactivated virus (**p < 0.005).
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).