Submitted:
20 March 2025
Posted:
21 March 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Plant Material & DNA Extraction
2.2. Molecular Analysis; PCR Amplification & SSR Markers
2.3. Data statistical Analysis
2.3.1. Variation Within Population
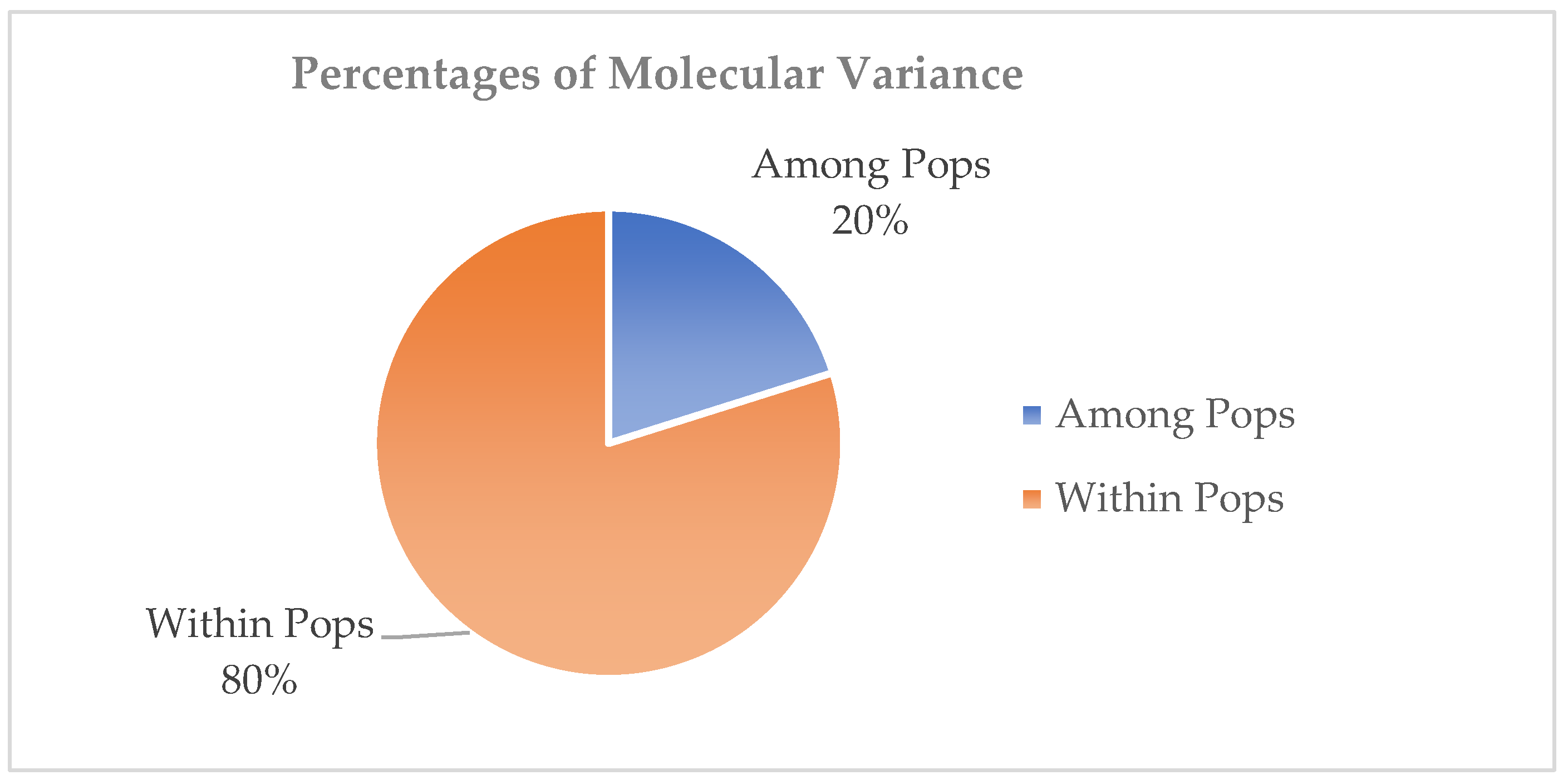
2.3.2. Variation Among Populations
3. Results
3.1. Genetic Variation Within Populations
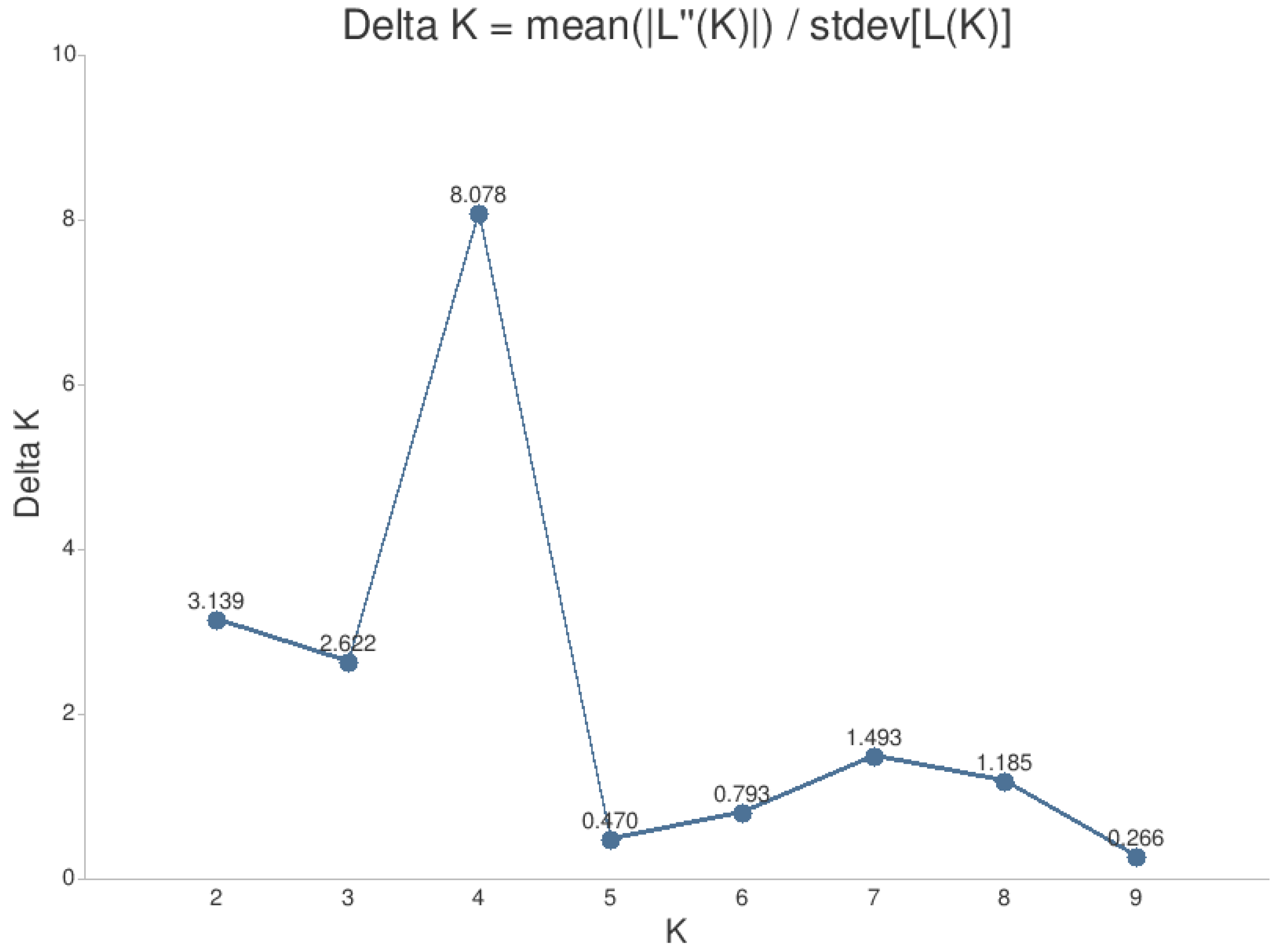
3.2. Genetic Variation Between and Within Populations
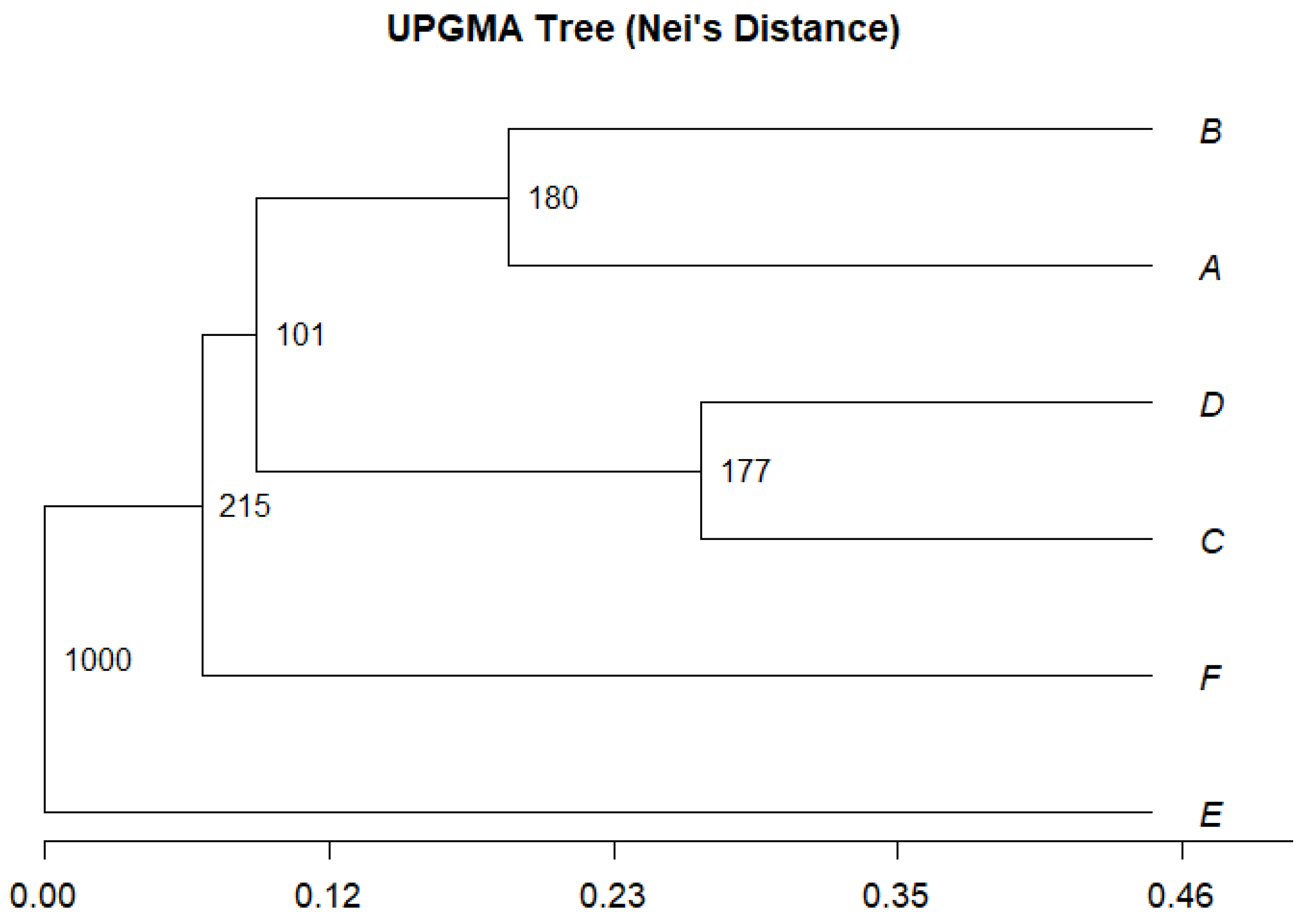
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments

Conflicts of Interest
References
- Arias, T.; Pires, J.C. A fully resolved chloroplast phylogeny of the brassica crops and wild relatives (Brassicaceae: Brassiceae): Novel clades and potential taxonomic implications. Taxon 2012, 61, 980–988. [Google Scholar] [CrossRef]
- Lysak, M.A.; Koch, M.A.; Pecinka, A.; Schubert, I. Chromosome triplication found across the tribe Brassiceae. Genome research 2005, 15, 516–525. [Google Scholar] [CrossRef] [PubMed]
- Schranz, M.E.; Lysak, M.A.; Mitchell-Olds, T. The ABC’s of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends in plant science 2006, 11, 535–542. [Google Scholar] [CrossRef] [PubMed]
- Tatout, C.; Warwick, S.; Lenoir, A.; Deragon, J.-M. SINE insertions as clade markers for wild crucifer species. Molecular Biology and Evolution 1999, 16, 1614–1614. [Google Scholar] [CrossRef]
- Eiseley, L.C.; Grote, A. Charles Darwin, Edward Blyth, and the Theory of Natural Selection. Proceedings of the American Philosophical Society 1959, 103, 94–158. [Google Scholar]
- Mabry, M.E.; Turner-Hissong, S.D.; Gallagher, E.Y.; McAlvay, A.C.; An, H.; Edger, P.P.; Moore, J.D.; Pink, D.A.; Teakle, G.R.; Stevens, C.J. The evolutionary history of wild, domesticated, and feral Brassica oleracea (Brassicaceae). Molecular Biology and Evolution 2021, 38, 4419–4434. [Google Scholar] [CrossRef]
- Ndondo, J.T.K. Review of the Food and Agriculture Organisation (FAO) Strategic Priorities on Food Safety 2023. In Food Safety-New Insights; IntechOpen: 2023.
- Sciandrello, S.; Brullo, C.; Brullo, S.; Giusso Del Galdo, G.; Minissale, P.; Salmeri, C. A new species of Brassica sect. Brassica (Brassicaceae) from Sicily. Plant Biosystems-An International Journal Dealing with all Aspects of Plant Biology 2013, 147, 812–820. [Google Scholar] [CrossRef]
- Hodgkin, T. Cabbages, kales, etc. 76-82. In. J. Smartt andN. Simmonds (ed.). Evolution of crop plants. 1995.
- Maggioni, L.; von Bothmer, R.; Poulsen, G.; Lipman, E. Domestication, diversity and use of Brassica oleracea L., based on ancient Greek and Latin texts. Genetic Resources and Crop Evolution 2018, 65, 137–159. [Google Scholar] [CrossRef]
- Snogerup, S.; Gustafsson, M.; Von Bothmer, R. Brassica sect. Brassica (Brassicaceae) I. taxonomy and variation. Willdenowia 1990, 271–365. [Google Scholar]
- Gustafsson, M.; Bentzer, B.; Bothmer, R.v.; Snogerup, S. Meiosis in Greek Brassica of the oleracea group. 1976.
- Dixon, G. Origins and diversity of Brassica and its relatives. In Vegetable brassicas and related crucifers; CABI Wallingford UK: 2006; pp. 1–33.
- Lázaro, A.; Aguinagalde, I. Genetic Diversity in Brassica oleracea L. (Cruciferae) and Wild Relatives (2n= 18) using RAPD Markers. Annals of Botany 1998, 82, 829–833. [Google Scholar] [CrossRef]
- Widén, B.; Andersson, S.; Rao, G.Y.; Widén, M. Population divergence of genetic (co) variance matrices in a subdivided plant species, Brassica cretica. Journal of Evolutionary Biology 2002, 15, 961–970. [Google Scholar] [CrossRef]
- Grant, V. Modes and origins of mechanical and ethological isolation in angiosperms. Proceedings of the National Academy of Sciences 1994, 91, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Frankham, R. Inbreeding and extinction: a threshold effect. Conservation Biology 1995, 9, 792–799. [Google Scholar] [CrossRef]
- Hu, S.; Yu, C.; Zhao, H.; Sun, G.; Zhao, S.; Vyvadilova, M.; Kucera, V. Genetic diversity of Brassica napus L. Germplasm from China and Europe assessed by some agronomically important characters. Euphytica 2007, 154, 9–16. [Google Scholar]
- Mishra, P.; Singh, N. Allelic diversity among short duration maize (Zea mays L.) genotypes using SSR markers. Madras Agricultural Journal 2012, 99, 1. [Google Scholar] [CrossRef]
- Xu, Y.; Xing, M.; Song, L.; Yan, J.; Lu, W.; Zeng, A. Genome-wide analysis of simple sequence repeats in cabbage (Brassica oleracea L.). Frontiers in Plant Science 2021, 12, 726084. [Google Scholar] [CrossRef]
- Allender, C.; Allainguillaume, J.; Lynn, J.; King, G.J. Simple sequence repeats reveal uneven distribution of genetic diversity in chloroplast genomes of Brassica oleracea L. and (n= 9) wild relatives. Theoretical and Applied Genetics 2007, 114, 609–618. [Google Scholar] [CrossRef]
- Shi, J.; Huang, S.; Zhan, J.; Yu, J.; Wang, X.; Hua, W.; Liu, S.; Liu, G.; Wang, H. Genome-wide microsatellite characterization and marker development in the sequenced Brassica crop species. DNA research 2014, 21, 53–68. [Google Scholar] [CrossRef]
- Thakur, A.K.; Singh, K.H.; Singh, L.; Nanjundan, J.; Khan, Y.J.; Singh, D. SSR marker variations in Brassica species provide insight into the origin and evolution of Brassica amphidiploids. Hereditas 2018, 155, 1–11. [Google Scholar] [CrossRef]
- Kumar, M.; Choi, J.-Y.; Kumari, N.; Pareek, A.; Kim, S.-R. Molecular breeding in Brassica for salt tolerance: importance of microsatellite (SSR) markers for molecular breeding in Brassica. Frontiers in Plant Science 2015, 6, 688. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical bulletin 1987.
- Edh, K.; Widén, B.; Ceplitis, A. Nuclear and chloroplast microsatellites reveal extreme population differentiation and limited gene flow in the Aegean endemic Brassica cretica (Brassicaceae). Molecular Ecology 2007, 16, 4972–4983. [Google Scholar] [CrossRef] [PubMed]
- Peakall, R.; Smouse, P.E. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Kalinowski, S.T. hp-rare 1.0: a computer program for performing rarefaction on measures of allelic richness. Molecular eEcology Notes 2005, 5, 187–189. [Google Scholar] [CrossRef]
- Chapuis, M.-P.; Estoup, A. Microsatellite null alleles and estimation of population differentiation. Molecular Biology and Evolution 2007, 24, 621–631. [Google Scholar] [CrossRef]
- Nei, M. Analysis of gene diversity in subdivided populations. Proceedings of the national academy of sciences 1973, 70, 3321–3323. [Google Scholar] [CrossRef]
- Cavalli-Sforza, L.L.; Edwards, A.W. Phylogenetic analysis. Models and estimation procedures. American Journal of Human Genetics 1967, 19, 233. [Google Scholar]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Molecular ecology resources 2015, 15, 1179–1191. [Google Scholar] [CrossRef]
- Schliep, K.; Potts, A.A.; Morrison, D.A.; Grimm, G.W. Intertwining phylogenetic trees and networks; 2167-9843; PeerJ Preprints: 2016.
- Paradis, E.; Schliep, K. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 2019, 35, 526–528. [Google Scholar] [CrossRef]
- Higgins, S.I.; Lavorel, S.; Revilla, E. Estimating plant migration rates under habitat loss and fragmentation. Oikos 2003, 101, 354–366. [Google Scholar] [CrossRef]
- Neilson, R.P.; Pitelka, L.F.; Solomon, A.M.; Nathan, R.; Midgley, G.F.; Fragoso, J.M.; Lischke, H.; Thompson, K. Forecasting regional to global plant migration in response to climate change. Bioscience 2005, 55, 749–759. [Google Scholar] [CrossRef]
- Snogerup, S. The Mediterranean islands. Plant Conservation in the Mediterranean area 1985, 159–173. [Google Scholar]
- Bittkau, C.; Comes, H.P. Molecular inference of a Late Pleistocene diversification shift in Nigella s. lat. (Ranunculaceae) resulting from increased speciation in the Aegean archipelago. Journal of Biogeography 2009, 36, 1346–1360. [Google Scholar] [CrossRef]
- Runemark, H. Reproductive drift, a neglected principle in reproductive biology. 1969.
- El-Esawi, M.A.; Germaine, K.; Bourke, P.; Malone, R. Genetic diversity and population structure of Brassica oleracea germplasm in Ireland using SSR markers. Comptes rendus biologies 2016, 339, 133–140. [Google Scholar] [CrossRef]
- Maggioni, L.; von Bothmer, R.; Poulsen, G.; Härnström Aloisi, K. Survey and genetic diversity of wild Brassica oleracea L. germplasm on the Atlantic coast of France. Genetic Resources and Crop Evolution 2020, 67, 1853–1866. [Google Scholar] [CrossRef]
- Watson-Jones, S.; Maxted, N.; Ford-Lloyd, B. Population baseline data for monitoring genetic diversity loss for 2010: a case study for Brassica species in the UK. Biological Conservation 2006, 132, 490–499. [Google Scholar] [CrossRef]
- POWO. Plants of the world online. Facilitated by the Royal Botanic Gardens, Kew. Published on the Internet. 2023.
- Euro+ Med, E. Euro+ Med PlantBase–the information resource for Euro-Mediterranean plant diversity. Available online at: 2006.
- Rakow, G. Species origin and economic importance of Brassica. In Brassica; Springer: 2004; pp. 3–11.
- Kioukis, A.; Michalopoulou, V.A.; Briers, L.; Pirintsos, S.; Studholme, D.J.; Pavlidis, P.; Sarris, P.F. Intraspecific diversification of the crop wild relative Brassica cretica Lam. using demographic model selection. BMC genomics 2020, 21, 1–14. [Google Scholar] [CrossRef]
- Aguinagalde, I., Gómez-Campo, C., & Sanchez-Yelamo, M. D. A chemosystematic survey on wild relatives of Brassica oleracea L. Botanical journal of the Linnean Society 1992, 109, 57–67.
- Maggioni, L.; von Bothmer, R.; Poulsen, G.; Branca, F. Origin and Domestication of Cole Crops (Brassica oleracea L.): Linguistic and Literary Considerations1. Economic Botany 2010, 64, 109–123. [Google Scholar] [CrossRef]
- Palmgren, M.; Shabala, S. Adapting crops for climate change: regaining lost abiotic stress tolerance in crops. Frontiers in Plant Science 2024, 2, 1416023. [Google Scholar] [CrossRef]
- Batool, M.; El-Badri, A.M.; Hassan, M.U.; Haiyun, Y.; Chunyun, W.; Zhenkun, Y.; Jie, K.; Wang, B.; Zhou, G. Drought stress in Brassica napus: effects, tolerance mechanisms, and management strategies. Journal of Plant Growth Regulation 2022, 1–25. [Google Scholar] [CrossRef]
- Trunschke, J.; Junker, R.R.; Kudo, G.; Alexander, J.M.; Richman, S.K.; Till-Bottraud, I. Effects of climate change on plant-pollinator interactions and its multitrophic consequences. Alpine Botany 2024, 134, 115–121. [Google Scholar] [CrossRef]
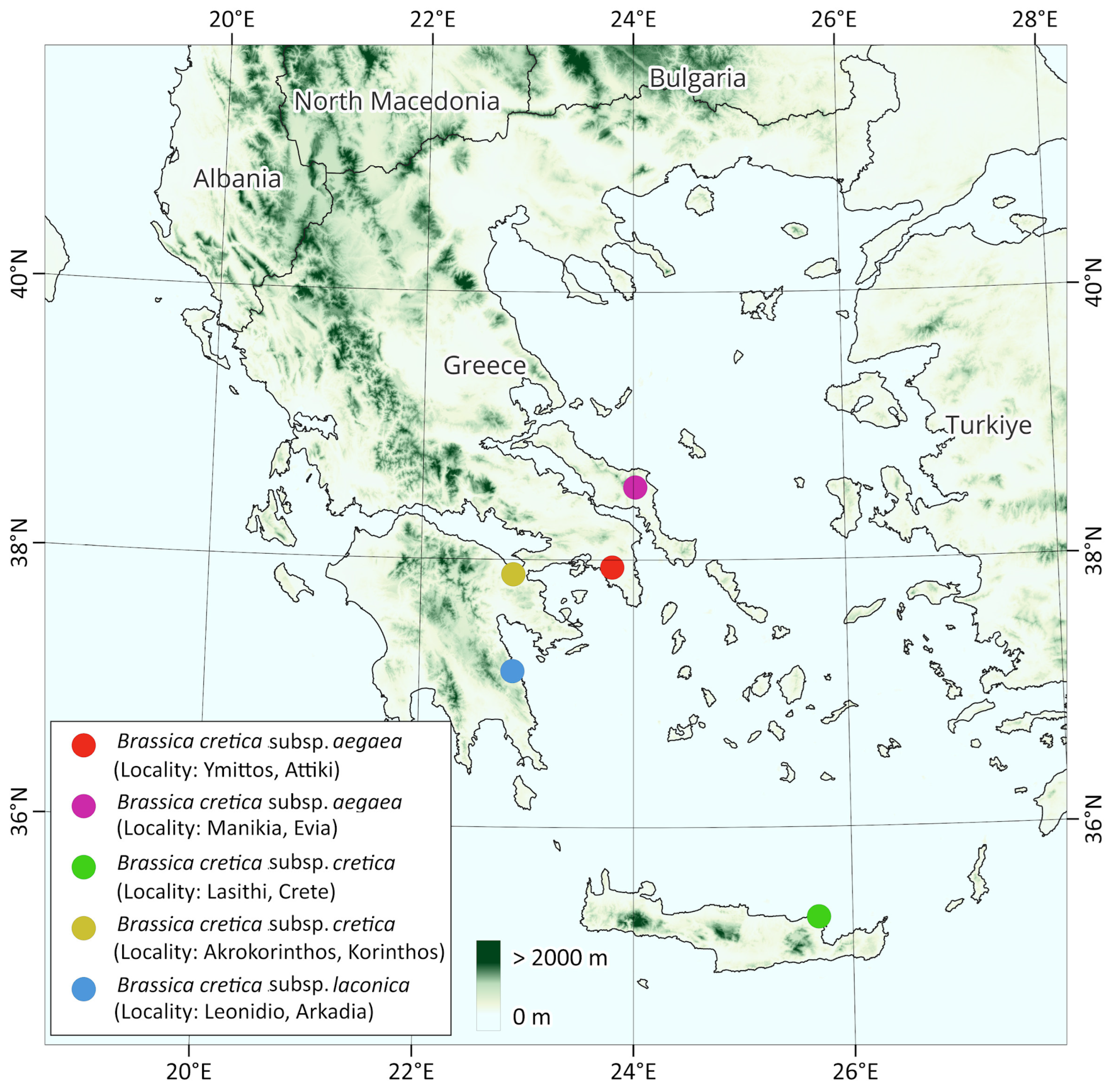

| CODE | Taxon | Locality | Country | Seed collection date | Latitude | Longitude |
|---|---|---|---|---|---|---|
| A | Brassica cretica subsp. aegaea | Manikia, Evia | Greece | June 2022 | 38ο 32.582′ | 24ο 1.052′ |
| B | Brassica cretica subsp. aegaea | Ymittos, Attiki | Greece | June 2022 | 37° 56.671′ | 23° 48.030′ |
| C | Brassica cretica subsp. cretica or nivea | Akrokorinthos, Korinthos | Greece | May 2022 | 37ο 53.363′ | 22ο 52.138′ |
| D | Brassica cretica subsp. cretica | Lasithi, Crete | Greece | May 2021 | 35ο 19.772′ | 25ο 41.458′ |
| E | Brassica cretica subsp. laconica | Leonidio, Arkadia | Greece | June 2022 | 37ο 9.940′ | 22ο 52.255′ |
| F | Brassica oleracea var Rubra | commercial variety of cabbage as outgroup |
| Locus | Forward & reverse primer sequences (5′→3′) | Repeat Motif | Size range (bp) | Ta (°C) |
|---|---|---|---|---|
| Ol10B11 | AAAATGTGAGGCTGTTTGGG TTTCGCAGCAGTAAACATGG |
(GA)25 | 76–180 | 52,5 |
| Ol10B01 | CCTCTTCAGTCGAGGTCTGG AATTTGGAAACAGAGTCGCC |
(GA)20 | 160–280 | 56 |
| Ol09A01 | TTCGAAGCTCATTATCGCAG CCGGGCTCTCTCTCTCTCTC |
(GA)75 | 120–340 | 56,5 |
| Ol10F11 | TTTGGAACGTCCGTAGAAGG CAGCTGACTTCGAAAGGTCC |
(GGC)7 | 139–184 | 56 |
| Ni4-B10 | GTCCTTGAGAAACTCCACCG CCGATCCCATTTCTAATCCC |
(CT)20 | 170–200 | 56 |
| sORA26 | TGTTTACCTGTTGGAGAT AACCCTAAGCATCTGCGA |
(GA)5 | 62–76 | 49 |
| BN12A | GCCGTTCTAGGGTTTGTGGGA GCCGTTCTAGGGTTTGTGGGA |
(GA)11(AAG)4 | 250–330 | 59 |
| Na10-F06 | CTCTTCGGTTCGATCCTCG TTTTTAACAGGAACGGTGGC |
(CCG)6 | 84–126 | 54,5 |
| nga111 | TGTTTTTTAGGACAAATGGCG CTCCAGTTGGAAGCTAAAGGG |
(GA)16 | 120–150 | 54,5 |
| MB4 | TGTTTTGATGTTTCCTACTG GAACCTGTGGCTTTTATTAC |
(TG)10 | 57–69 | 50 |
| Locus | Ol10B11 | Ol10B01 | Ol09A01 | Ol10F11 | Ni4-B10 | sORA26 | BN12A | Na10-F06 | nga111 | MB4 | MEAN |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Na | 27 | 30 | 32 | 19 | 19 | 18 | 18 | 16 | 20 | 20 | 21,9 |
| Ne | 3,606 | 3,985 | 4,107 | 2,489 | 2,844 | 2,759 | 2,629 | 2,413 | 2,805 | 2,859 | 3,049 |
| I | 1,374 | 1,438 | 1,418 | 0,977 | 1,081 | 1,052 | 1,005 | 0,907 | 1,082 | 1,114 | 1,145 |
| Ho | 0,075 | 0,35 | 0,308 | 0,308 | 0,175 | 0 | 0 | 0,05 | 0,017 | 0 | 0,128 |
| He | 0,720 | 0,734 | 0,705 | 0,583 | 0,638 | 0,634 | 0,61 | 0,568 | 0,631 | 0,645 | 0,647 |
| Population | Na | Ne | I | He | Ho | AR | pAR | FST |
|---|---|---|---|---|---|---|---|---|
| A | 4.400 | 3.519 | 1.279 | 0.677 | 0140 | 3.69 | 0.82 | 0.808 |
| B | 3800 | 3.191 | 1.218 | 0678 | 0.095 | 3.44 | 0.86 | 0.862 |
| C | 3200 | 2.917 | 1.091 | 0645 | 0.060 | 3.05 | 0.62 | 0.886 |
| D | 3700 | 3.043 | 1.131 | 0.636 | 0.125 | 3.25 | 0.09 | 0.840 |
| E | 3.300 | 2.747 | 1.075 | 0.629 | 0.130 | 3.05 | 0.60 | 0.792 |
| F | 3500 | 2.879 | 1.075 | 0.617 | 0.220 | 3.09 | 0.72 | 0.682 |
| Mean | 3.650 | 3.050 | 1.145 | 0.647 | 0.128 | 3.26 | 0.62 | 0.812 |
| A | B | C | D | E | F | |
|---|---|---|---|---|---|---|
| 0.000 | 0.081 | 0.147 | 0.094 | 0.145 | 0.125 | A |
| 0.122 | 0.000 | 0.128 | 0.123 | 0.156 | 0.148 | B |
| 0.203 | 0.172 | 0.000 | 0.074 | 0. 179 | 0,177 | C |
| 0.131 | 0.175 | 0.106 | 0.000 | 0.151 | 0.129 | D |
| 0.200 | 0.212 | 0.234 | 0.212 | 0.000 | 0.154 | E |
| 0.172 | 0.199 | 0.236 | 0.179 | 0.215 | 0.000 | F |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





