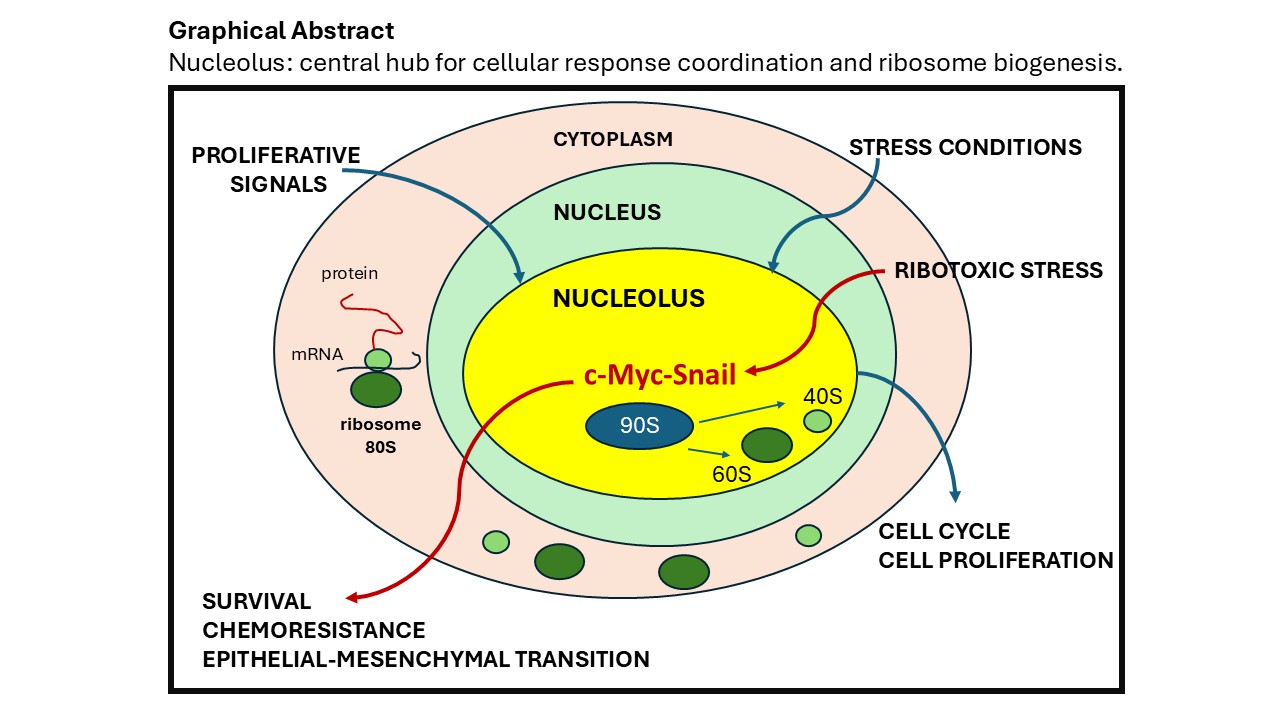
1. Introduction
The nucleolus, a structure located within the nucleus of eukaryotic cells, plays a crucial role in producing and assembling ribosomal subunits, which are crucial for protein synthesis [
1]. Proliferating cells, including rapidly dividing cancer cells, must achieve sufficient mass and size to support their high protein demand before division. Thus, nucleoli perform a key function in maintenance of homeostasis in cells, and they can directly influence cell cycle progression, cell growth, and proliferation [
2]. They synthesize half of all transcript pool and utilizes up to 80% of energetic and material resources of the cell [
3]. These processes involve several hundred protein trans-acting factors and small nucleolar RNAs (snoRNA), which serve to guide the specificity of ribosomal RNA (rRNA) chemical modifications, ribosomal precursor RNA (pre-rRNA) folding and cleavage. Once ribosomal precursors are released from the nucleolar structure, they undergo further maturation in the nucleolus and nucleoplasm prior to becoming fully functional ribosomal subunits in the cytoplasm, ready to engage in translating mRNAs into proteins. Ribosomal subunits are the structural components of ribosomes, which are responsible for synthesizing proteins in cells. They are composed of two subunits, called the large (60S) and small subunits (40S), which form the functional ribosomes (80S). The large subunit contains three rRNAs 28S, 5.8S, 5S and 50 ribosomal proteins, (RPL) while the small subunit contains one rRNA 18S and 33 ribosomal proteins (RPS) [
4]. The rRNAs provide the structural framework for the ribosome, while the proteins help to stabilize the structure and facilitate interactions with other molecules involved in protein synthesis. The transcribed rRNA molecule is used only once, being immediately assembled with ribosomal proteins to produce over 2,000–10,000 ribosomes per minute [
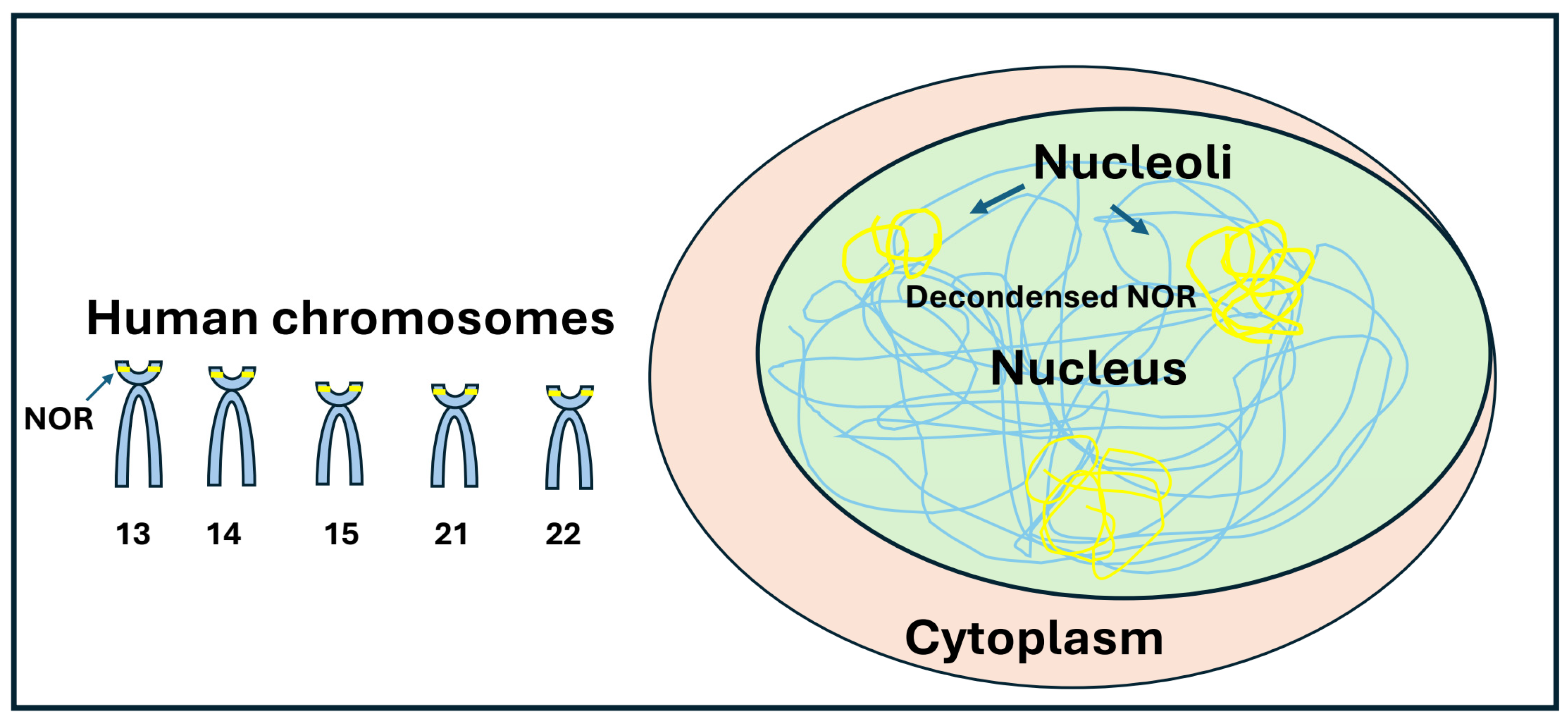
5]. The nucleolus forms around nucleolus organizer regions (NORs), which consist of rDNA repeat clusters typically distributed across multiple chromosomes. The number of repeats varies ranging between a total of 100–600 copies in a diploid genome Ribosomal DNA represent with its repetitive sequence a region particularly prone to DNA damage and rearrangements during repair. The segregation of damaged ribosomal DNA into nucleolar caps has been suggested to serve as prevention of inter-chromosomal recombination and to promote ribosomal DNA repair. High-resolution microscopy studies have shown that the repression of RNA polymerase I transcription following rDNA double-strand breaks (DSBs) depends on the DNA repair kinases ATM and ATR. Additionally, the nucleolar protein TCOF1 (Treacle) is crucial for nucleolar cap formation in response to rDNA damage. TCOF1 depletion prevents nucleolar cap formation, blocks rDNA transcription silencing, and results in reduced cell viability, increased apoptosis, and genomic instability. Furthermore, TCOF1 facilitates the recruitment of key DNA repair factors, including TOPBP1 and the MRE11-RAD50-NBS1 (MRN) complex, to nucleolar caps. Recent research suggests that the covalent attachment of the ubiquitin-fold modifier 1 (UFM1) protein to target proteins (UFMylation), plays a role in rDNA repair [
6,
7]. The rDNA unit contains DNA that codes for ribosomal RNA, and small nucleolar RNA molecules. In humans, NORs are present in the short arm of five acrocentric chromosomes (13, 14, 15, 21 and 22) (
Figure 1). The fundamental repeat unit consists of a rDNA core that encodes the 47S precursor of rRNA (47S pre-rRNA) and intergenic spacers (IGS). The IGS (IGS1 and IGS2) may contain regulatory elements, and other RNAs transcribed by RNA Polymerase II [
8]. The 47S pre-rRNA is processed into the three mature rRNAs, 18S, 5.8S and 28S rRNA [
9]. By contrast, the 5S rRNA gene is transcribed by RNA Polymerase III [
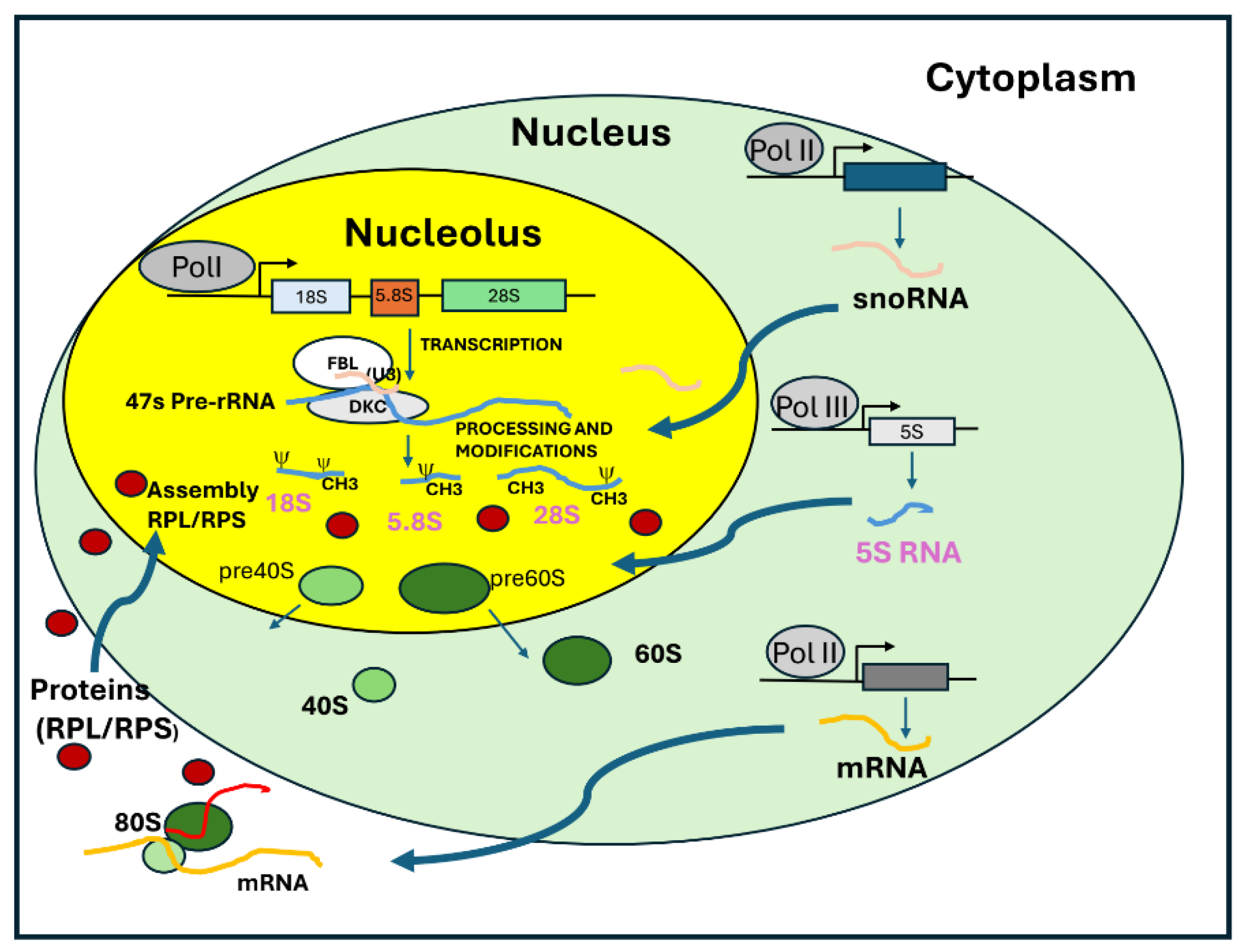
10]. In humans, the 5S RNA genes are tandemly repeated on chromosome 1 and are localized in close to nucleoli. 47S pre-rRNA processing involves a series of specific endo- and exonucleolytic cleavages to remove transcribed sequences that are not part of the mature rRNAs. This process is accompanied by the chemical modification of approximately 200 nucleotides within rRNA sequences. These post-transcriptional modifications (
Figure 2) are orchestrated by ribonucleoprotein complexes containing snoRNAs of two types: C/D box snoRNAs and H/ACA box snoRNAs. These snoRNA are associated with fibrillarin, which catalyses 2′-O-ribose methylation, dyskerin, which catalyses pseudo-uridylation and other enzymes involved in rRNA modification. These modifications impact interactions between rRNAs, tRNAs and mRNAs, and some are known to fine tune translation rates and efficiency. Ribose 2′-O-methylation is the most abundant rRNA chemical modification and displays a complex pattern in rRNA. These modifications occur early in ribosomal RNA maturation, and although their precise function remains unclear, snoRNAs play a crucial role in directing the modification machinery to specific target sites. These modifications target nucleotides in functionally critical regions of the ribosome, including the peptidyl transferase center, which is essential for peptide bond formation during protein synthesis. The U3 C/D box snoRNP plays a crucial role in 18S rRNA maturation by base-pairing with both the 5′-ETS and 18S rRNA. It coordinates the formation of the 18S rRNA pseudoknot while facilitating early cleavages in the 5′-ETS and ITS1. This essential function makes U3 snoRNA indispensable for 18S rRNA production and, consequently, for cell viability [
11]. Like U3, other snoRNPs such as U8, U14, U17, and U22 play a crucial role in multiple pre-rRNA processing steps by chaperoning pre-rRNAs. The folding and processing of pre-rRNA are tightly regulated by both ribosomal proteins (RPs) and ribosomal assembly factors (RAFs). Most ribosomal proteins are essential for ribosomal subunit synthesis, with their incorporation closely coordinated with pre-rRNA folding and RAF recruitment. Indeed, depletion of small-subunit ribosomal proteins blocks pre-rRNA maturation. RAFs transiently associate with forming ribosomal subunits, contributing to enzymatic, structural, and regulatory functions. Synthesis of the 5S rRNA requires a specific regulatory factor called transcription factor IIIA (TFIIIA). This factor associates with the general class III initiation factors TFIIIB and TFIIIC on the 5S gene promoter and stimulates transcription. The promoter element necessary for 5S rRNA gene transcription is in the transcribed region. After processing of the 3 ends, the 5S rRNA is associated with the ribosomal protein L5 (RPL5). A specialized importin called Syo1 mediates nuclear import of the ribosomal proteins RPL5 and RPL11 and likely chaperones the assembly with the 5S RNA [
12]. This complex is then addressed to the nucleolus where it is incorporated into the 60S particles. Also, the incorporation of the 5S ribonuclear protein (RNP) into pre-60S particles depends on association with ribosomal assembly factors. In mammalian cells, the free 5S-RPL5 has ribosome-independent functions in cell cycle regulation, as shown by its capacity to modulate the p53 activity. Under nucleolar stress, the 5S RNP in a free pool can sequester the ubiquitin ligase Mdm2, which stabilizes p53 levels and can cause cell cycle arrest and apoptosis [
12].
2. Nucleolus Organization
The nucleolar function is accompanied by organization of the nucleolus into distinct sub-compartments. Transmission electron microscopy has revealed three major structures within nucleoli: fibrillar centers (FC), dense fibrillar components (DFC), and the granular component (GC) [
13,
14,
15]. rDNA transcription units are found in the FC and consist of tandem repeats of these genes. rRNAs are harbored within the DFC and are processed there. Fibrillarin (FBL) and Nucleophosmin (NPM1/B23) are canonical domain markers for the DFC and the GC, respectively. FBL and NPM1 play successive roles in ribosome biogenesis. Later stages of rRNA processing take place in the GC. Thus, the processing of rRNA is spatially arranged in accordance with the ultrastructure of these compartments. One of the most intriguing aspects of nucleolar function is the continuous flux of its components [
16]. Significantly rRNA at various stages of processing moves through the nucleolus, while precursor ribosomal subunits are constantly exported from the nucleoplasm to the cytoplasm for further assembly. This is often described as a vectorial process where the ribosome biogenesis steps such as the precursor rRNA transcription must occur before rRNA modification and binding by ribosomal proteins. Recent studies highlight that the directional flux within the nucleolus may be driven by the forces such as described by the liquid-liquid phase separation model [
16]. Proteomic analysis revealed over 500 proteins that localize to the nucleolus. These proteins are involved in cell cycle control, DNA processing, DNA damage response and repair, in addition to the many proteins connected with ribosome subunit production [
17,
18]. Ribosome levels can vary across different cell types and stages. A recent study found that fluctuations in ribosome synthesis during the circadian rhythm are linked to liver metabolism. In hepatocytes, protein production peaks at night are followed by equivalent proteins degradation during the day. This rhythmic is controlled by the Target of Rapamycin pathway (TOR) which is activated by nutrient mainly amino acids. While the impact of circadian and feeding rhythms on transcription is well-documented, the rhythmic coordination of mRNA translation and ribosome biogenesis has only recently been discovered [
19].
3. rDNA and Epigenetic Modifications
The nucleolus exhibits significant structural diversity during development and across different tissues, along with a dynamic organization that involves the assembly and disassembly of its components throughout the cell cycle, DNA repair, and in response to stress. The number of active rRNA genes at the ribosomal DNA gene repeat is regulated by epigenetic mechanisms. During differentiation, the nucleosome remodeling complex (NoRC) recruits DNA methyltransferases to active rDNA genes and establishes silent heterochromatin at the nucleolus. These silenced rDNA genes are important for genome stability and form hubs around which non-ribosomal DNA organizes into nucleolus-associated domains (NADs). The rDNA of actively transcribed genes exists in a euchromatin configuration that is characterized by DNA hypomethylation, H4ac, and H3K4me2 [
20,
21]. The rDNA methylation and nucleolar size vary across individuals and are associated with age and longevity [
22]. Genomic screens for DNA methylation markers of age across non-rDNA segments identified 353 and 90 CpG sites with the ability to predict chronological age in the human and mouse genomes. Dysfunction of the NORs can lead to abnormalities in ribosome production, which can have a variety of consequences for cellular function. Several nucleolar proteins, transcription regulators and chromatin modulators contribute to determining nucleolus structures [
23]. The epigenetic state of rDNA regulates not only ribosome biogenesis, but also the spatial organization and transcriptional activity of the genome. The ribosomal DNA gene is found in three different transcriptional states: inactive, pending and active [
24]. Inactive NORs localize outside the nucleolus while the pending NORs localizes in the nucleolus and has silenced units that could be easily activated. The active NORs localise in the nucleolus and the epigentic status of rDNA is controlled by multiple factors. One of these factors is upstream binding factor (UBF), without which NORs become inactive and unbound to nucleoli [
23]. UBF is a multi-HMGB-box protein that acts both as an epigenetic factor to establish the open conformation of the chromatin on the ribosomal genes and as a basal transcription factor in RNA Polymerase I transcription [
25,
26,
27]. It plays an essential role in the aggregation of nucleolar proteins resulting in nucleososome-like structures. Active rDNA genes are nucleosome-free and bound by UBF and the transcription intermediary factor B (TIF-1B), which initiate transcription by RNA polymerase I. In contrast, the promoters of silent rRNA genes are methylated and their coding regions are packed by nucleosomes with repressive histone marks.
In addition to binding at the rDNA promoter, UBF also binds throughout the rDNA coding region and the IGS sequence. It displaces linker histone H1 and contributes to the decondensed state of the euchromatic rDNA [
28,
29]. The rDNA of silent genes exists in a closed heterochromatin state, characterized by H3K9me, H3K20me, and CpG methylation. Approximately half of the rRNA genes are maintained in an active state.
A key regulator of rRNA transcription is transcription termination factor 1 (TTF-I), a multifunctional nucleolar protein that binds to terminator elements downstream of rDNA [31,32]. Structurally, TTF-I consists of three main domains: a C-terminal DNA-binding domain, essential for recognizing terminator elements, a central domain, required for transcription termination, transcriptional activation, and replication fork arrest and a N-terminal negative regulatory domain (NRD), which inhibits DNA binding. Interactions between the NRD and the C-terminal domain mask the DNA-binding domain, thereby modulating TTF-I activity. TTF-1 recognises a consensus sequence known as the ‘Sal box’, an 11-base pair motif that can be repeated up to 10 times downstream of the 3′ end of the pre-rRNA sequence. Binding of TTF-I to the Sal box is crucial for halting RNA polymerase I elongation, thereby facilitating pre-rRNA synthesis termination through the formation of a replication fork arrest. TTF-I plays a dual role in rDNA regulation: it terminates ribosomal gene transcription, mediates replication fork arrest but also regulates RNA polymerase I transcription. In addition to these terminator elements, another TTF-I binding site is typically located ~170 base pairs upstream of the transcription start site, where it plays a crucial role in transcriptional regulation.
It can trigger nucleosome remodeling and to antagonize repression of ribosomal gene transcription on chromatin templates. TTF1 interacts with two critical components, the cockayne syndrome B protein (CSB) and the nucleolar remodeling complex (NoRC). NoRC has been shown to play an essential role in rDNA silencing. However, interaction with TIP5, a subunit of NoRC, recovers DNA-binding activity and facilitates both DNA methylation and histone deacetylation, resulting in the silencing of rDNA [33,34]. Conversely, when TTF1 binds to CSB, it activates chromatin through remodeling and epigenetic modifications, thereby promoting ribosomal gene transcription. The activity of TTF1 is further regulated by nucleolin (C23), which prevents the recruitment of TIP5 and histone deacetylase 1 (HDAC1), key factors in establishing a repressive heterochromatin state. Nucleolin depletion results in increased heterochromatin marks (H3K9me2) and reduced euchromatin marks (H4K12Ac, H3K4me3), underscoring its role in maintaining an active chromatin state.
4. RNA Polymerase I and Transcriptional Factors
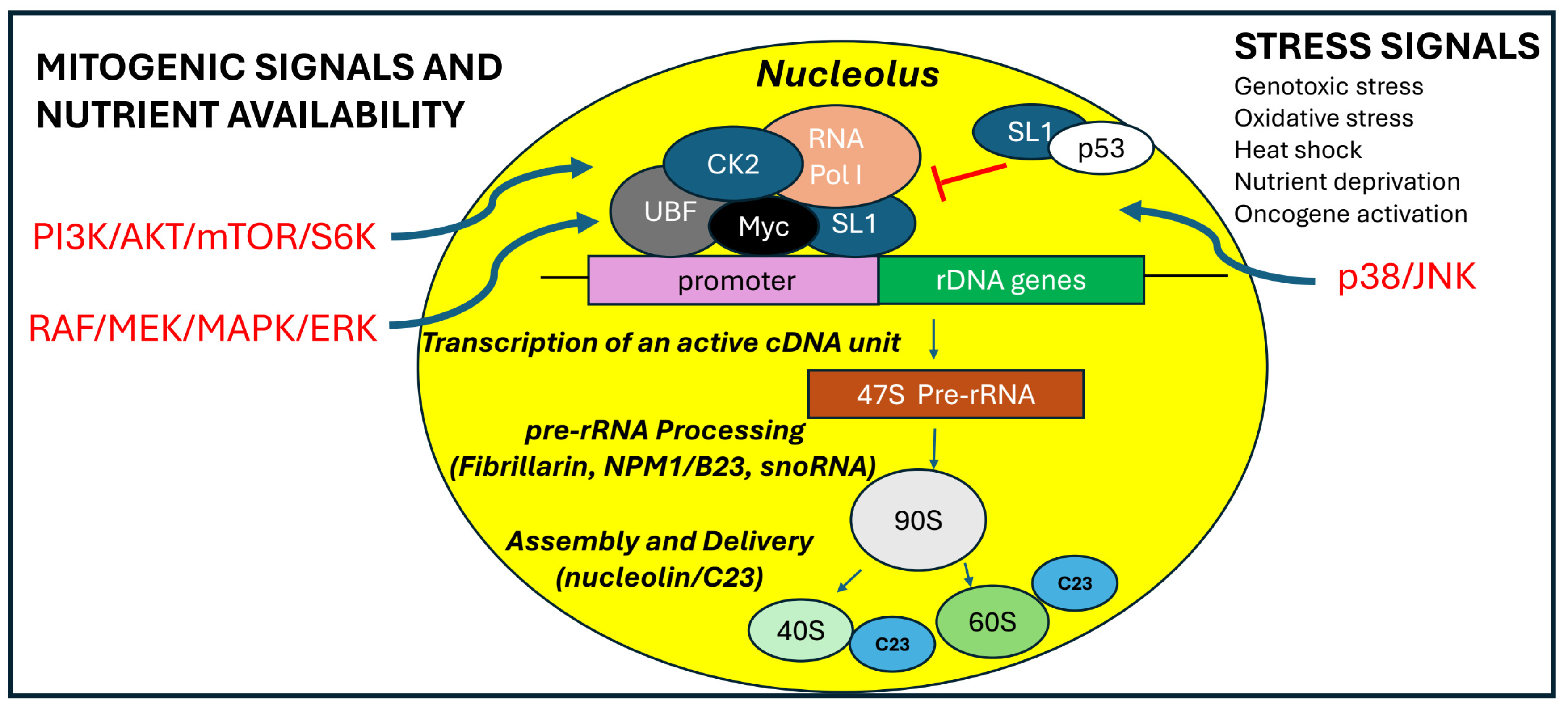
The eukaryotic ribosomal rRNA genes 18S, 28S and 5.8S are transcribed by RNA polymerase I. The recruitment of RNA Pol I to the transcription start site is the result of a series of interactions between specific transcription factors and rDNA promoter. The promoter of the ribosomal gene contains two critical elements: the core element and the upstream control element (UCE). Two transcription factors are required to efficiently activate the rDNA promoter: the selective factor 1 (SL1) and the upstream binding factor (UBF). The RNA polymerase I can interact with both UBF and SL1. The cooperative interaction between these factors creates a stable template, with two UBF molecules forming a dimer bound to the upstream promoter element and at least one SL1 molecule bound to the core promoter element. The UBF binding to promoter elements induces a structural conformation that mimics a nucleosome fold, facilitating the initiation of transcription. [35]. In addition, the retinoblastoma protein (pRb), interacts with UBF and prevents UBF-dependent activation of rDNA transcription. On the contrary the SV40 large T antigen, activates RNA Polymerase I transcription by interacting with SL1. Both SL1 and UBF are subject to regulation via phosphorylation and acetylation [36,37]. UBF recruits and activates selectivity factor 1 (SL-1) (
Figure 3) that consists of a TATA-binding protein (TBP) and five additional factors: TAFI110, TAFI48, TAFI63, TAFI12, and TAFI41 [38,39,40]. In mammalian cells, RNA polymerase I activated complex were found to contain core RNA polymerase I subunits and the preinitiation factor RRN3. Both genetic and biochemical experiments have demonstrated that RRN3 is essential for rDNA transcription. Current models suggest that RRN3 acts as a bridge between RNA polymerase I and the committed rDNA promoter [41]. RNA Polymerase I contains four peripheral subunits unique to this enzyme: A43 (human RPA43), A14, A49, and A34.5. Preinitiation factor RRN3 binds to the A43-A14 stalk and recruits the RRN3-Pol I complex to the rDNA promoter. RRN3 directly associates with the A43 subunit of RNA Polymerase I, enabling the enzyme for transcription initiation [42]. Bioinformatic approach of the sequence of RRN3 identified a domain with weak identity to the DNA binding domain of heat shock transcription factor 2 [43]. Similarly, the association of RNA polymerase I-specific transcription initiation factor RRN3 with RPA43 prevents the enzyme dimerization and maintains RNA Polymerase I in its monomeric form [44]. The subunits TAFI63 and TAFI110 interact with RRN3 on the RRN3-Pol I complex and recruit RNA Polymerase I to the rDNA promoter. Together, these proteins form the preinitiation complex (PIC) that bind the promoter favourable for transcription initiation. RNA Polymerase I transitions from its open complex, where it is bound to DNA, into its elongation complex, actively synthesizing rRNA. During the elongation, in the active site, two magnesium cations in the catalytic domain coordinate a NTP condensation reaction. Moreover, when RNA polymerase I encounters a DNA lesion or a mis incorporated nucleotide, it undergoes a transcriptional pause, awaiting the activation of the cell’s DNA damage repair mechanisms. During the transcription process, the RNA polymerase I can catalyse RNA cleavage. This activity requires the homologous subunits A12.2, Rpb9, and C11. Inefficient RNA cleavage further leads to proofreading errors [45,46]. The transcription termination elements are positioned on two separate sites on the rDNA gene repeat at the 3 end of the transcribed region and upstream of the transcription start site. Transcription termination factor I (TTF-I) binds to the termination element at the 3 ends of the transcribed region and triggers the RNA polymerase I to pause. The subunit A12.2 is required for the RNA Pol I release from the DNA template. External factors, such as zinc availability and temperature, also influence RNA Polymerase I activity. Conditions that harm cell growth, including stress, nutrient starvation, down-regulate transcription of rDNA genes whereas agents that stimulate growth and proliferation upregulate rDNA transcription. TIF-IA, the RNA Pol I-associated transcription factor that transmits external signals to the nucleolar transcription machinery, is targeted by a variety of protein kinases that phosphorylate TIF-IA at multiple sites. Energy deficits and a high AMP/ATP leading to the phosphorylation and inactivation of RRN3 [47]. Moreover, rDNA transcription fluctuates during the cell cycle, being low in early G
1-phase, reaching highest levels in S- and G
2-phase, and being shut off in mitosis [48]. Cell cycle-dependent transcription of RNA Polymerase I is also achieved by post-translational modifications of the transcription factors SL-1 and UBF. During the S and G2 phases, both SL-1 and UBF are fully active, contributing to transcription initiation during these stages [48]. During mitosis, SL-1 is phosphorylated, preventing its interaction with UBF and impairing transcription initiation. SL-1 is dephosphorylated at the end of mitosis, but UBF remains inactive until late G1. In proliferating cells, UBF phosphorylation enhances its interaction with SL-1 and RNA Pol I, whereas in quiescent cells, UBF remains hypophosphorylated and inactive. Additionally, UBF acetylation, regulated by the acetyltransferase CBP, promotes rDNA transcription by counteracting the repressive effects of tumor suppressors [48]. Notably, the retinoblastoma protein (pRb) has been shown to suppress ribosomal transcription both in vitro and in vivo by directly interacting with UBF.
RNA Polymerase I and Myc. Myc is the most powerful inducer of ribosome biogenesis. It stimulates the expression of RNA Polymerase I-associated transcription factors and other nucleolar proteins, enabling high RNA Polymerase I transcription rates and increased ribosome biogenesis [49,50]. Nucleophosmin (NPM1 or B23), a highly abundant nucleolar protein that interacts with Myc, and directs its nucleolar localization. The hyperactivation of nucleoli, which can be triggered by oncogene activation plays a critical role in carcinogenesis and cancer progression. The transition from cellular quiescence to cell cycle entry and proliferation is driven by the Myc-dependent attachment of ribosomal DNA to the nuclear matrix via the non-transcribed intergenic spacer (IGS) region of rDNA [51]. Oncogenic Myc physically associates with rDNA remodels the chromatin. Similarly, the Myc inhibitor reduces induction of pre-rRNA levels as well as the level of matrix attachment in growing cells. The 10058-F4 Myc inhibitor inhibits interaction of Myc with its hetero-dimerization partner, Max, suggesting the possibility that Myc/Max could bind to E-box sites that are found throughout the IGS sequence [52]. Additionally, genes encoding protein components of the ribosomes displayed an increased mRNA expression upon Myc activation [53]. Myc enhances rRNA upregulation by facilitating the binding of DNA consensus elements to selectivity factor 1 (SL1), which subsequently recruits RNA Polymerase I and stimulates rRNA transcription, driving cell cycle entry. Notably, Myc is frequently overexpressed in various cancers [54].
RNA Polymerase I and p53. In normal cells, surveillance systems based on tumor suppressors have evolved to counteract excessive changes in ribosome biosynthesis and inhibit cell growth. Ribosome biogenesis is regulated by several tumor suppressors, including the Alternative Reading Frame (ARF), p53, phosphatase and tensin homolog (PTEN), and retinoblastoma protein (pRB). The ARF protein has been shown to regulate the cell cycle through both p53-dependent and p53-independent pathways [55]. ARF is also recognized as a negative regulator of rRNA transcription and maturation. Specifically, ARF binds to and inhibits the phosphorylation of the upstream binding transcription factor (UBF) [56]. Additionally, ARF promotes the sumoylation of various interacting proteins, such as Topoisomerase I, MDM2, p53, and the Early Growth Response protein (EGR1) [57,58]. The ARF-mediated sumoylation of EGR1 is essential for PTEN activation, which directly regulates cell size and protein synthesis [59,60]. Tumor suppressors like pRB and p53 further inhibit RNA polymerase I activity and disrupt the assembly of the transcriptional machinery on the rDNA promoter. Under stress conditions, ARF sequesters MDM2, an E3 ubiquitin ligase, thereby stabilizing p53 levels in the nucleolus. In the nucleolus, p53 inhibits RNA polymerase I activity by sequestering the SL1 factor. Another regulator of RNA polymerase I is the early growth response 1 (EGR1) protein. Activated by stress signals, it functions as a negative regulator of RNA polymerase I. EGR1 localizes to the nucleolus, where its function is closely associated with the expression of the nucleolar proteins nucleophosmin (NPM or B23) and ARF [59].
5. RNA Polymerase I and Cell Signaling
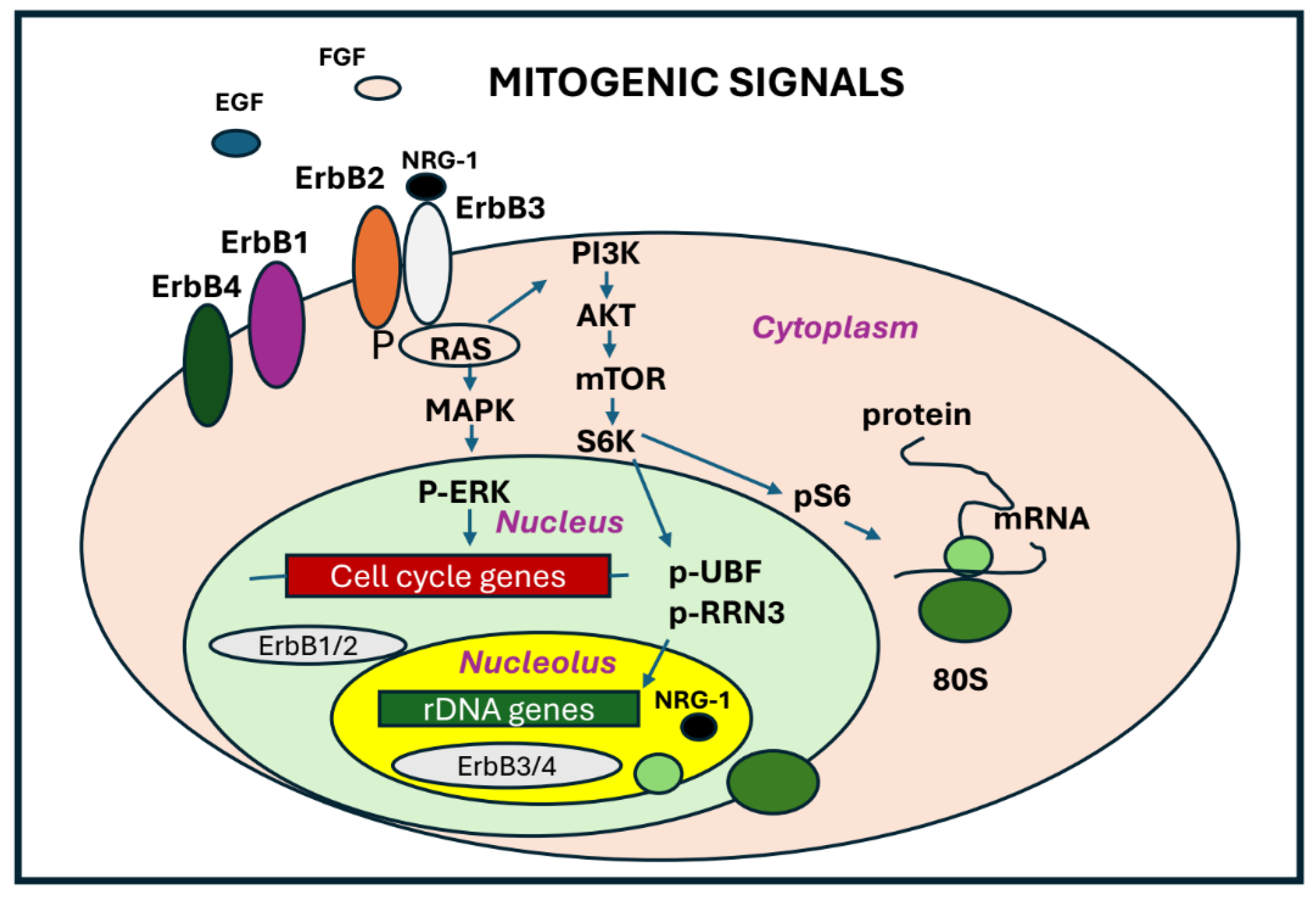
The cellular response to stress is a crucial adaptation mechanism to environmental changes. This response is characterized by significant alterations in gene expression, enabling the cell to maintain homeostasis and survival. Nucleolar metabolism is influenced by the interaction between pathways activated from extracellular signals to coordinate ribosome synthesis and cell proliferation [61]. The regulation of RNA Polymerase I is governed by post-translational modifications in response to external stimuli. One of the key regulatory for this process is the mammalian target of rapamycin (mTOR) protein. The mTOR signaling pathway integrates a wide range of signals to control genes involved in the control of the cellular growth and nutrient response. A major effect of mTOR signaling is the upregulation of RNA Polymerase I transcription, mediated through the mTOR complex 1 (mTORC1) and its downstream activation of the kinase S6K1 in response to increased nutrient availability [62]. S6K1 activation leads to the phosphorylation of the initiation factors RRN3 and UBF [63], promoting RNA Polymerase I activity (
Figure 4). At the same time, the increased nucleotide demand driven by increased RNA Pol I activity is achieved by mTORC1-dependent stimulation of purine and pyrimidine synthesis pathways. During an increased cell growth, mTOR and protein kinase 2 (CK2) further phosphorylate UBF, thereby increasing the RNA Polymerase I activity [64]. RNA Polymerase I activity can also be enhanced by growth stimuli transmitted via the MAPK signaling pathway. In response to growth factor signaling, Ras-GTP activates the MAPK pathway, which triggers the nuclear import of ERK. Subsequently, ERK phosphorylates and activates RRN3, thereby enhancing RNA Pol I activity [65]. Growth factors that activate the ERK pathway promote UBF phosphorylation, enhancing its interaction with rDNA and increasing RNA Pol I transcription [66]. However, the level of 47S pre-rRNA decreases to varying extents depending on the specific stress-inducing agents. Stress conditions activate the p38-MAPK and c-Jun N-terminal kinase (JNK) pathways, which collaborate via shared upstream and downstream effectors to regulate gene expression in response to environmental challenges. This coordinated response, known as the environmental stress response (ESR), is triggered by factors such as osmotic and oxidative stress, inflammatory cytokines, and ribotoxic agents. Notably, ribosomal rRNA synthesis is more susceptible to oxidative stress than overall mRNA synthesis. JNKs, classified as stress-activated protein kinases (SAPKs), play a pivotal role in the cellular response to environmental stress, balancing pro-survival signals with pro-apoptotic pathways. Their activity is induced by alkylating agents, hyperosmotic shock, proinflammatory cytokines, and oxidative stress. Under such conditions, RNA polymerase I activity is impaired due to TIF-IA inactivation, mediated by JNK-dependent phosphorylation at threonine 200. This modification disrupts TIF-IA’s interaction with RNA Pol I and TIF-IB/SL1, preventing transcription initiation complex formation and causing TIF-IA translocation from the nucleolus to the nucleoplasm. Various stress stimuli also induce nucleolar protein relocalization. Tumor suppressor proteins like p53 are stabilized by nucleolar sequestration, while nucleolin (C23) and nucleophosmin (B23) are translocated to the nucleoplasm, where they contribute to cell cycle arrest [67]. Additionally, under nutrient deprivation, cells downregulate energy-intensive processes, such as ribosome biogenesis and protein synthesis. Low ATP levels suppress rRNA gene transcription, whereas elevated ATP levels enhance it, reflecting the tight coupling of ribosomal RNA synthesis with cellular energy availability. AMP-activated protein kinase (AMPK) plays a crucial role in translating changes in energy levels into adaptive cellular responses. The activation of AMPK by glucose deprivation or treatment with the AMP-mimetic drug AICAR (5-amino-4-imidazolecarboxamide ribonucleotide) leads to inactivation of TIF-IA both in vivo and in vitro. AMPK phosphorylates TIF-IA at serine 635 and this phosphorylation impairs the interaction of TIF-IA with the TBP-containing promoter selectivity factor SL1. Consequently, recruitment of RNA Pol I to the rDNA promoter and transcription complex formation is impaired. Recently, members of the EGFR family have also been identified in the nucleolus of both normal and cancer cells [68]. These tyrosine kinase receptors have a large glycosylated extracellular domain, a hydrophobic transmembrane region and a cytoplasmic domain, which carry the tyrosine kinase activity [69]. The ErbB receptors are expressed in various tissues of epithelial, mesenchymal and neuronal origin. They have been implicated in various aspects of neural development, including circuit generation, axon lining, neurotransmission, and synaptic plasticity. Substantial evidence supports the involvement of ErbB family in the development and progression of various types of cancer [70]. In numerous cells, multiple members of the ErbB receptor family are co-expressed. Generally, these receptors function on the cell membrane, where they bind to growth factors or specific ligands. Upon binding, they undergo homo or heterodimerization, activating their intrinsic tyrosine kinase activity and recruiting adaptor proteins that trigger downstream signaling pathways [71]. This signaling generates crosstalk with multiple cellular processes, such as proliferation and cell motility. Additionally, mitogenic growth factors, including neuregulin-1 and its isoform NRG1-β3, ligands of ErbB3 receptor have been identified in the nucleus and nucleolus of the cell. The receptors ErbB1 and ErbB2 have been identified in the nucleus as full-length proteins, while ErbB3 and ErbB4 have been found in the nucleus and nucleolus as truncated isoforms [72,73,74]. Two shorter ErbB3 variants have been identified in the nucleus and nucleolus of human cells. The first variant, an 80-kDa ErbB3, is involved in regulating the transcription of the cyclin D1 gene [75,76]. The second variant, a 50-kDa ErbB3, colocalizes with fibrillarin in the nucleoli of various tumor cell lines and primary glioblastoma cells [77] and relocates from the nucleolus to the cytoplasm, when the levels of the NRG-1 ligand increase, promoting pre-rRNA synthesis. The 50-kDa variant of ErbB3 found in glioblastoma nucleoli also interacts with key ribosome biogenesis factors, such as the upstream binding factor (UBF). Silencing ErbB3 in glioma cell lines blocks the cell cycle and reduces proliferation. This effect is disrupted by actinomycin D, which causes the nuclear accumulation of ErbB3 in association with the nucleolar protein nucleolin (C23). Nucleolin is an abundantly expressed nucleolar phosphoprotein of exponentially growing cells. It is present in abundance at the dense fibrillar and granular regions of nucleolus. This nucleolar protein is involved in the control of transcription of ribosomal RNA (rRNA), in ribosome assembly, and in nucleocytoplasmic delivery of ribosomal subunits. Recently it was demonstrated that nucleolin, is also present on the surface of several cells and its overexpression has been correlated with tumor grade, suggesting a its crucial role in tumor [78]. Despite these findings, the nuclear functions of ErbBs remain unclear, warranting further investigation into its impact on ribosome biogenesis and tumor progression.
7. The Role of the Nucleolus in Cancer
The nucleolus plays a crucial role as a sensor of various cellular stresses, including genotoxic and oxidative stress, nutrient deprivation, and oncogene activation [79]. Nucleolar morphology, including its shape, size, and number per nucleus, can be profoundly remodelled in disease. A well-established positive correlation exists between nucleolar size and the intensity of ribosome biosynthesis, particularly in cells undergoing neoplastic transformation and during cancer progression [80,81]. Notably, dysregulated synthesis of pre-rRNA transcripts has been associated with poor cancer prognosis [84,85]. Additionally, prominent nucleoli have frequently been linked to worse cancer outcomes [85]. In various tumors, intense staining of the short arms of chromosomes corresponding to nucleolar organizer regions (NORs) correlates with increased expression of ribosomal RNA genes [86]. Beyond cancer, nucleolar dysfunction has been implicated in a range of human diseases, particularly ribosomopathies. These disorders can be classified into two groups, with one major category being inherited bone marrow failure syndromes (IBMFS) and the Treacher Collins Syndrome. Notably, IBMFS-related ribosomopathies predispose patients to cancer development [87,88,89].
Fibrillarin and ribosome heterogeneity. The enlarged nucleoli and increased cell proliferation are observed along with more intensive rDNA transcriptional activity which is frequently associated with enhanced expression of factors involved in various stages of ribosome biosynthesis as upstream-binding factor (UBF), DNA topoisomerase I, fibrillarin (FBL), nucleolin (C23) and nucleophosmin (NMPI or B23), as well as small nucleolar RNAs (snoRNAs) and ribosomal proteins (RPs). Increased cancer cell aggressiveness has been linked not only to enhanced rRNA biosynthesis but also to the activation of specific pre-rRNA maturation pathways. These pathways introduce new post-transcriptional modifications into rRNA, leading to elevated ribosome production and altered translational functionality [90]. This alternative rRNA maturation process during ribosome biogenesis assigns new regulatory roles to rRNAs within the ribosome. Beyond serving as structural components, rRNAs actively modulate mRNA decoding during translation. Ribosome heterogeneity, driven by changes in rRNA post-transcriptional modifications, ribosomal protein stoichiometry, and post-translational modifications of ribosomal proteins, plays a crucial role in cancer development [91]. These alterations result in specialized ribosomes that selectively enhance the translation of specific proteins, potentially promoting tumor growth and progression [92]. A key nucleolar protein involved in post-transcriptional modifications of ribosomal precursor rRNAs is fibrillarin. As a C/D box snoRNA-guided methyltransferase, fibrillarin plays a critical role in rRNA 2′-O-methylation, pre-rRNA processing, and ribosome assembly. Recent studies have demonstrated that rRNA 2′-O-methylation is dynamic, influencing ribosomal translational capabilities and potentially altering protein synthesis in cancer [93,94]. Fibrillarin not only facilitates rRNA methylation but also regulates RNA polymerase I activity through the methylation of rDNA gene promoters. Specifically, it modulates the methylation of a glutamine residue on histone H2A at the glutamine 104, thereby influencing RNA Pol I transcription [95]. Fibrillarin expression is highly regulated in various physiological and pathological contexts, including development, stem cell differentiation, viral infections, and cancer. In cancer models, the nucleolar protein fibrillarin modulation influences tumor progression, with sustained expression prolonging the pluripotent state of mouse embryonic stem cells [96]. In breast cancer cells, altered fibrillarin expression correlates with changes in rRNA 2′-O-methylation, affecting translational accuracy and the initiation of mRNAs containing internal ribosome entry site (IRES) elements [97,98]. In the recent studies fibrillarin knockdown leads to site-specific alterations in 2′-O-methylation, including at critical ribosomal positions, selectively modifying ribosome function. These findings highlight the extensive modulation potential of rRNA 2′-O-methylation patterns. The site-specific effects of fibrillarin knockdown suggest that regulating common components of the methylation machinery can fine-tune 2′-O-methylation patterns, thereby influencing ribosome function.
The observed downregulation of fibrillarin during neurogenesis and stem cell differentiation may directly impact rRNA modification and ribosome activity [99]. Conversely, fibrillarin overexpression in tumors could increase 2′-O-methylation at specific sites, altering translational control in cancer cells.
Nucleolus and epithelial-mesenchymal transition (EMT). The induction of rDNA transcription is also associated with cellular plasticity, dedifferentiation, and stemness. The nucleolus has been identified as a localization site for the transcriptional factor Snail, a key inducer of epithelial-mesenchymal transition (EMT). EMT is a crucial process during embryonic development. During this process the epithelial cells acquire fibroblast-like characteristics, decreases intercellular adhesion and increases the cell motility [100,101]. Additionally, Snail contributes to tumor progression by downregulating EMT-associated genes, including E-cadherin (E-cad) and claudin. E-cadherin is a member of cadherin family and component of adhesion junctions and the primary organizer of the epithelial phenotype [102,103,104]. The loss of E-cad expression is associated with tumor metastasis and induced expression of E-cad in cancer cells can prevent tumor progression and invasion [105,106,107]. Additionally, an increased Snail levels contribute to tumor resistance against various chemotherapeutic drugs [108]. Snail activity also induces the expression of genes associated with an invasive phenotype, such as fibronectin and the metalloprotease 9 (MMP9). The expression of Snail is regulated by a complex signaling network that includes integrin-linked kinase (ILK), phosphatidylinositol 3-kinase (PI3K), mitogen-activated protein kinases (MAPKs), glycogen synthase kinase 3-beta (GSK-3β), and NF-κB. The TGF-β/Smad pathway, which induce EMT in hepatocytes, epithelial cells, and mesothelial cells, activates Snail expression. Additionally, Notch signaling employs distinct mechanisms that act synergistically to regulate Snail synthesis. Snail’s subcellular localization is regulated by phosphorylation, particularly through p21-activated kinase 1 (PAK1). This enzyme phosphorylates Snail at serine 246, promoting its nuclear localization and enhancing its transcriptional activity. The stable and active form of Snail localizes only in the nucleus of the cell. In the cytoplasm, Snail has a short half-life due to ubiquitin-mediated proteasomal degradation. Several E3 ubiquitin ligases, including FBXW7 and FBXL14 promote Snail degradation via the proteasome [109]. Conversely the deubiquitinates enzyme such as USP3 and USP11 play a crucial role in regulating Snail stability removing the ubiquitin subunits from the protein.
Nucleolar accumulation of Snail is induced specifically by ribotoxic stress characterised by the activation of the nucleolar deubiquitinase USP36 that consequent increases the Snail1 stability. The nucleolar localization of Snail is positively correlated with an increase in 47S pre-rRNA level and the silencing of USP36 abrogates this effect [110]. Conversely the Snail K157R mutant is unable to accumulate in the nucleolus, and Snail knockdown results in a significant reduction in 47S pre-rRNA expression. Furthermore, the USP36 knockdown also strongly inhibits 47S pre-rRNA expression. All this finding has address to identification of new USP36/Snail nucleolar axis that promotes ribosome biogenesis. The increasing of the expression of ribosomal precursor 47-pre-rRNA induced by Snail, improves ribosome biogenesis and support cancer cell survival under stress conditions. Also, chemotherapeutic agents such as doxorubicin, translation inhibitors like cycloheximide, and well-known ribotoxic stress inducers such as puromycin and blasticidin disrupt ribosome function by activating the p38/JNK signaling pathways, leading to increase of the nucleolar USP36 and Snail1 protein expression. In contrast, rapamycin, an mTOR inhibitor, fails to activate JNK signaling or upregulate USP36 and Snail. The nucleolar USP36-Snail axis includes another crucial player component, the ribosomal regulatory protein1 (RR1) [111]. RR1 regulates ribosome biogenesis by recruiting 5S ribonucleoprotein (RNP) to form the pre-60S ribosomal subunit with ribosomal production factor 2 (Rpf2). Its depletion results in the accumulation of the pre-60 subunit in the nucleoplasm and stalling of the ribosome synthesis. The RR1 is also component of a second important axis, the RPL11-c-Myc-Snail axis. This axis is involved in the regulation of the invasion and metastasis of cancer cells. The accumulation of the ribosomal protein RPL11 in the nucleoplasm inhibited c-Myc-dependent transcription of EMT-related genes. USP36 is also a crucial regulator of Myc [112]. It directly binds to Myc and interacts specifically with tumor suppressor Fbw7γ in the nucleolus, but not with Fbw7α present in the nucleoplasm [113]. The downregulation of USP36 reduce significantly the levels of Myc and inhibited cell proliferation. Furthermore, the knockdown of USP36 abolished the c-Myc induction following serum stimulation demonstrating that USP36 plays a critical role in c-Myc stabilization in response to growth signals. The USP36 overexpression was also observed in many human cancers such as breast and lung cancers, implying its oncogenic nature.
8. Conclusions
The nucleolus has long been considered as a distinct subnuclear compartment exclusively dedicated to ribosome biogenesis. However, its spatial segregation from key receptors and extracellular signaling pathways has contributed to an underestimation of its function as cell sensor. However emerging evidence indicates that the nucleolus acts as a dynamic integrative hub where crucial signaling pathways converge with the aim to regulate cellular proliferation and survival. Beyond its primary role in ribosome production, the nucleolus constantly modulates the rate of ribosome biogenesis in response to cellular metabolic demands. The nucleolar sequestration of proto-oncogenes, such as MYC, and tumor suppressors, including p53, plays a pivotal role in the control of ribosome biogenesis, protein synthesis, and cell cycle progression. In cancer this sophisticated control is deregulated with the scope to induce abnormal synthesis of the ribosome to support the high level of cell proliferation. However, under specific cell growth condition, the nucleolus can influence differential post-transcriptional modifications of 47S pre-rRNA with the production of ribosome heterogeneity. Recent evidence attribute to the nucleolus a role of critical regulator of chemoresistance. Under ribosome stress condition of growth, it can counteract the repression of 47S pre-rRNA synthesis recruiting the transcription factors Snail, a key driver of epithelial-mesenchymal transition (EMT) inducing cell survival. All these finding open a new function of the nucleolus as a central player in tumor biology and cellular adaptation and tumor biology. Targeting nucleolar metabolism may provide novel therapeutic strategies by unveiling previously unrecognized molecular mechanisms that play a crucial cell fate decision.
Author Contributions
Conceptualization, D.P.; writing and review, D.P.; funding acquisition, D.P. The author has read and agreed to the published version of the manuscript.
Conflicts of Interest
The author declares no conflicts of interest.
Abbreviations
| ErbB |
Receptor tyrosine-protein kinase |
| UBF |
Upstream binding factor |
| ARF |
Alternate reading frame protein |
| EGR1 |
Early growth response 1 |
| ActD |
Actinomycin D |
| ErbB3 |
v-erb-b erythroblastic leukemia viral oncogene homolog |
| SL-1 |
Selectivity factor 1 |
| mTOR |
Mammalian target of rapamycin |
| IGS |
Intergenic spacers |
| PTEN |
Phosphatase and tensin homolog |
| pRB |
retinoblastoma protein |
| NORs |
nucleolus organizer regions |
| FC |
Fibrillar centers |
| DFC |
Dense fibrillar components |
| GC |
Granular component |
| rRNA |
Ribosomal RNA |
| rDNA |
Ribosomal DNA |
| NPM |
Nucleophosmin |
| B23 |
Nucleophosmin |
| TTF-1 |
Transcription termination factor I |
| FBL |
Fibrillarin |
| C23 |
Nucleolin |
| CK2 |
Protein kinase 2 |
References
- Hurt∙ E., Cheng J., ∙ Baβler∙J., Iwasa∙J., Beckmann R. SnapShot: Eukaryotic ribosome biogenesis I v186, 10 p2282-2282.e1; 2023. [CrossRef]
- Yuan F., Xu C., Li G., Tong T. Nucleolar TRF2 attenuated nucleolus stress-induced HCC cell-cycle arrest by altering rRNA synthesis. Cell Death Dis. 2018. [CrossRef]
- Shore, D., Albert B. Ribosome biogenesis and the cellular energy economy. Curr Biol. 2022; 32:R611-R617. [CrossRef]
- Dubois, M.L., Boisvert F.M. The Nucleolus: Structure and Function. The Functional Nucleus. 2016: 23:29–49. [CrossRef]
- Lewis, J.D. and Tollervey D. Like attracts like: getting RNA processing together in the nucleus. Science. 2000; 288:1385–9. [PubMed: 10827942]).
- PanichnantakulP., LisbethC., Aguilar L.,Evan Daynard E., Mackenzie Guest M., Colten Peters C., Jackie Vogel J., Oeffinger M. Protein UFMylation regulates early events during ribosomal DNA-damage response. 2024; 43:9114738.
- Nelson, J.O., Watase G.J, Warsinger-Pepe N., Yamashita Y.M. Mechanisms of rDNA Copy Number Maintenance. Trends Genet. 2019;35:734-742. [CrossRef]
- Lindström MS, et al., Nucleolus as an emerging hub in maintenance of genome stability and cancer pathogenesis. Oncogene, 2018; 37:2351–2366. [PubMed: 29429989.
- Schöfer C and Weipoltshammer K. Nucleolus and chromatin. Histochem Cell Biol. 2018; 150:209–225. [PubMed: 30046888].
- Yu, S. and Lemos B. The long-range interaction map of ribosomal DNA arrays. PLoS Genet. 2018; 14: p. e1007258. [PubMed: 29570716].
- Sklias A, Cruciani S, Marchand V, Spagnuolo M, Lavergne G, Bourguignon V, Brambilla A, Dreos R, Marygold SJ, Novoa EM, Motorin Y, Roignant JY. Comprehensive map of ribosomal 2’-O-methylation and C/D box snoRNAs in Drosophila melanogaster. Nucleic Acids Res. 2024 Apr 12;52(6):2848-2864. [CrossRef]
- Castillo Duque de Estrada NM, Thoms M, Flemming D, Hammaren HM, Buschauer R, Ameismeier M, Baßler J, Beck M, Beckmann R, Hurt E. Structure of nascent 5S RNPs at the crossroad between ribosome assembly and MDM2-p53 pathways. Nat Struct Mol Biol. 2023 30(8):1119-1131. [CrossRef]
- King, M.R., Ruff K.M., Pappu R.V. Emergent microenvironments of nucleoli. Nucleus. 2024; 15:2319957. [CrossRef]
- Lavering, E.D., Gandhamaneni M., Weeks D.L. Intrinsically disordered regions are not sufficient to direct the compartmental localization of nucleolar proteins in the nucleus. PLoS Biol. 2023; 21: e3002378. [CrossRef]
- Tsekrekou, M., Stratigi K., Chatzinikolaou G. The Nucleolus: In Genome Maintenance and Repair. Int J Mol Sci. 2017; 18:1411. [CrossRef]
- Lafontaine, D.L.J., Riback J.A., Bascetin R., Brangwynne C.P. The nucleolus as a multiphase liquid condensate. Nat Rev Mol Cell Biol. 2021; 22:165-182. [CrossRef]
- Moore, H.M., Bai B., Boisvert F.M., Latonen L., Rantanen V., Simpson J.C., Pepperkok R., Lamond A.I., Laiho M. Quantitative proteomics and dynamic imaging of the nucleolus reveal distinct responses to UV and ionizing radiation. Mol Cell Proteomics. 2011; 10:M111.009241. [CrossRef]
- Andersen JS, Lam YW, Leung AK, Ong SE, Lyon CE, Lamond AI, Mann M. Nucleolar proteome dynamics. Nature. 2005 Jan 6;433(7021):77-83. [CrossRef] [PubMed]
- Aitken S, Semple CA. The circadian dynamics of small nucleolar RNA in the mouse liver. J R Soc Interface. 2017 May;14(130):20170034. [CrossRef] [PubMed] [PubMed Central]
- Xie W, Ling T, Zhou Y, Feng W, Zhu Q, Stunnenberg HG, Grummt I, Tao W. The chromatin remodeling complex NuRD establishes the poised state of rRNA genes characterized by bivalent histone modifications and altered nucleosome positions. Proc Natl Acad Sci U S A. 2012 May 22;109(21):8161-6. [CrossRef]
- Salifou K, Ray S, Verrier L, Aguirrebengoa M, Trouche D, Panov KI, Vandromme M. The histone demethylase JMJD2A/KDM4A links ribosomal RNA transcription to nutrients and growth factors availability. Nat Commun. 2016; 7:10174. [CrossRef]
- Wang M, Lemos B. Ribosomal DNA harbors an evolutionarily conserved clock of biological aging. Genome Res. 2019 Mar;29(3):325-333. [CrossRef] [PubMed] [PubMed Central]
- Moss, T., LeDoux M.S., and Crane-Robinson C. HMG-boxes, ribosomopathies and neurodegenerative disease. Front. Genet. 2023; 14:1225832. [CrossRef]
- Sanij E, Hannan RD. The role of UBF in regulating the structure and dynamics of transcriptionally active rDNA chromatin. Epigenetics. 2009;4(6):374-82. [CrossRef]
- Cerqueira, A. V., Lemos, B. Ribosomal DNA and the Nucleolus as Keystones of Nuclear Architecture, Organization, and Function Trends in Genetics. 2019; 35:710-723; doi.org/10.1016/j.tig.2019.07.
- Mais C, Wright JE, Prieto JL, Raggett SL, McStay B. UBF-binding site arrays form pseudo-NORs and sequester the RNA polymerase I transcription machinery. Genes Dev. 2005;19(1):50-64. [CrossRef]
- Dousset T, Wang C, Verheggen C, Chen D, Hernandez-Verdun D, Huang S. Initiation of nucleolar assembly is independent of RNA polymerase I transcription. Mol Biol Cell. 2000;11(8):2705-17. [CrossRef]
- Ueshima S, Nagata K, Okuwaki M. Internal Associations of the Acidic Region of Upstream Binding Factor Control Its Nucleolar Localization. Mol Cell Biol. 2017; 37(22): e00218-17. [CrossRef]
- Chen, J., Teo B.H.D., Cai Y., Wee S.Y.K., Lu J. The linker histone H1.2 is a novel component of the nucleolar organizer regions. J. Biol. Chem. 2018; 293:2358-2369. [CrossRef]
- Hamdane N, Stefanovsky VY, Tremblay MG, Németh A, Paquet E, Lessard F, Sanij E, Hannan R, Moss T. Conditional inactivation of Upstream Binding Factor reveals its epigenetic functions and the existence of a somatic nucleolar precursor body. PLoS Genet. 2014; 10(8):e1004505. [CrossRef]
- Santoro, R. The silence of the ribosomal RNA genes. Cell Mol Life Sci. 2005;62: 2067-79. [CrossRef]
- Vinod Tiwari, Beverly A Baptiste, Mustafa N Okur, Vilhelm A Bohr, Current and emerging roles of Cockayne syndrome group B (CSB) protein, Nucleic Acids Research. 2021; 49:2418–2434. [CrossRef]
- Akamatsu, Y., Kobayashi T. The Human RNA Polymerase I Transcription Terminator Complex Acts as a Replication Fork Barrier That Coordinates the Progress of Replication with rRNA Transcription Activity. Mol Cell Biol. 2015; 35:1871-81. [CrossRef]
- Németh, A., Strohner R., Grummt I., Längst G. The chromatin remodeling complex NoRC and TTF-I cooperate in the regulation of the mammalian rRNA genes in vivo. Nucleic Acids Res. 2004; 32:4091-9. [CrossRef]
- Voit R, Grummt I. Phosphorylation of UBF at serine 388 is required for interaction with RNA polymerase I and activation of rDNA transcription. Proc Natl Acad Sci U S A. 2001; 98(24):13631-6. [CrossRef]
- Cavanaugh AH, Hempel WM, Taylor LJ, Rogalsky V, Todorov G, Rothblum LI. Activity of RNA polymerase I transcription factor UBF blocked by Rb gene product. Nature. 1995;374(6518):177-80. [CrossRef]
- O’Mahony DJ, Xie WQ, Smith SD, Singer HA, Rothblum LI. Differential phosphorylation and localization of the transcription factor UBF in vivo in response to serum deprivation. In vitro dephosphorylation of UBF reduces its transactivation properties. J Biol Chem. 1992;267(1):35-8. [PubMed]
- Pitts, S., Laiho M. Regulation of RNA Polymerase I Stability and Function. Cancers. 2022; 14:5776. [CrossRef]
- Panov, K.I, Friedrich J.K, Russell J., Zomerdijk J.C. UBF activates RNA polymerase I transcription by stimulating promoter escape. EMBO J. 2006; 25:3310-22. [CrossRef]
- Gorski, J.J., Pathak S., Panov K., Kasciukovic T., Panova T., Russell J., Zomerdijk J.C. A novel TBP-associated factor of SL1 functions in RNA polymerase I transcription. EMBO J. 2007:1560-8. [CrossRef]
- Sadian, Y., Baudin F., Tafur L., Murciano B., Wetzel R., Weis F., Müller C.W. Molecular insight into RNA polymerase I promoter recognition and promoter melting. Nat Commun. 2019; 10:5543. [CrossRef]
- Peyroche G, Milkereit P, Bischler N, Tschochner H, Schultz P, Sentenac A, Carles C, Riva M. The recruitment of RNA polymerase I on rDNA is mediated by the interaction of the A43 subunit with Rrn3. EMBO J. 2000; 19(20):5473-82. [CrossRef]
- Stepanchick A, Zhi H, Cavanaugh AH, Rothblum K, Schneider DA, Rothblum LI. DNA binding by the ribosomal DNA transcription factor rrn3 is essential for ribosomal DNA transcription. J Biol Chem. 2013;288(13):9135-44. [CrossRef]
- Pilsl M, Crucifix C, Papai G, Krupp F, Steinbauer R, Griesenbeck J, Milkereit P, Tschochner H, Schultz P. Structure of the initiation-competent RNA polymerase I and its implication for transcription. Nat Commun. 2016;7:12126. [CrossRef]
- Schwank, K.,∙ Schmid ∙C., Tobias Fremter T.,∙ Philipp Milkereit P., Joachim Griesenbeck J. Herbert Tschochner H. RNA polymerase I (Pol I) lobe-binding subunit Rpa12.
- Ruan W, Lehmann E, Thomm M, Kostrewa D, Cramer P. Evolution of two modes of intrinsic RNA polymerase transcript cleavage. J Biol Chem. 2011;286(21):18701-7. [CrossRef]
- Hoppe S, Bierhoff H, Cado I, Weber A, Tiebe M, Grummt I, Voit R. AMP-activated protein kinase adapts rRNA synthesis to cellular energy supply. Proc Natl Acad Sci U S A. 2009; 106(42):17781-6. [CrossRef]
- Meraner J, Lechner M, Loidl A, Goralik-Schramel M, Voit R, Grummt I, Loidl P. Acetylation of UBF changes during the cell cycle and regulates the interaction of UBF with RNA polymerase I. Nucleic Acids Res. 2006;34(6):1798-806. [CrossRef]
- Arabi, A., Wu S., Ridderstråle K., Bierhoff H., Shiue C., Fatyol K., Fahlén S., Hydbring P., Söderberg O., Grummt I., Larsson L.G., Wright A.P. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat Cell Biol. 2005; 7:303-10. [CrossRef]
- van Riggelen, J., Yetil, A. & Felsher, D. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer 10, 301–309 (2010). [CrossRef]
- Chiou-Nan Shiue, Amir Nematollahi-Mahani, Anthony P.H. Wright, Myc-induced anchorage of the rDNA IGS region to nucleolar matrix modulates growth-stimulated changes in higher-order rDNA architecture, Nucleic Acids Research, Volume 42, Issue 9, 14 May 2014, Pages 5505–5517. [CrossRef]
- Grandori C. Mac J. Siebelt F. Ayer D.E. Eisenman R.N. Myc-Max heterodimers activate a DEAD box gene and interact with multiple E box-related sites in vivo. 19096; EMBO J: 15: 4344-4357.
- Destefanis, F., Manara V., Bellosta P. Myc as a Regulator of Ribosome Biogenesis and Cell Competition: A Link to Cancer. Int J Mol Sci. 2020; 21:4037. [CrossRef]
- Moon, H., Park H., Ro S.W. c-Myc-driven Hepatocarcinogenesis. Anticancer Res. 2021;41: 4937-4946. [CrossRef]
- Boone, D.N., Qi Y., Li Z., Hann S.R. Egr1 mediates p53-independent c-Myc-induced apoptosis via a non-canonical ARF-dependent transcriptional mechanism. 2011; Proc Natl Acad Sci USA; 108: 632–637. [CrossRef]
- Ayrault, O., Andrique, L., Fauvin, D. et al. Human tumor suppressor p14ARF negatively regulates rRNA transcription and inhibits UBF1 transcription factor phosphorylation. Oncogene 25, 7577–7586 (2006). [CrossRef]
- Dimitris P Xirodimas, June Chisholm, Joana M.S Desterro, David P Lane, Ronald T Hay, P14ARF promotes accumulation of SUMO-1 conjugated (H)Mdm2; 2002; FEBS Letters; 528:207-211. [CrossRef]
- Chen, L., Chen J. MDM2-ARF complex regulates p53 sumoylation. Oncogene. 2003; 22:5348–57.
- Ponti, D., Bellenchi G.C., Puca R., Bastianelli D., Maroder M., Ragona G., et al. The transcription factor EGR1 localizes to the nucleolus and is linked to suppression of ribosomal precursor synthesis. PLoS One. 2014. [CrossRef]
- Ponti, D., Bastianelli D., Rosa P., Pacini L., Ibrahim M., Rendina E.A. et al. The expression of B23 and EGR1 proteins is functionally linked in tumor cells under stress conditions. BMC Cell Biol. 2015; 16:27.
- Stefanovsky, V., Langlois F., Gagnon-Kugler T., Rothblum L.I., Moss T. Growth factor signaling regulates elongation of RNA polymerase I transcription in mammals via UBF phosphorylation and R-chromatin remodeling. Mol Cell. 2006; 21:629–39.
- Hannan, K.M., Brandene perburger Y., Jenkins A., Sharkey K., Cavanaugh A., Rothblum L., Moss T., Poortinga G., McArthur G.A., Pearson R.B., Hannan R.D. mTOR-dependent regulation of ribosomal gene transcription requires S6K1 and is mediated by phosphorylation of the carboxy-terminal activation domain of the nucleolar transcription factor UBF. Mol Cell Biol. 2003;23(23):8862-77. [CrossRef]
- Hannan, K.M., Brandenburger Y., Jenkins A., Sharkey K., Cavanaugh A., Rothblum L., Moss T., Poortinga G., McArthur G.A., Pearson R.B., Hannan R.D. mTOR-dependent regulation of ribosomal gene transcription requires S6K1 and is mediated by phosphorylation of the carboxy-terminal activation domain of the nucleolar transcription factor UBF. Mol Cell Biol. 2003;8862-77. [CrossRef]
- Lin, C.Y., Navarro S., Reddy S., Comai L. CK2-mediated stimulation of Pol I transcription by stabilization of UBF-SL1 interaction. Nucleic Acids Res. 2006; 34:4752-66. [CrossRef]
- Blattner, C., Jennebach S., Herzog F., Mayer A., Cheung A.C., Witte G., Lorenzen K., Hopfner K.P., Heck A.J., Aebersold R., Cramer P. Molecular basis of Rrn3-regulated RNA polymerase I initiation and cell growth. Genes Dev. 2011; 25:2093-105. [CrossRef]
- Lavoie H., Gagnon J., Therrien M. ERK signalling: a master regulator of cell behaviour, life and fate. Nat Rev Mol Cell Biol. 2020; 21:607–632. [CrossRef]
- Mayer C, Bierhoff H, Grummt I. The nucleolus as a stress sensor: JNK2 inactivates the transcription factor TIF-IA and down-regulates rRNA synthesis. Genes Dev. 2005;19:933-41. [CrossRef]
- Andrique L, Fauvin D, Maassarani M, Colasson H, Vannier B, Séité P. ErbB3 80kDa, a nuclear variant of the ErbB3 receptor, binds to the Cyclin D1 pro moter to activate cell proliferation but is negatively controlled by p14ARF. Cell Signal. 2012; 24:1074–85.
- Wang Z. ErbB receptors and cancer. In: Wang Z, editor. ErbB receptor signaling. Methods in molecular biology, vol 1652. New York: Humana Press; 2017. [CrossRef]
- Kruspig B, Monteverde T, Neidler S, Hock A, Kerr E, Nixon C, et al. The Erbb network facilitates KRAS-driven lung tumorigeneis. Sci Transl Med. 2018. [CrossRef]
- Wang, Z. ErbB receptors and cancer. In: Wang Z, editor. ErbB receptor signaling. Methods in molecular biology, vol 1652. New York: Humana Press; 2017. [CrossRef]
- Wang YN, Yamaguchi H, Hsu JM, Hung MC. Nuclear trafficking of the epidermal growth factor receptor family membrane proteins. Oncogene. 2010; 29:3997-4006. [CrossRef]
- Giri DK, Ali-Seyed M, Li LY, Lee DF, Ling P, Bartholomeusz G, Wang SC, Hung MC. Endosomal transport of ErbB-2: mechanism for nuclear entry of the cell surface receptor. Mol Cell Biol. 2005;25:11005-18. [CrossRef]
- Ni CY, Murphy MP, Golde TE, Carpenter G. gamma -Secretase cleavage and nuclear localization of ErbB-4 receptor tyrosine kinase. Science. 2001; 294:2179-81. [CrossRef]
- Andrique L, Fauvin D, El Maassarani M, Colasson H, Vannier B, Séité P. ErbB3(80 kDa), a nuclear variant of the ErbB3 receptor, binds to the Cyclin D1 promoter to activate cell proliferation but is negatively controlled by p14ARF. Cell Signal. 2012; 24:1074-85. [CrossRef]
- Reif, R., Adawy A., Vartak N., Schröder J., Günther G., Ghallab A., Schmidt M., Schormann W., Hengstler J.G. Activated ErbB3 Translocates to the Nucleus via Clathrin-independent Endocytosis, Which Is Associated with Proliferating Cells. J Biol Chem. 2016; 291:3837-47. [CrossRef]
- Tagliaferro, M., Rosa P., Bellenchi G.C., Bastianelli D., Trotta R., Tito C., Fazi F., Calogero A., Ponti D. Nucleolar localization of the ErbB3 receptor as a new target in glioblastoma. BMC Mol Cell Biol. 2022;23:13. [CrossRef]
- Balça-Silva, J., do Carmo A., Tão H., Rebelo O., Barbosa M., Moura-Neto V., Sarmento-Ribeiro A.B., Lopes M.C., Moreira J.N. Nucleolin is expressed in patient-derived samples and glioblastoma cells, enabling improved intracellular drug delivery and cytotoxicity. Exp Cell Res. 2018; 370:68-77. [CrossRef]
- González-Arzola, K. The nucleolus: Coordinating stress response and genomicstability.2024;1867;195029. [CrossRef]
- Derenzini M, Trerè D, Pession A, Montanaro L, Sirri V, Ochs RL. Nucleolar function and size in cancer cells. Am J Pathol. 1998;152(5):1291-7.
- Montironi R, Braccischi A, Scarpelli M, Matera G, Alberti R. Value of quantitative nucleolar features in the preoperative cytological diagnosis of follicular neoplasias of the thyroid. J Clin Pathol. 1991;44:509-14. [CrossRef]
- Belin S, Beghin A, Solano-Gonzàlez E, Bezin L, Brunet-Manquat S, Textoris J, Prats AC, Mertani HC, Dumontet C, Diaz JJ. Dysregulation of ribosome biogenesis and translational capacity is associated with tumor progression of human breast cancer cells. PLoS One. 2009;4(9):e7147. [CrossRef]
- Williamson D, Lu YJ, Fang C, Pritchard-Jones K, Shipley J. Nascent pre-rRNA overexpression correlates with an adverse prognosis in alveolar rhabdomyosarcoma. Genes Chromosomes Cancer. 2006;45(9):839-45. [CrossRef]
- Gamel JW, McLean IW. Computerized histopathologic assessment of malignant potential. II. A practical method for predicting survival following enucleation for uveal melanoma. Cancer. 1983; 52:1032-8.
- van Diest PJ, Mouriquand J, Schipper NW, Baak JP. Prognostic value of nucleolar morphometric variables in cytological breast cancer specimens. J Clin Pathol. 1990 Feb;43(2):157-9. [CrossRef]
- Elhamamsy AR, Metge BJ, Alsheikh HA, Shevde LA, Samant RS. Ribosome Biogenesis: A Central Player in Cancer Metastasis and Therapeutic Resistance. Cancer Res. 2022;82(13):2344-2353. [CrossRef]
- Goodpasture C, Bloom SE. Visualization of nucleolar organizer regions im mammalian chromosomes using silver staining. Chromosoma. 1975; 53(1):37-50. [CrossRef]
- Aspesi A, Ellis SR. Rare ribosomopathies: insights into mechanisms of cancer. Nat Rev Cancer. 2019;19(4):228-238. [CrossRef]
- Venturi G, Montanaro L. How Altered Ribosome Production Can Cause or Contribute to Human Disease: The Spectrum of Ribosomopathies. Cells. 2020;9(10):2300. [CrossRef]
- Mensah MA, Niskanen H, Magalhaes AP, Basu S, Kircher M, Sczakiel HL, Reiter AMV, Elsner J, Meinecke P, Biskup S, Chung BHY, Dombrowsky G, Eckmann-Scholz C, Hitz MP, Hoischen A, Holterhus PM, Hülsemann W, Kahrizi K, Kalscheuer VM, Kan A, Krumbiegel M, Kurth I, Leubner J, Longardt AC, Moritz JD, Najmabadi H, Skipalova K, Snijders Blok L, Tzschach A, Wiedersberg E, Zenker M, Garcia-Cabau C, Buschow R, Salvatella X, Kraushar ML, Mundlos S, Caliebe A, Spielmann M, Horn D, Hnisz D. Aberrant phase separation and nucleolar dysfunction in rare genetic diseases. Nature. 2023 Feb;614(7948):564-571. [CrossRef]
- Babaian A, Rothe K, Girodat D, Minia I, Djondovic S, Milek M, Spencer Miko SE, Wieden HJ, Landthaler M, Morin GB, Mager DL. Loss of m1acp3Ψ Ribosomal RNA Modification Is a Major Feature of Cancer. Cell Rep. 2020; 31:107611. [CrossRef]
- Li D, Wang J. Ribosome heterogeneity in stem cells and development. J Cell Biol. 2020; 219:e202001108. [CrossRef]
- Xue M, Dong L, Zhang H, Li Y, Qiu K, Zhao Z, Gao M, Han L, Chan AKN, Li W, Leung K, Wang K, Pokharel SP, Qing Y, Liu W, Wang X, Ren L, Bi H, Yang L, Shen C, Chen Z, Melstrom L, Li H, Timchenko N, Deng X, Huang W, Rosen ST, Tian J, Xu L, Diao J, Chen CW, Chen J, Shen B, Chen H, Su R. METTL16 promotes liver cancer stem cell self-renewal via controlling ribosome biogenesis and mRNA translation. J Hematol Oncol. 2024;17(1):7. [CrossRef]
- Sklias A, Cruciani S, Marchand V, Spagnuolo M, Lavergne G, Bourguignon V, Brambilla A, Dreos R, Marygold SJ, Novoa EM, Motorin Y, Roignant JY. Comprehensive map of ribosomal 2’-O-methylation and C/D box snoRNAs in Drosophila melanogaster. Nucleic Acids Res. 2024;52(6):2848-2864. [CrossRef]
- Barros-Silva D, Klavert J, Jenster G, Jerónimo C, Lafontaine DLJ, Martens-Uzunova ES. The role of OncoSnoRNAs and Ribosomal RNA 2’-O-methylation in Cancer. RNA Biol. 2021;61-74. [CrossRef]
- Iyer-Bierhoff A, Krogh N, Tessarz P, Ruppert T, Nielsen H, Grummt I. SIRT7-Dependent Deacetylation of Fibrillarin Controls Histone H2A Methylation and rRNA Synthesis during the Cell Cycle. Cell Rep. 2018;25(11):2946-2954.e5. [CrossRef]
- Sun X, Gao C, Xu X, Li M, Zhao X, Wang Y, Wang Y, Zhang S, Yan Z, Liu X, Wu C. FBL promotes cancer cell resistance to DNA damage and BRCA1 transcription via YBX1. EMBO Rep. 2023;24(9): e56230. [CrossRef]
- Schlatter S, Senn C, Fussenegger M. Modulation of translation-initiation in CHO-K1 cells by rapamycin-induced heterodimerization of engineered eIF4G fusion proteins. Biotechnol Bioeng. 2003; 83:210-25. [CrossRef]
- Erales J, Marchand V, Panthu B, Gillot S, Belin S, Ghayad SE, Garcia M, Laforêts F, Marcel V, Baudin-Baillieu A, Bertin P, Couté Y, Adrait A, Meyer M, Therizols G, Yusupov M, Namy O, Ohlmann T, Motorin Y, Catez F, Diaz JJ. Evidence for rRNA 2’-O-methylation plasticity: Control of intrinsic translational capabilities of human ribosomes. Proc Natl Acad Sci U S A. 2017;114(49):12934-12939. [CrossRef]
- Han Z, Zhang Q, Zhu Y, Chen J, Li W. Ribosomes: An Exciting Avenue in Stem Cell Research. Stem Cells Int. 2020; 2020:8863539. [CrossRef]
- Brombin A, Joly JS, Jamen F. New tricks for an old dog: ribosome biogenesis contributes to stem cell homeostasis. Curr Opin Genet Dev. 2015; 34:61-70. [CrossRef]
- Aigner K, Dampier B, Descovich L, Mikula M, Sultan A, Schreiber M, Mikulits W, Brabletz T, Strand D, Obrist P, Sommergruber W, Schweifer N, Wernitznig A, Beug H, Foisner R, Eger A. The transcription factor ZEB1 (deltaEF1) promotes tumour cell dedifferentiation by repressing master regulators of epithelial polarity. Oncogene. 2007;26(49):6979-88. [CrossRef]
- Gumbiner, BM. Regulation of cadherin-mediated adhesion in morphogenesis. Nat Rev Mol Cell Biol. 2005 Aug;6(8):622-34. [CrossRef] [PubMed]
- Moody SE, Perez D, Pan TC, Sarkisian CJ, Portocarrero CP, Sterner CJ, Notorfrancesco KL, Cardiff RD, Chodosh LA. The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell. 2005;8(3):197-209. [CrossRef]
- De Craene B, Berx G. Snail in the frame of malignant tumor recurrence. Breast Cancer Res. 2006;8(4):105. [CrossRef]
- Whiteman EL, Liu CJ, Fearon ER, Margolis B. The transcription factor snail represses Crumbs3 expression and disrupts apico-basal polarity complexes. Oncogene. 2008;27(27):3875-9. [CrossRef]
- Peinado H, Ballestar E, Esteller M, Cano A. Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/ HDAC2 complex. Mol Cell Biol 2004; 24:306-19.
- Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol 2006; 7:131-42.
- Qin K, Yu S, Liu Y, Guo R, Guo S, Fei J, Wang Y, Jia K, Xu Z, Chen H, Li F, Niu M, Dai MS, Dai L, Cao Y, Zhang Y, Xiao ZJ, Yi Y. USP36 stabilizes nucleolar Snail1 to promote ribosome biogenesis and cancer cell survival upon ribotoxic stress. Nat Commun. 2023;14:6473. [CrossRef]
- Yu Q, Zhou BP, Wu Y. The regulation of snail: on the ubiquitin edge. Cancer Cell Microenviron. 2017;4(2):e1567.
- Qin, K., Yu, S., Liu, Y. et al. USP36 stabilizes nucleolar Snail1 to promote ribosome biogenesis and cancer cell survival upon ribotoxic stress. Nat Commun 14, 6473 (2023). [CrossRef]
- Wang R, Peng C, Song J, Hua Y, Wu Q, Deng L, Cao Y, Zhang J, Zhang L, Wu L, Hou L. Downregulated RRS1 inhibits invasion and metastasis of BT549 through RPL11-c-Myc-SNAIL axis. Int J Oncol. 2022;60(3):33. [CrossRef]
- Sun XX, Sears RC, Dai MS. Deubiquitinating c-Myc: USP36 steps up in the nucleolus. Cell Cycle. 2015;14(24):3786-93. [CrossRef]
- Sun XX, He X, Yin L, Komada M, Sears RC, Dai MS. The nucleolar ubiquitin-specific protease USP36 deubiquitinates and stabilizes c-Myc. Proc Natl Acad Sci U S A. 2015;112(12):3734-9. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).