Submitted:
13 September 2024
Posted:
15 September 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
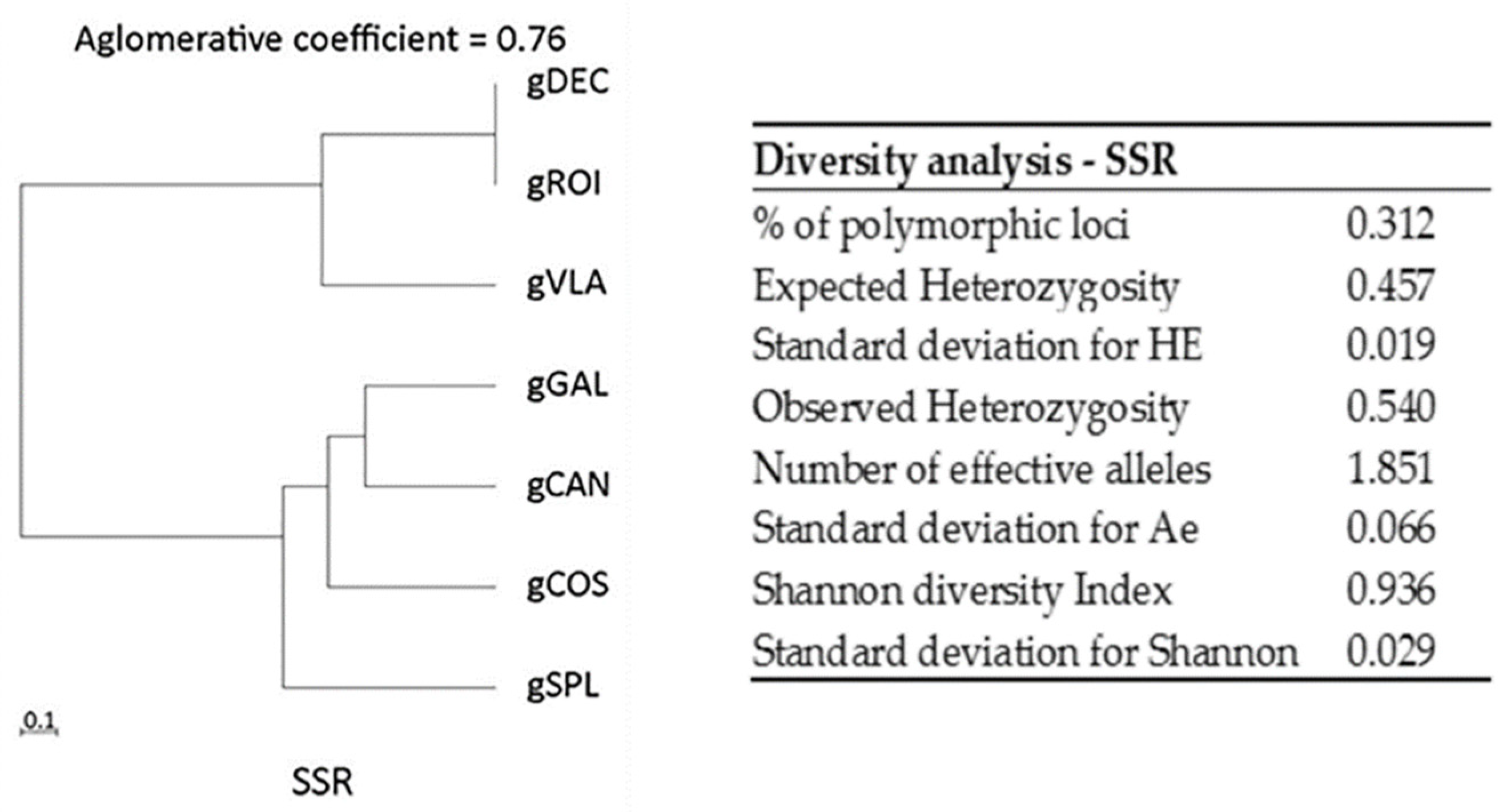
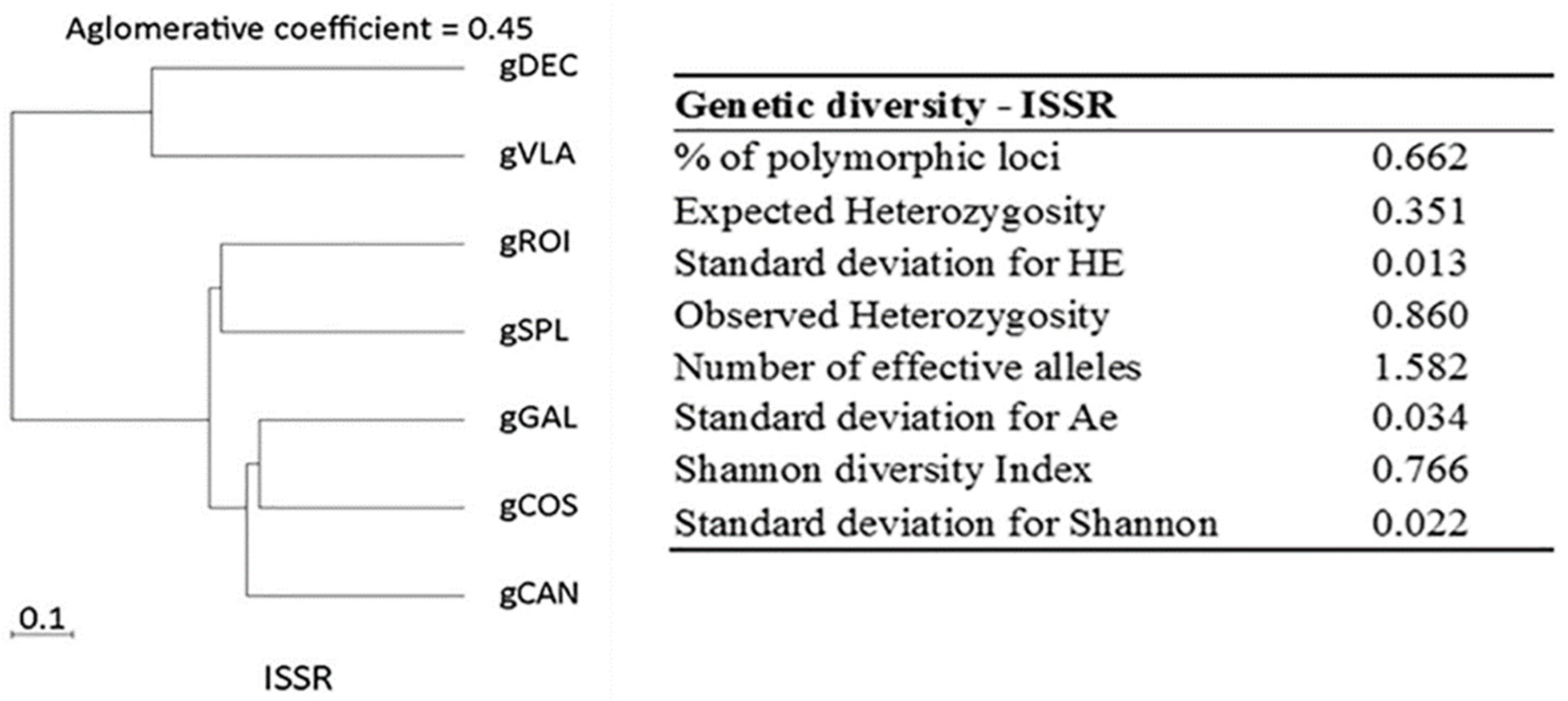
2.1. Genetic Diversity Analysis
2.1.1. SSR Analysis
2.1.2. ISSR Analysis
2.2. NGS Data Analysis and Quality Control
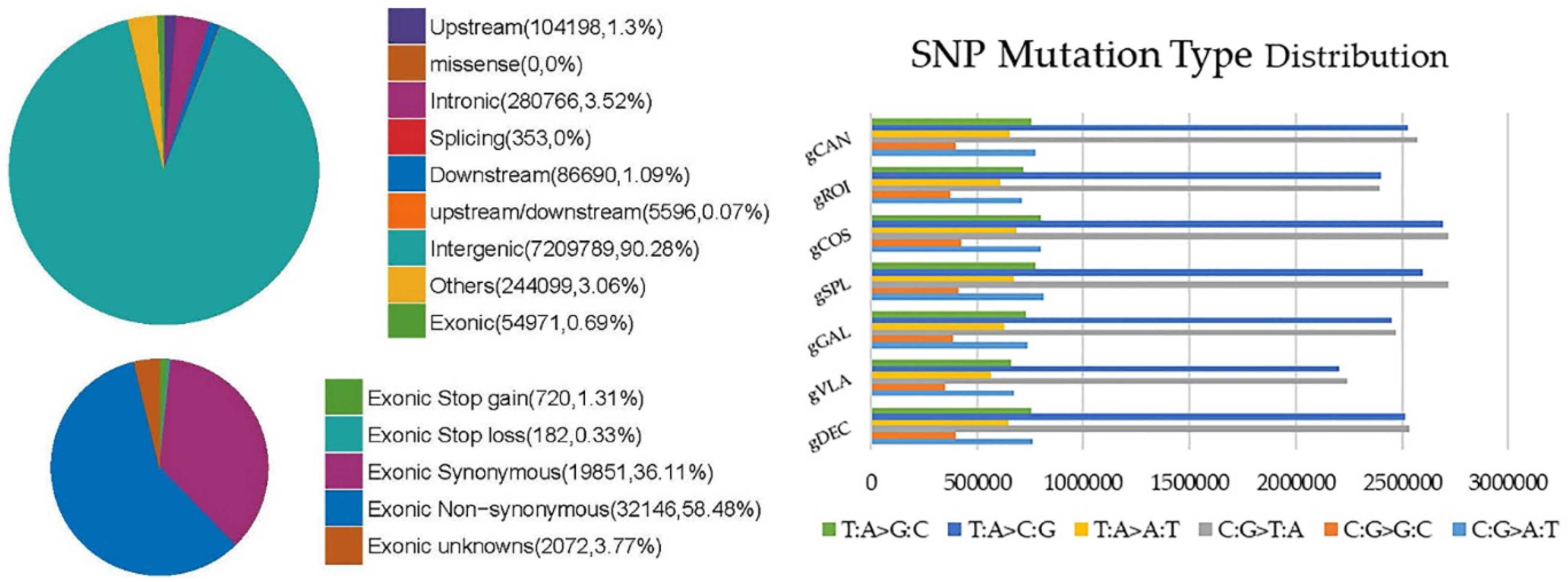
2.2.1. Single Nucleotide Polymorphism (SNP) Detection and Annotation
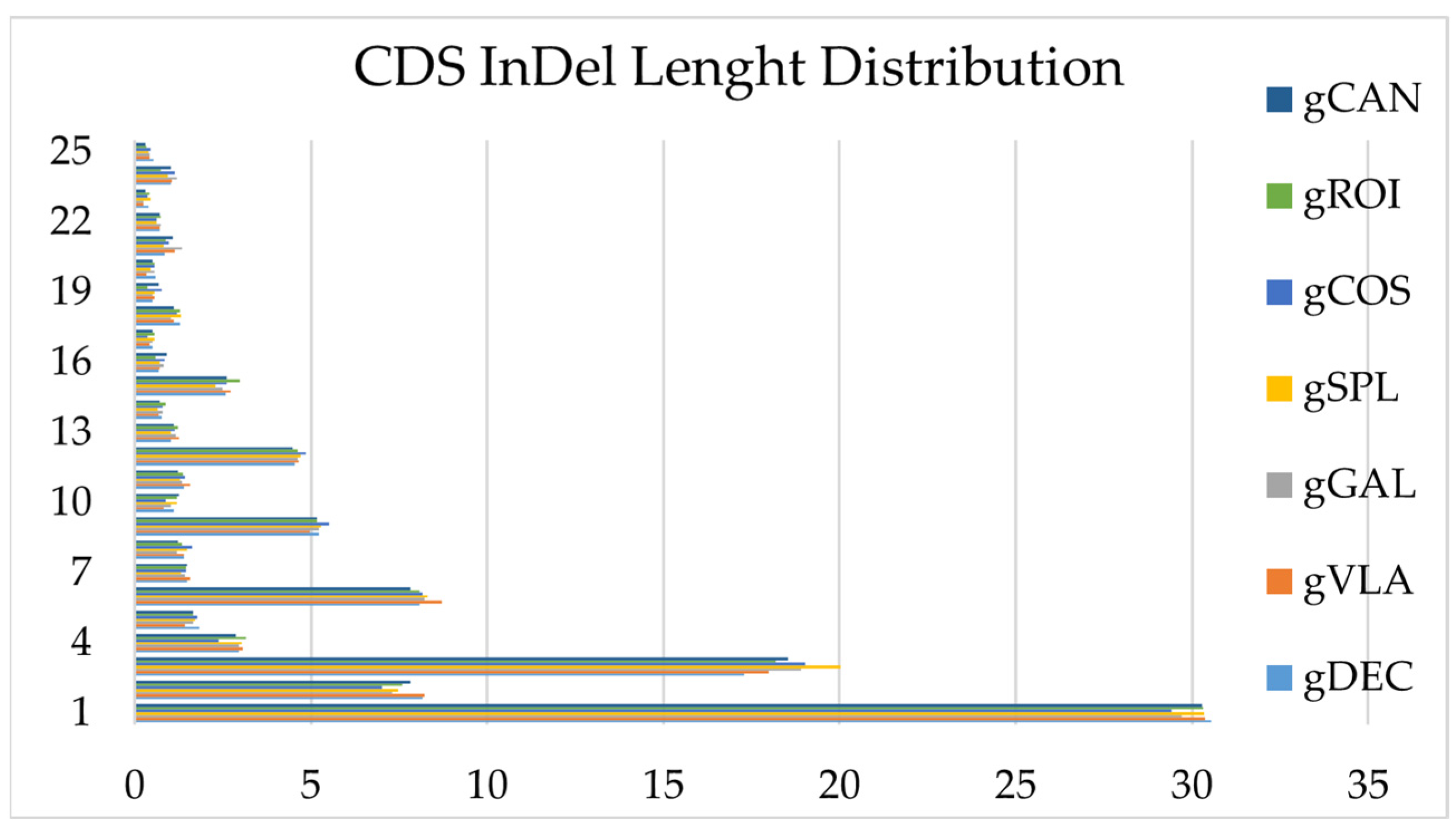
2.2.2. Insertion/Deletion (InDel) Detection and Annotation
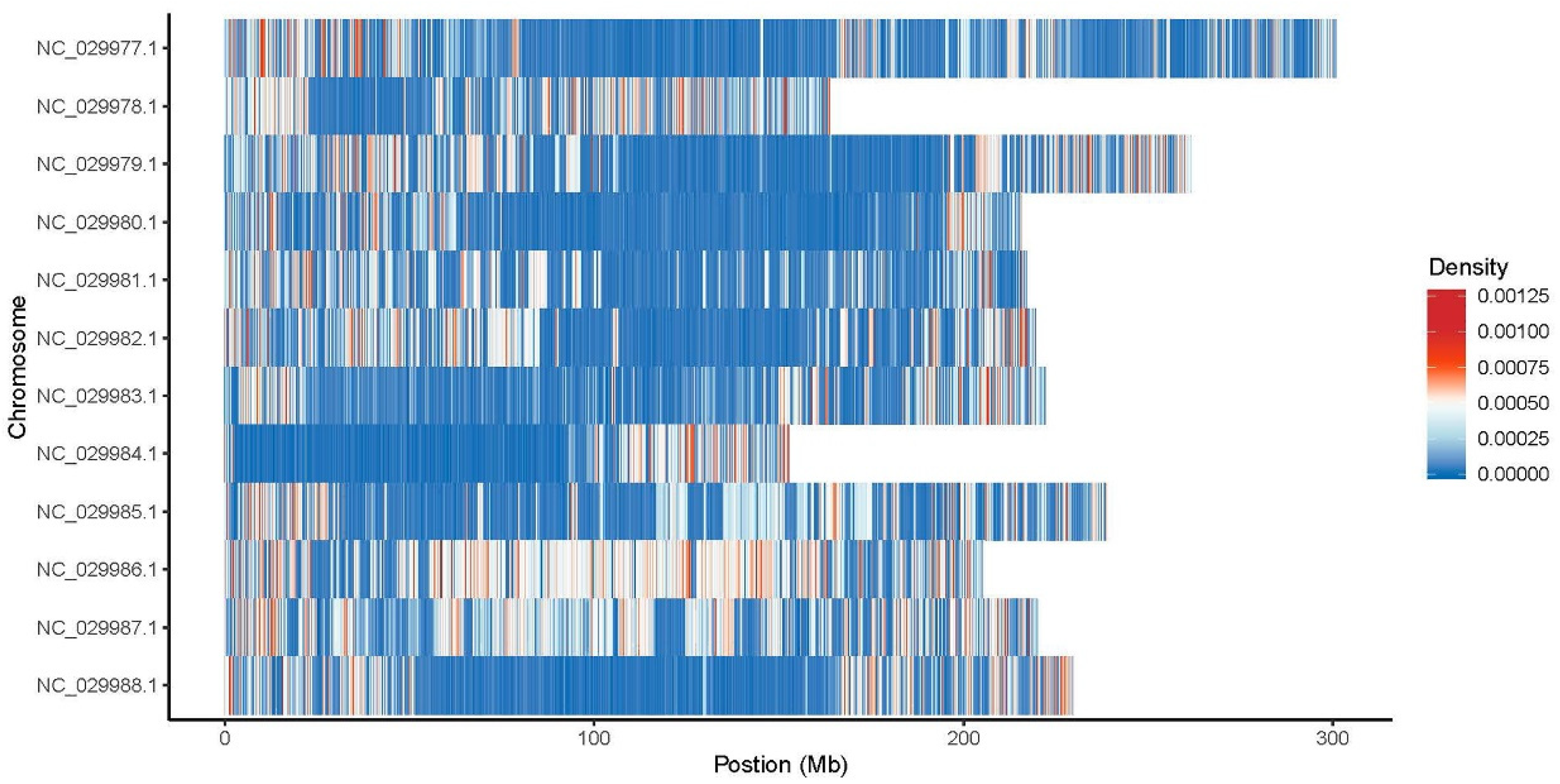
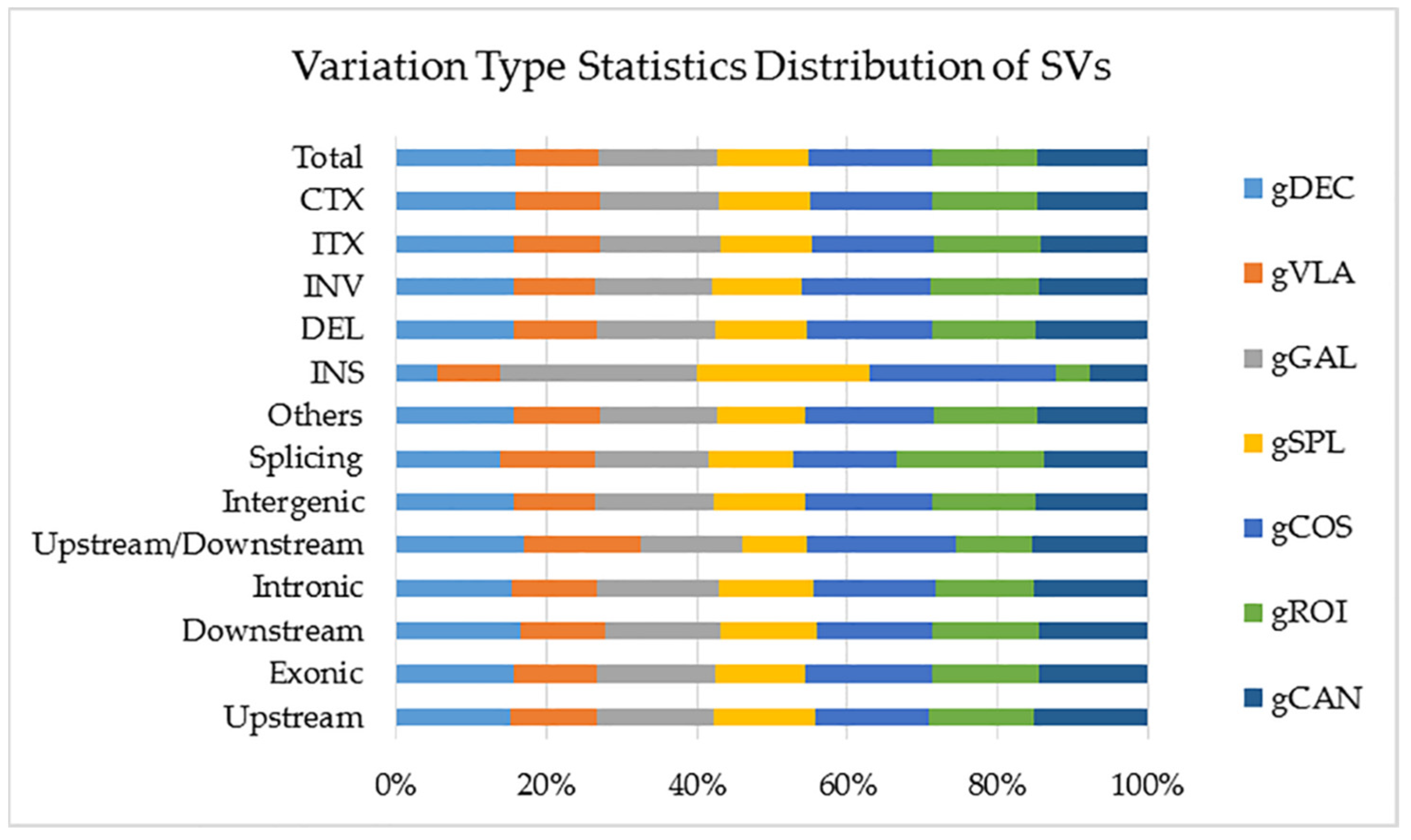
2.2.3. Structural Variant (SV) Detection and Annotation
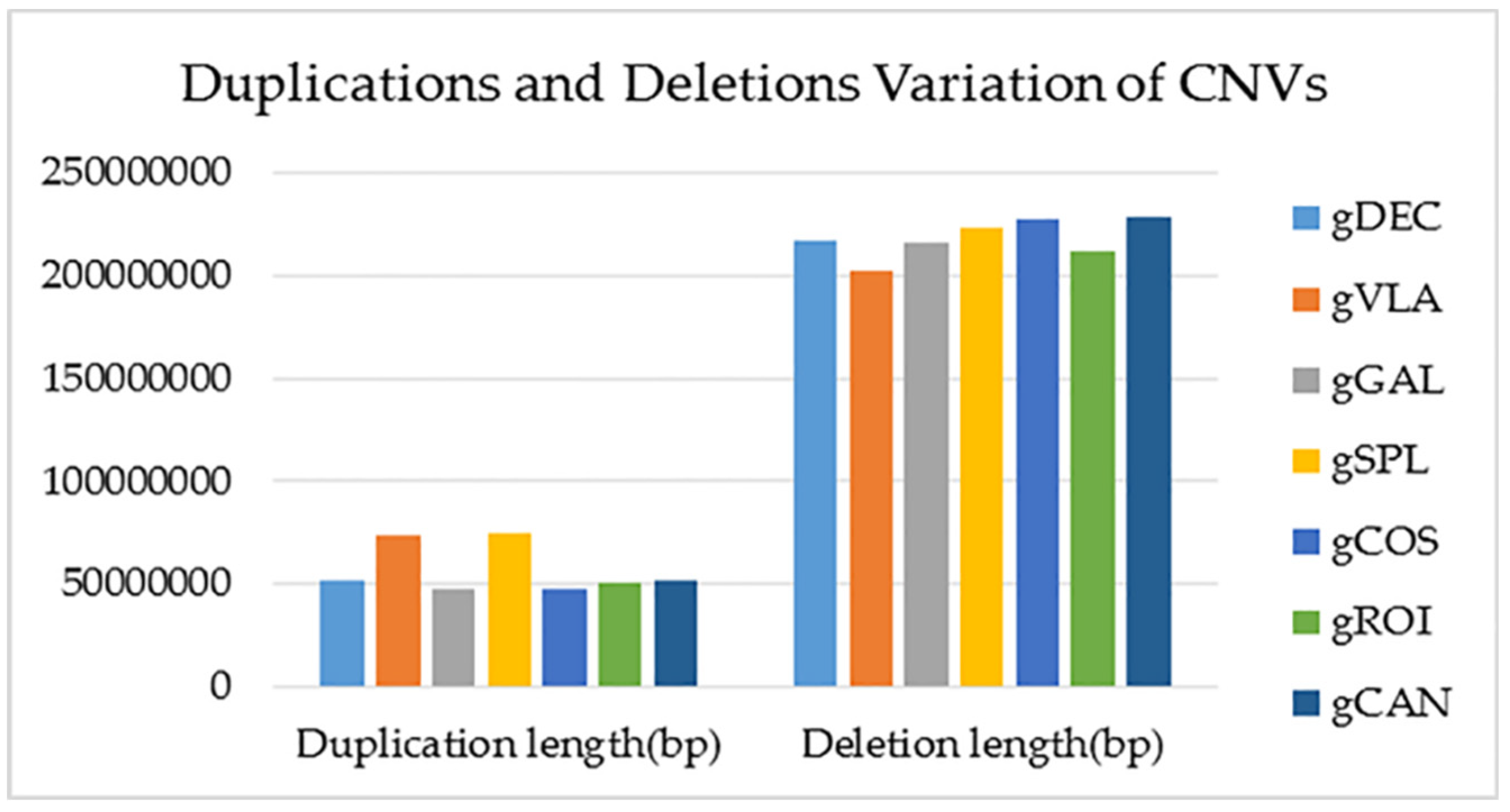
2.2.4. Copy-Number Variation (CNV) Detection and Annotation
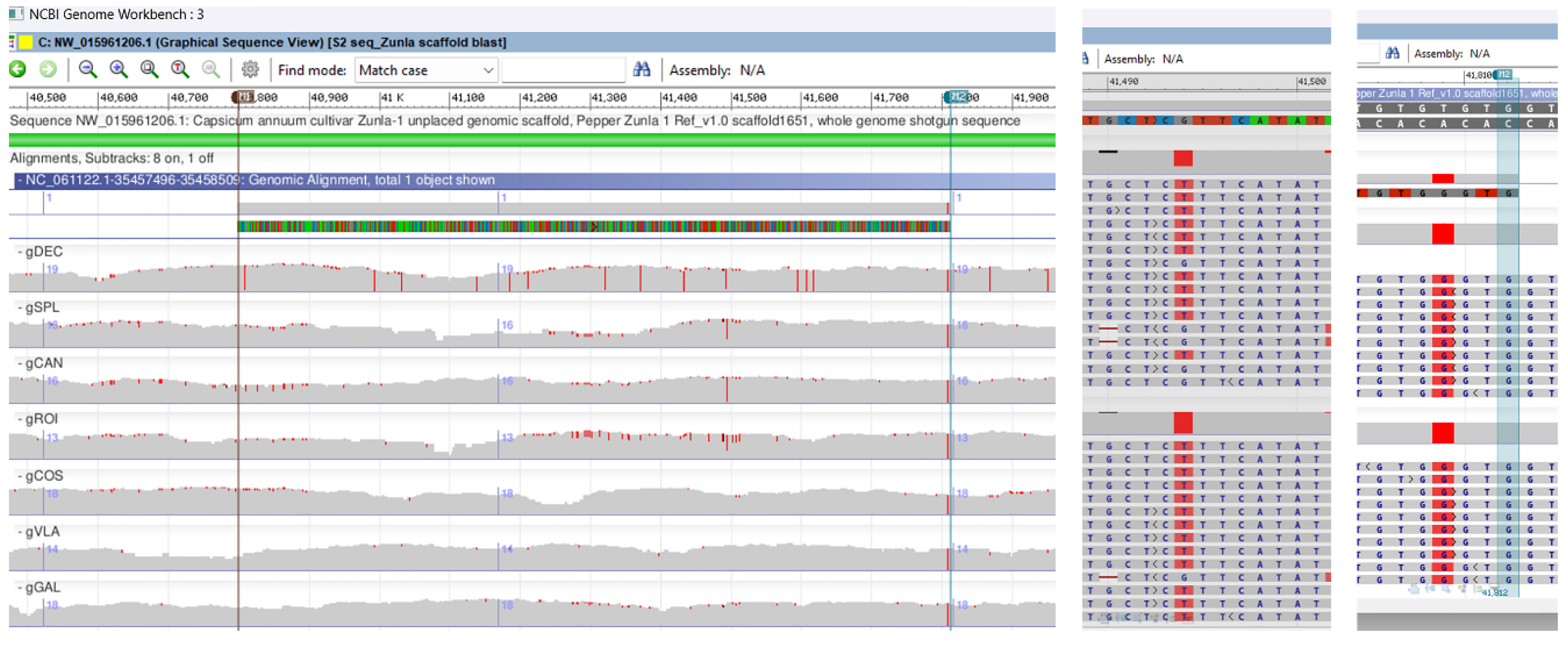
2.3. Sanger Sequencing and Multiple Genomic Alignments
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. DNA Extraction
4.3. ISSR Analysis
4.4. SSR Analysis
4.5. Molecular Markers Data Analysis
4.6. NGS, Data Processing and Sequencing Analysis
4.7. Cloning and Sanger Sequencing
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Paran, I., & van der Knaap, E. Genetic and molecular regulation of fruit and plant domestication traits in tomato and pepper. Journal of Experimental Botany, 2007; 58(14), 3841–3852. [CrossRef]
- Van Zonneveld, M.; Ramirez, M.; Williams, D. E.; Petz, M.; Meckelmann, S.; Avila, T.; Bejarano, C.; Ríos, L.; Peña, K.; Jäger, M.; Libreros, D.; Amaya, K.; Scheldeman, X. Screening genetic resources of Capsicum peppers in their primary center of diversity in Bolivia and Peru. PloS one, 2015; Volume 10(9), e0134663. [CrossRef]
- Barcanu-Tudor, E.; Drăghici, E. M. & Vînătoru, C. New Variety of Sweet Pepper (Capsicum annuum var. Grossum) Obtained at VRDS Buzău. Bulletin UASVM Horticulture, 2018; Volume 75(1). [CrossRef]
- Olatunji, T. L., & Afolayan, A. J. Evaluation of genetic relationship among varieties of Capsicum annuum L. and Capsicum frutescens L. in West Africa using ISSR markers. Heliyon, 2019; Volume 5(5), e01700. [CrossRef]
- Lam, H. M.; Xu, X.; Liu, X.; Chen, W.; Yang, G.; Wong, F. L.; Li, M.W.; He, W.; Qin, N.; Wang, B.; Li, J. & Zhang, G. Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat. Genet., 2010; Volume 42(12), pp. 1053-1059. [CrossRef]
- Solomon, A. M.; Han, K.; Lee, J. H.; Lee, H. Y.; Jang, S. & Kang, B. C. Genetic diversity and population structure of Ethiopian Capsicum germplasms. PloS one, 2019; Volume 14(5), e0216886. [CrossRef]
- Thul, S. T.; Darokar, M. P.; Shasany, A. K. & Khanuja, S. P. Molecular profiling for genetic variability in Capsicum species based on ISSR and RAPD markers. Mol. Biotech., 2012; Volume 51(2), pp. 137-147. [CrossRef]
- Portis, E.; Nagy, I.; Sasvári, Z.; Stágel, A.; Barchi, L. & Lanteri, S. The design of Capsicum spp. SSR assays via analysis of in silico DNA sequence, and their potential utility for genetic mapping. Plant Sci., 2007; Volume 172(3), pp. 640-648. [CrossRef]
- Rai, M. K.; Phulwaria, M. & Shekhawat, N. S. Transferability of simple sequence repeat (SSR) markers developed in guava (Psidium guajava L.) to four Myrtaceae species. Mol. Biol. Rep., 2013; Volume 40(8), pp. 5067-5071. [CrossRef]
- Tsaballa, A.; Ganopoulos, I.; Timplalexi, A.; Aliki, X.; Bosmali, I.; Irini, N. O.; Tsaftaris A. & Madesis, P. Molecular characteri-zation of Greek pepper (Capsicum annuum L) landraces with neutral (ISSR) and gene-based (SCoT and EST-SSR) molecular markers. Biochem. Syst. Ecol., 2015; Volume 59, pp. 256-263. [CrossRef]
- Ince, A. G.; Karaca, M. & Turgut, K. Development of new set of EST-SSR primer pairs for celery (Apium graveolens L.). Planta Med., 2010; Volume 76(12), P036. [CrossRef]
- Zhao, Y., Gui, L., Hou, C., Zhang, D., & Sun, S. GwasWA: A GWAS one-stop analysis platform from WGS data to variant effect assessment. Computers in Biology and Medicine, 2024; 169, 107820. [CrossRef]
- Choudhury, A., Ramsay, M., Hazelhurst, S., Aron, S., Bardien, S., Botha, G.,…& Pepper, M. S. Whole-genome sequencing for an enhanced understanding of genetic variation among South Africans. Nature communications, 2017; 8(1), 2062. [CrossRef]
- Ou, L., Li, D., Lv, J., Chen, W., Zhang, Z., Li, X., ... & Zou, X. Pan-genome of cultivated pepper (Capsicum) and its use in gene presence–absence variation analyses. New Phytologist, 2018; 220(2), 360-363. [CrossRef]
- Park, M., & Choi, D. The structure of pepper genome. Genetics, Genomics and Breeding of Peppers and Eggplants, 2013; 122-126. [CrossRef]
- The official catalogue of cultivated plant varieties in Romania for 2020 (ISTIS). https://istis.ro/image/data/download/catalog-oficial/CATALOG%202020.pdf.
- Lee JM, Nahm SH, Kim YM, Kim BD. Characterization and molecular genetic mapping of microsatellite loci in pepper. Theor. Appl Genet. 2004; 108 (4):619-27. [CrossRef]
- Ibarra-Torres, P.; Valadez-Moctezuma, E.; Pérez-Grajales, M.; Rodríguez-Campos, J. & Jaramillo-Flores, M. E. Inter-and intra-specific differentiation of Capsicum annuum and Capsicum pubescens using ISSR and SSR markers. Sci. Hortic., 2015; Volume 181, pp. 137-146. [CrossRef]
- Sbîrciog, G.; Buzatu, A.; Mândru, I. & Scurtu, I. Achievements in pepper breeding at Research Development Institute for Veg-etable and Flower Growing-Vidra. Curr.Trend. Nat. Sci., 2016; Volume 5(10), pp. 33-37.
- Drăghici, M. C.; Cristea G. M.; Popa E. E.; Miteluț C. A.; Popescu A. P.; Tylewicz U.; Rosa D. M.; Popa E. M. Research on blanching pretreatment and freezing technology effect on selected vegetables. AgroLife Sci. J., 2023; 12(2), 69–76. [CrossRef]
- Agapie, O. L.; Florin, S.; Costel, V.; Bianca, T.; Elena, B.; Geanina, N. & Ion, G. Description of valuable genotypes from germplasm collection of hot peppers set by directions of use. Bulletin UASVM Horticulture, 2020; Volume 77(2), pp. 117-121.
- González M. X.R.; Vicente O. Agrobiodiversity: conservation, threats, challenges, and strategies for the 21st century. AgroLife Sci. J., 2023; 12(1), 174–185. [CrossRef]
- Agapie O.L., Barcanu E. A Brief Description of Cultivated Chili Peppers. Sci. Papers. Series B, Horticulture, 2024; Vol. LXVIII, Issue 1, Print ISSN 2285-5653, 375-380.
- Iordăchescu, M.; Udriște, A. A.; Popa, V. & Bădulescu, L. Seed germination survey of Romanian tomato and pepper varieties. Res. J. Agric. Sci., 2020; Volume 52(2).
- Vintilă, M. & Niculescu, F. A. Technical aspects concerning the preservation of peppers in different storage conditions. Sci. Papers Ser. B Hortic., 2015; Volume 59, pp. 281-284.
- Hoble, A.; Dirja, M.; Luca, E.; Luca, L. & Salagean, T. Technology Elements for Irrigated Pepper (Capsicum annuum L.) growth in Field Cultivation Conditions. Bulletin UASVM Horticulture, 2010; Volume 67(2). [CrossRef]
- Scaeteanu V.G.; Săndulescu E.B.; Alistar C.F.; Croitoru C.M.; Madjar R.A.; Alistar A.; Gîlea G.C.; Stavrescu M. A short note on water quality and some biodiversity components in Gurban valley, Giurgiu County. AgroLife Sci.J., 2023; 12(2), 167–180. [CrossRef]
- Dimitrova K.; Kartalska Y.; Panayotov N. Effect of application of biostimulant Protifert LN 6.5 on the epiphytic and rhizosphere bacteria of pepper seedlings. Sci. Papers. Series B, Horticulture, 2024; Vol. LXVIII, Issue 1, Print ISSN 2285-5653, 438-443.
- Stoica V.; Hoza D. Research on the influence of organic fertilizers on the agrochemical indicators of the soil. Sci. Papers. Series B, Horticulture, 2024; Vol. LXVIII, Issue 1, Print ISSN 2285-5653, 188-193.
- Uleanu, F. Results on the effect of different types of Romanian native peat bio composites pots on seedling growth. Curr.Trend. Nat. Sci., 2013; Volume 2(3), pp. 92-95.
- Iordăchescu, M.; Udriște, A. A.; Jerca, O.; Bădulescu, L. Seedling Emergence Comparison of Several Romanian Tomato and Pepper Varieties. Bulletin of University of Agricultural Sciences and Veterinary Medicine Cluj-Napoca. Horticulture, 2021; Volume 78(1):76. [CrossRef]
- Finnegan, D. J. Retrotransposons. Current Biology, 2012; 22(11), R432-R437. [CrossRef]
- de Assis, R., Baba, V. Y., Cintra, L. A., Gonçalves, L. S. A., Rodrigues, R., & Vanzela, A. L. L. Genome relationships and LTR-retrotransposon diversity in three cultivated Capsicum L.(Solanaceae) species. BMC genomics, 2020; 21, 1-14. [CrossRef]
- Park, M., & Choi, D. The structure of pepper genome. Genetics, Genomics and Breeding of Peppers and Eggplants, 2013; 122-126. [CrossRef]
- Park, M., Park, J., Kim, S., Kwon, J.-K., Park, H. M., Bae, I. H., Yang, T.-J., Lee, Y.-H., Kang, B.-C., & Choi, D. Evolution of the large genome in Capsicum annuum occurred through accumulation of single-type long terminal repeat retrotransposons and their derivatives. The Plant Journal, 2012; 69(6), 1018–1029. [CrossRef]
- Wang, Y., Tang, X., Cheng, Z., Mueller, L., Giovannoni, J., & Tanksley, S. D. Euchromatin and Pericentromeric Heterochromatin: Comparative Composition in the Tomato Genome. Genetics, 2006; 172(4), 2529–2540. [CrossRef]
- Meyers, B. C., Tingey, S. V., & Morgante, M. Abundance, Distribution, and Transcriptional Activity of Repetitive Elements in the Maize Genome. Genome Research, 2001; 11(10), 1660–1676. [CrossRef]
- Galindo-González, L., Mhiri, C., Deyholos, M. K., & Grandbastien, M.-A. LTR-retrotransposons in plants: Engines of evolution. Gene, 2017; 626, 14–25. [CrossRef]
- Islam, M. M., El-Sappah, A. H., Ali, H. M., Zandi, P., Huang, Q., Soaud, S. A., ... & Liang, Y. Pathogenesis-related proteins (PRs) countering environmental stress in plants: A review. South African Journal of Botany, 2023; 160, 414-427. [CrossRef]
- Ali, S., Ganai, B. A., Kamili, A. N., Bhat, A. A., Mir, Z. A., Bhat, J. A., ... & Grover, A. Pathogenesis-related proteins and peptides as promising tools for engineering plants with multiple stress tolerance. Microbiological research, 2018; 212, 29-37. [CrossRef]
- Anisimova, O. K., Shchennikova, A. V., Kochieva, E. Z., & Filyushin, M. A. Pathogenesis-related genes of PR1, PR2, PR4, and PR5 families are involved in the response to Fusarium infection in garlic (Allium sativum L.). International journal of molecular sciences, 2021; 22(13), 6688. [CrossRef]
- Dos Santos, C., & Franco, O. L. Pathogenesis-related proteins (PRs) with enzyme activity activating plant defense responses. Plants, 2023; 12(11), 2226. [CrossRef]
- Yang, J., Zhang, J., Du, H., Zhao, H., Li, H., Xu, Y., ... & Wen, C. The vegetable SNP database: an integrated resource for plant breeders and scientists. Genomics, 2022; 114(3), 110348. [CrossRef]
- Wang, S., Yang, X., Xu, M., Lin, X., Lin, T., Qi, J., ... & Huang, S. A rare SNP identified a TCP transcription factor essential for tendril development in cucumber. Molecular Plant, 2015; 8(12), 1795-1808. [CrossRef]
- Zhou, H., Liu, Q., Li, J., Jiang, D., Zhou, L., Wu, P., ... & Zhuang, C. Photoperiod-and thermo-sensitive genic male sterility in rice are caused by a point mutation in a novel noncoding RNA that produces a small RNA. Cell research, 2012; 22(4), 649-660. [CrossRef]
- Jia, J. I. A., Huan, W. A. N. G., Yang, X. M., Bo, C. H. E. N., Wei, R. Q., Cheng, Y. B., & Hai, N. I. A. N. Identification of the long InDels through whole genome resequencing to fine map of qIF05-1 controlling seed isoflavone content in soybean (Glycine max L. Merr.). Journal of Integrative Agriculture. 2023. [CrossRef]
- Fliege C E, Ward R A, Vogel P, Nguyen H, Quach T, Guo M, Viana J, Santos L B, Specht J E, Clemente T E, Hudson M E, Diers B W. Fine mapping and cloning of the major seed protein quantitative trait loci on soybean chromosome 20. Plant Journal, 2022; 110, 114-128. [CrossRef]
- Moghaddam, S. M., Song, Q., Mamidi, S., Schmutz, J., Lee, R., Cregan, P., Osorno, J. M., & McClean, P. E. Developing market class specific InDel markers from next generation sequence data in Phaseolus vulgaris L. Frontiers in Plant Science, 2014; 5, 185. [CrossRef]
- Guo, G., Zhang, G., Pan, B., Diao, W., Liu, J., Ge, W., Gao, C., Zhang, Y., Jiang, C., & Wang, S. Development and Application of InDel Markers for Capsicum spp. Based on Whole-Genome Re-Sequencing. Scientific Reports, 2019; 9(1), 3691. [CrossRef]
- Britten, R. J., Rowen, L., Williams, J., & Cameron, R. A. Majority of divergence between closely related DNA samples is due to indels. Proceedings of the National Academy of Sciences, 2003; 100(8), 4661–4665. [CrossRef]
- Delmore, K. E., Van Doren, B. M., Ullrich, K., Curk, T., van der Jeugd, H. P., & Liedvogel, M. Structural genomic variation and migratory behavior in a wild songbird. Evolution Letters, 2023; 7(6), 401-412. [CrossRef]
- Blue, Y. A., & Satake, A. Analyses of gene copy number variation in diverse epigenetic regulatory gene families across plants: Increased copy numbers of BRUSHY1/TONSOKU/MGOUN3 (BRU1/TSK/MGO3) and SILENCING DEFECTIVE 3 (SDE3) in long-lived trees. Plant Gene, 2022; 32, 100384. [CrossRef]
- Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., ... & Marra, M. A. Circos: an information aesthetic for comparative genomics. Genome research, 2009; 19(9), 1639-1645. [CrossRef]
- Pacheco, Á.; Alvarado, G.; Rodríguez, F.; Crossa, J.; Burgueño, J. BIO-R (Biodiversity analysis with R for Windows) Version 3.0, 2020; International Maize and Wheat Improvement Center.
- Laval, G.; San Cristobal, M. & Chevalet, C. Measuring genetic distances between breeds: use of some distances in various short term evolution models. Genet. Sel. Evol., 2002; Volume 34(4), pp. 1-27. [CrossRef]
- Nei, M. Molecular evolutionary genetics. Columbia University Press, 1987.
- Serrote, C. M. L.; Reiniger, L. R. S.; Silva, K. B.; dos Santos Rabaiolli, S. M. & Stefanel, C. M. Determining the Polymorphism Information Content of a molecular marker. Gene, 2020; Volume 726, 144175. [CrossRef]
- Botstein, D.; White, R. L.; Skolnick, M. & Davis, R. W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet., 1980; Volume 2(3), pp. 314.
- Roldàn-Ruiz, I.; Dendauw, J.; Van Bockstaele, E.; Depicker, A. & De Loose, M. AFLP markers reveal high polymorphic rates in ryegrasses (Lolium spp.). Molec. Breed., 2000; Volume 6(2), pp. 125-134. http://hdl.handle.net/1854/LU-133034.
- De Riek, J.; Calsyn, E.; Everaert, I.; Van Bockstaele, E. & De Loose, M. AFLP based alternatives for the assessment of distinct-ness, uniformity and stability of sugar beet varieties. Theor. Appl. Genet., 2001; Volume 103(8), pp. 1254-1265. [CrossRef]
- Cock, P.J.A., Fields, et al. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Research. 2010; 38(6):1767-1771. [CrossRef]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25(14):1754-1760. [CrossRef]
- Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25(16):2078-2079. [CrossRef]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Research. 2010; 38(16):e164. [CrossRef]
- Chen, K, et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nature Methods. 2009; 6:677-681. [CrossRef]
- Abyzov A, Urban A E, Snyder M, et al. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome research. 2011; 21(6):974-984. [CrossRef]

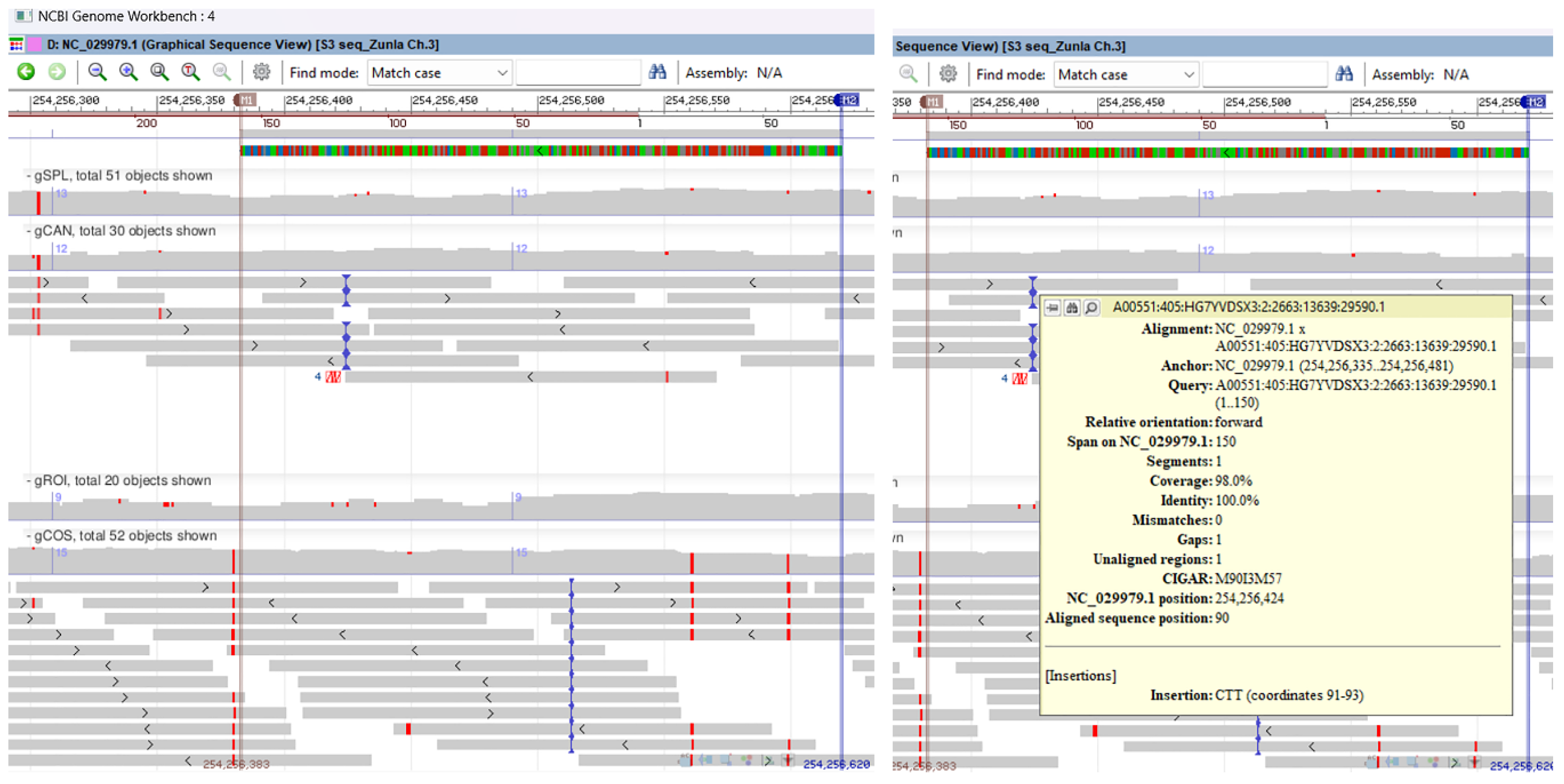
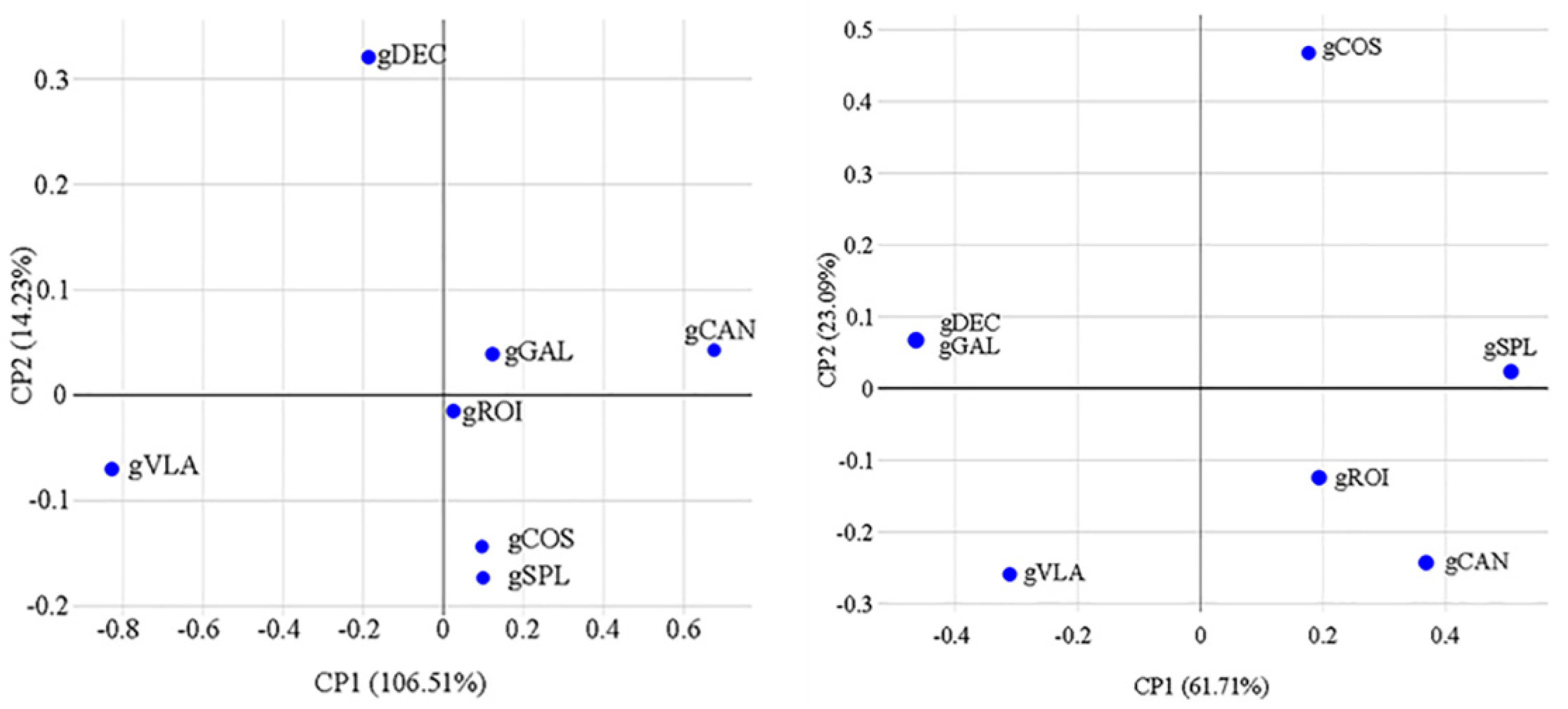
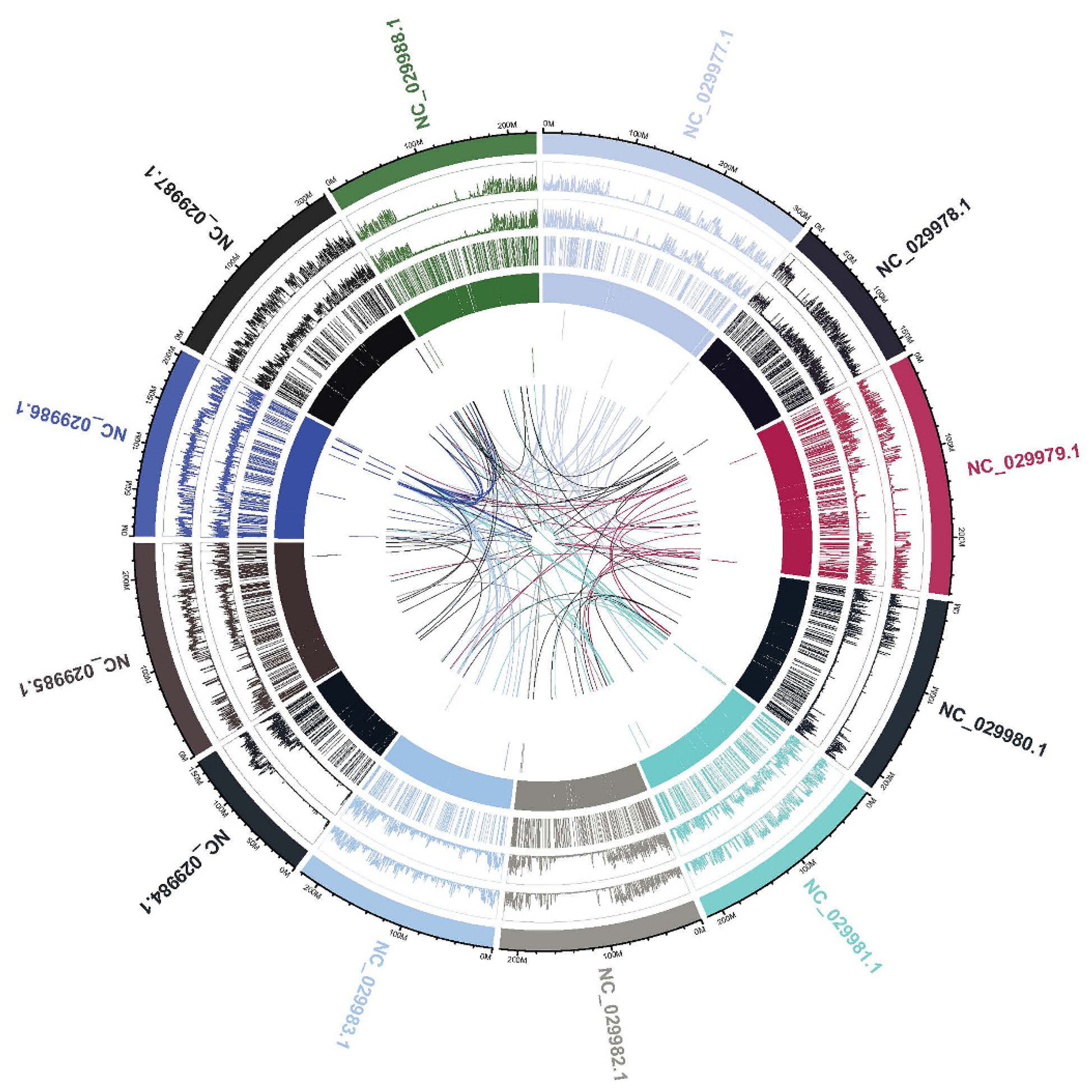
| ID | Locus | Primer (forward/ reverse) | Size (bp) | Tm (ºC) | Total alleles | PIC value |
|---|---|---|---|---|---|---|
| SSRP3P4 | AF244121 | 5'TACCTCCTCGCCAATCCTTCTG 3'/ 5'TTGAAAGTTCTTTCCATGACAACC 3' |
200-400 bp | 45 | 3 | 0.63 |
| SSRP5P6 | HpmS 1-148 | 5'GGCGGAGAAGAACTAGACGATTAGC3'/ 5'TCACCCAATCCACATAGACG 3' |
150-250 bp | 45 | 4 | 0.72 |
| SSRP9P10 | HpmS 1_1 | 5'TCAACCCAATATTAAGGTCACTTCC3'/ 5'CCAGGCGGGGATTGTAGATG3' |
260 pb | 49 | NA | NA |
| SSRP11P12 | HpmS 1_274 | 5'TCCCAGACCCCTCGTGATAG3'/ 5'TCCTGCTCCTTCCACAACTG 3' |
190-530 bp | 47 | 4 | 0.71 |
| SSRP19P20 | HpmS 1_172 | 5'GGGTTTGCATGATCTAAGCATTTT3'/ 5'CGCTGGAATGCATTGTCAAAGA3' |
230-420 bp | 48 | 3 | 0.66 |
| ID | Primer | Tm (ºC) | Total bands (TB) | Range of the amplification product (bp) | PIC value |
|---|---|---|---|---|---|
| P21 | 5’ACGACAGACAGACAGACA3’ | 51 | 38 | 850-4000 bp | 0.08 |
| P22 | 5’ACACACACACACACACCTG3’ | 50 | 28 | 500-2800 bp | NA |
| P23 | 5'GCAGACAGACAGACAGACGC3' | 50 | 68 | 500-4000 bp | 0.28 |
| P24 | 5'GAGAGAGAGAGAGAGACTC 3' | 50 | 56 | 800-3800 bp | 0.23 |
| P25 | 5'GAGAGAGAGAGAGAGACTC3' | 50 | 80 | 550-3100 bp | 0.29 |
| P26 | 5'CACACACACACACACAAGT 3' | 51 | 27 | 1000-2500 bp | 0.26 |
| P27 | 5'GACAGACAGACAGACAGT3' | 51 | 70 | 380-4000 bp | 0.20 |
| P28 | 5'TCCTCCTCCTCCTCCAGCT3' | 50 | 34 | 350-2700 bp | 0.29 |
| Genotype | gDEC | gVLA | gGAL | gSPL | gCOS | gROI | gCAN |
|---|---|---|---|---|---|---|---|
| Upstream | 98681 | 93968 | 97182 | 104198 | 100293 | 95159 | 98084 |
|
Exonic Stop gain |
645 | 644 | 635 | 720 | 664 | 657 | 634 |
|
Exonic Stop loss |
164 | 162 | 166 | 182 | 175 | 163 | 160 |
|
Exonic Synonymous |
17898 | 17757 | 17668 | 19851 | 18012 | 17321 | 18229 |
|
Exonic Non-synonymous |
29071 | 28736 | 29115 | 32146 | 29349 | 28540 | 29504 |
| Intronic | 255395 | 240657 | 249296 | 280766 | 262290 | 246344 | 260011 |
| Splicing | 307 | 286 | 308 | 353 | 303 | 295 | 319 |
| Downstream | 80621 | 77439 | 79503 | 86690 | 82171 | 77640 | 80567 |
|
Upstream/ Downstream |
5010 | 4943 | 5126 | 5596 | 5365 | 4728 | 5058 |
| Intergenic | 6899284 | 6009182 | 6706315 | 7209789 | 7383834 | 6528591 | 6955410 |
| Others | 229351 | 213191 | 222529 | 244099 | 236949 | 214605 | 233623 |
| ts | 5051787 | 4442276 | 4923303 | 5313137 | 5408262 | 4796597 | 5099410 |
| tv | 2565834 | 2245868 | 2485560 | 2672644 | 2712300 | 2418530 | 2583426 |
| ts/tv | 1.969 | 1.978 | 1.981 | 1.988 | 1.994 | 1.983 | 1.974 |
| Het. rate | 0.162 | 0.636 | 0.126 | 0.764 | 0.13 | 0.116 | 0.121 |
| Total | 7617621 | 6688144 | 7408863 | 7985781 | 8120562 | 7215127 | 7682836 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





