Submitted:
19 March 2025
Posted:
20 March 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Sequence Alignments
2.2. Evolutionary Conservation of Amino Acids
2.3. Protein Structure Loop Modeling
2.4. HGH-Receptor Interaction Analysis
2.5. Disease-Causing Mutations
2.6. Prediction of Protein Stability Upon Mutation Using Site-Directed Mutagenesis Tool (SDM)
2.7. Analysis of HGH Allele Frequencies from Genome Sequencing
3. Results
3.1. Analysis of GH Sequence Across Species
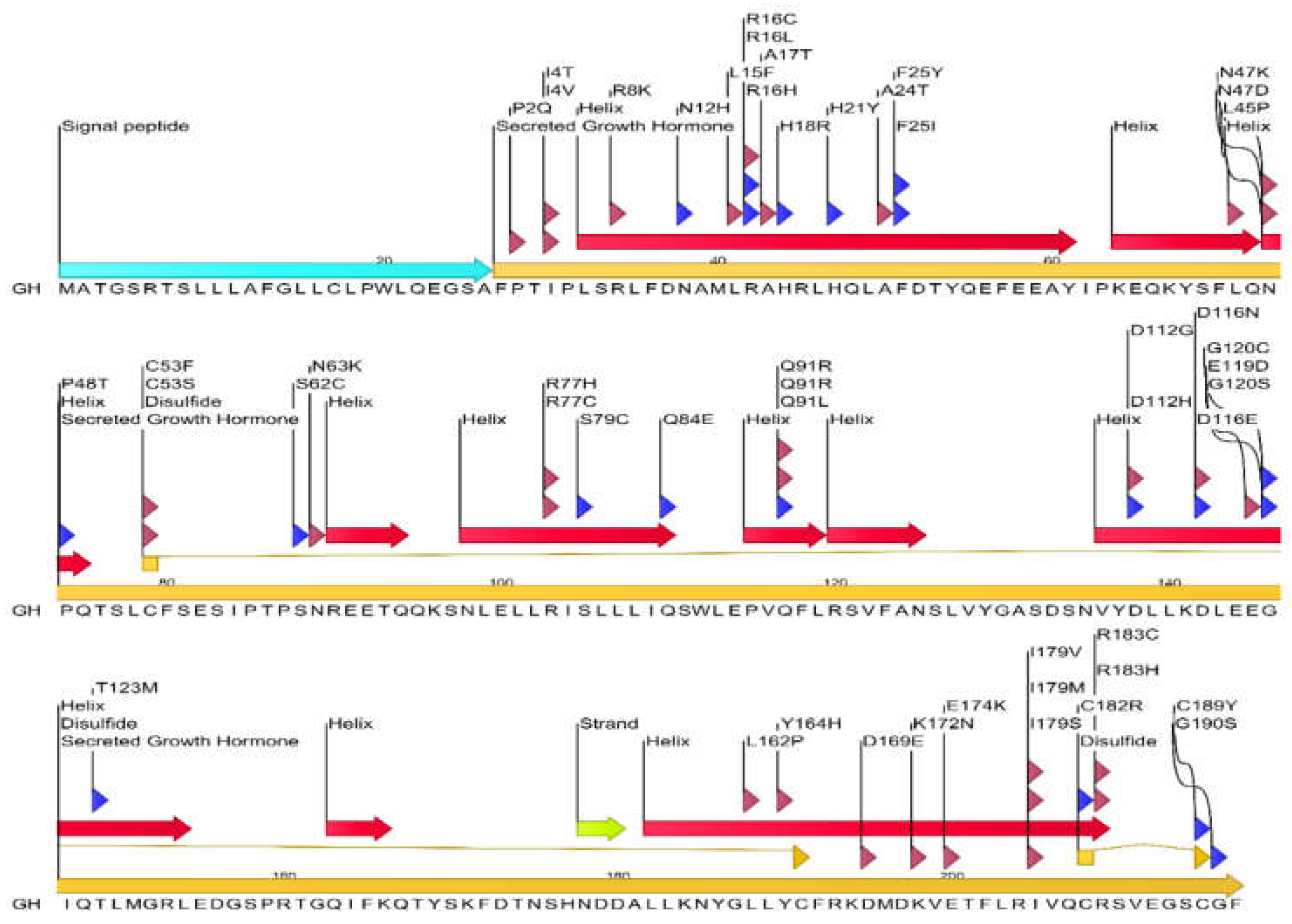
3.1.1. Analysis of Human GH Mutations and Their Locations in GH Protein
3.1.2. Comparison of Human GH Protein Sequence with Diverse GH Homologues Across Species
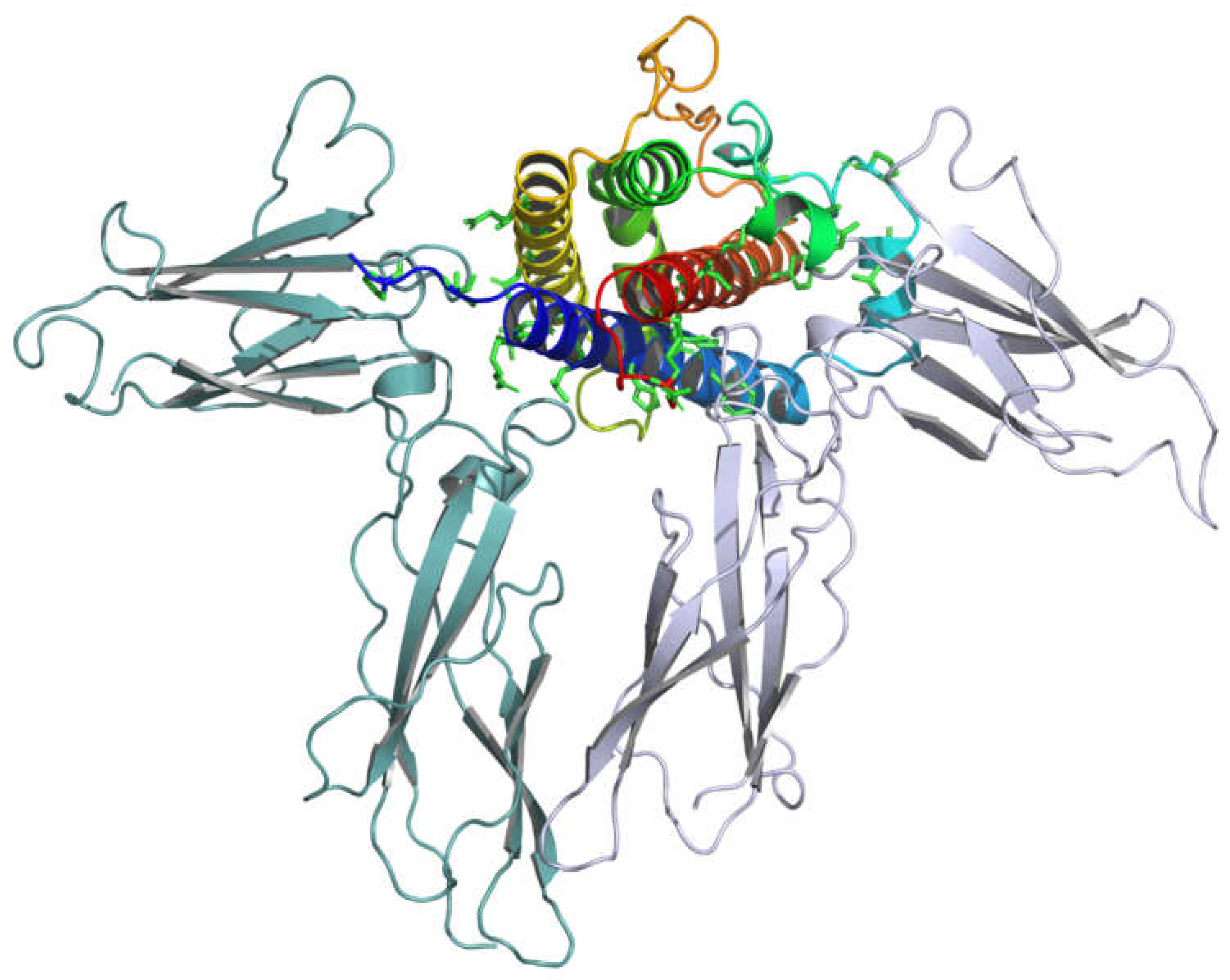
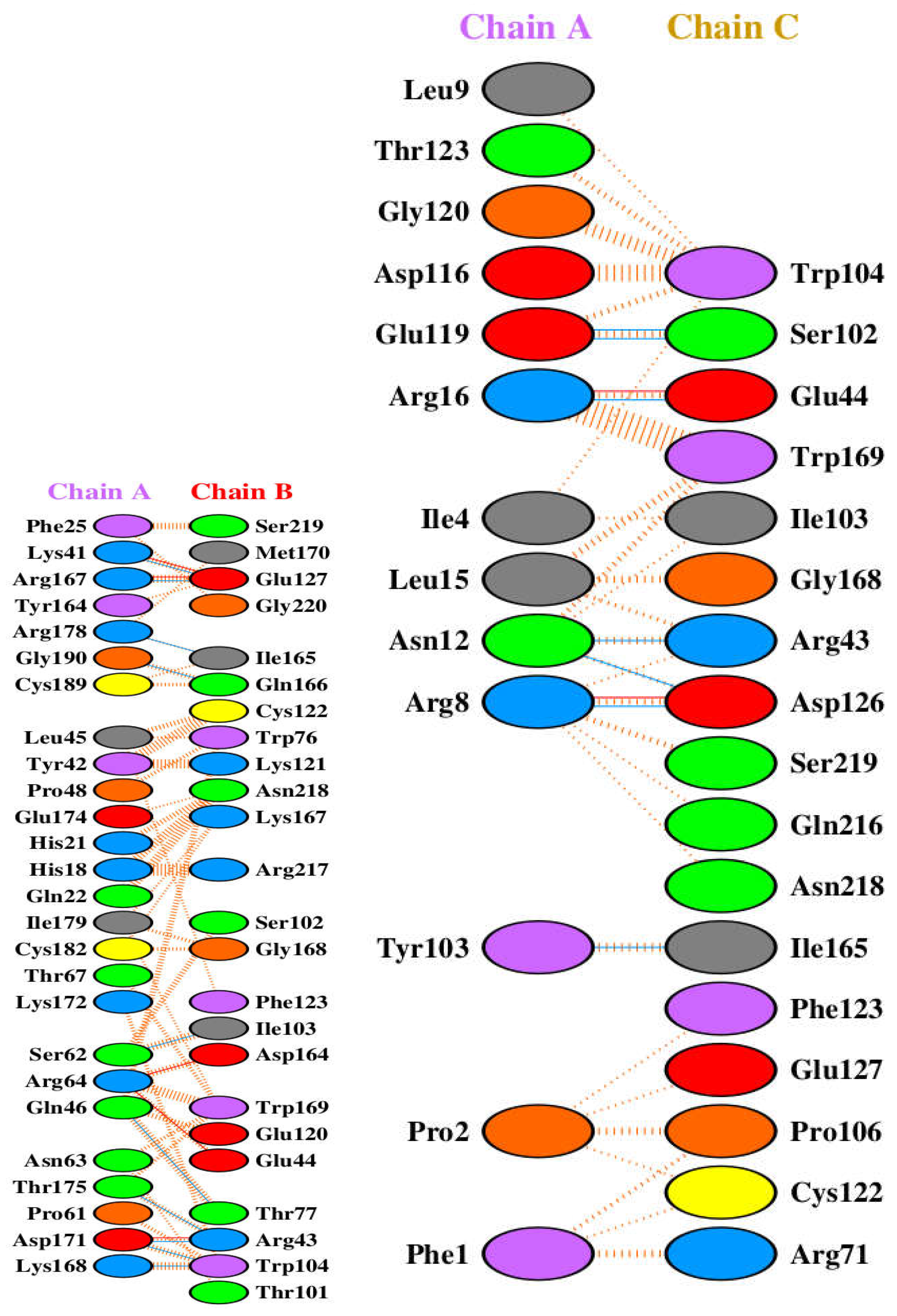
3.2. Analysis of GH-GHR Contacts
Structural Analysis of GH-GHR Contacts
- His18: The mutation H18R is at an interface residue interacting with Arg217 of the GHR.
- His21: The mutation H21Y is at an interface residue interacting with Glu44 of the GHR.
- Phe25: Mutations F25Y and F25I are located at a key interface residue forming both hydrogen bonds and hydrophobic contacts with Ser219 of the GHR.
- Lys41: Lys41 forms interactions with Met170 of the GHR.
- Leu45: The mutation L45P is at an interface residue interacting with Trp76 of the GHR.
- Pro48: The mutation P48T is located at a residue involved in interactions with Asn218 of the GHR.
- Ile179: Mutations I179V, I179M, and I179S are located at a residue interacting with Ser102 of the GHR.
- Cys182: The mutation C182R is at an interface residue forming interactions with Gly168 of the GHR.
- Lys172: The mutation K172N is at a residue interacting with Phe123 of the GHR.
- Ser62: The mutation S62C is at an interface residue interacting with Asp164 of the GHR.
- Asn63: The mutation N63K is at an interface residue interacting with Glu44 of the GHR.
- Pro61: Pro61 interacts with Thr77 of the GHR.
- Lys168: Lys168 interacts with Trp104 of the GHR.
- Tyr164: The mutation Y164H is at a key interface residue interacting with Gly220 of the GHR.
- Glu174: The mutation E174K is at a residue interacting with Lys167 of the GHR.
- Gly190: The mutation G190S is at an interface residue interacting with Ile165 of the GHR.
- Cys189: The mutation C189Y is at an interface residue interacting with Gln166 of the GHR.
- Arg16: Mutations R16C, R16L, and R16H are located in close proximity to the interface and might indirectly affect binding.
- Phe1: Phe1 interacts with Arg71 of GHR chain C.
- Pro2: The mutation P2Q is located at an interface residue interacting with Pro106 of GHR chain C.
- Ile4: Mutations I4T and I4V are at interface residues interacting with Ile103 of GHR chain C.
- Arg8: The mutation R8K is located at a residue interacting with Asp126 of GHR chain C.
- Asn12: The mutation N12H is at an interface residue interacting with Arg43 of GHR chain C.
- Leu15: The mutation L15F is at an interface residue interacting with Gly168 of GHR chain C.
- Arg16: Mutations R16C, R16L, and R16H are located at a key interface residue interacting with Glu44 of GHR chain C.
- Leu9: Leu9 interacts with Asp126 of GHR chain C.
- Gly120: Mutations G120C and G120S are at interface residues interacting with Ser102 of GHR chain C.
- Asp116: Mutations D116N and D116E are at interface residues interacting with Trp104 of GHR chain C.
- Glu119: The mutation E119D is at an interface residue interacting with Ser102 of GHR chain C.
- Thr123: The mutation T123M is at an interface residue interacting with Trp104 of GHR chain C.
- Tyr103: Tyr103 interacts with Ile165 of GHR chain C.
- Arg16 forms a hydrogen bond with Glu44 on chain C and is in close proximity to the interface with chain B
- Gly120 forms a hydrogen bond with Ser102 on chain C and is likely involved in interactions within the first binding site.
- Asp116 forms a hydrogen bond with Trp104 on chain C and is likely involved in interactions within the first binding site.
- Glu119 forms a hydrogen bond with Ser102 on chain C and is likely involved in interactions within the first binding site.
- Thr123 forms a hydrogen bond and hydrophobic contact with Trp104 on chain C and is likely involved in interactions within the first binding site.
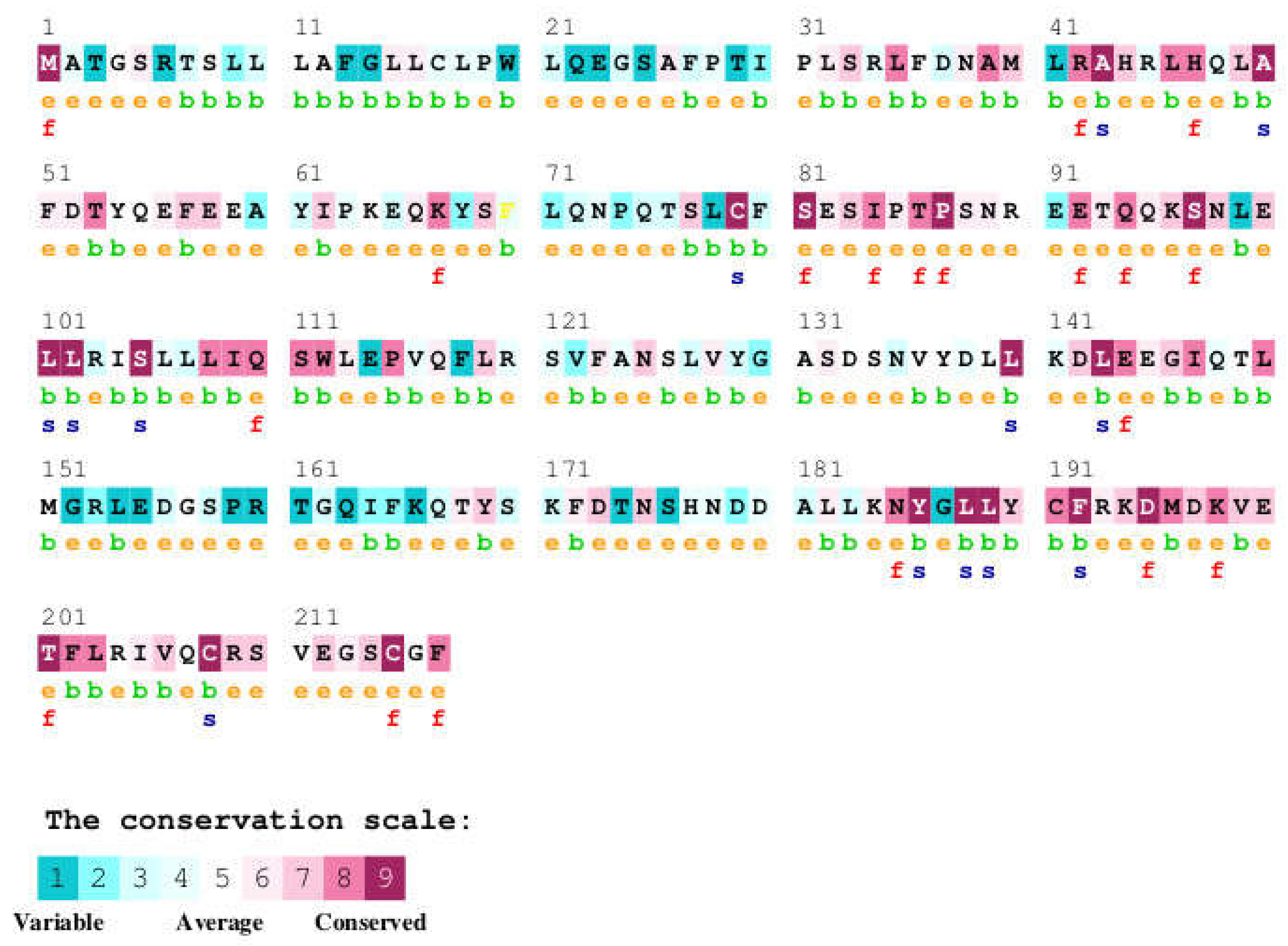
3.3. Analysis of Sequence Conservation Pattern in Growth Hormone at GH-GHR Contact Points
3.4. Stability Analysis of GH Mutations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| GH | Growth hormone |
| GHR | Growth hormone receptor |
| GHD | Growth hormone deficiency |
| GHBP | Growth hormone binding protein |
References
- Rojas Velazquez, M. N.; Noebauer, M.; Pandey, A. V. , Loss of Protein Stability and Function Caused by P228L Variation in NADPH-Cytochrome P450 Reductase Linked to Lower Testosterone Levels. International journal of molecular sciences 2022, 23, 10141. [Google Scholar] [CrossRef] [PubMed]
- Prado, M. J.; Singh, S.; Ligabue-Braun, R.; Meneghetti, B. V.; Rispoli, T.; Kopacek, C.; Monteiro, K.; Zaha, A.; Rossetti, M. L. R.; Pandey, A. V. , Characterization of Mutations Causing CYP21A2 Deficiency in Brazilian and Portuguese Populations. Int J Mol Sci 2021, 23. [Google Scholar]
- Parween, S.; Rojas Velazquez, M. N.; Udhane, S. S.; Kagawa, N.; Pandey, A. V. , Variability in Loss of Multiple Enzyme Activities Due to the Human Genetic Variation P284T Located in the Flexible Hinge Region of NADPH Cytochrome P450 Oxidoreductase. Frontiers in Pharmacology 2019, 10. [Google Scholar]
- Parween, S.; DiNardo, G.; Baj, F.; Zhang, C.; Gilardi, G.; Pandey, A. V. , Differential effects of variations in human P450 oxidoreductase on the aromatase activity of CYP19A1 polymorphisms R264C and R264H. J Steroid Biochem Mol Biol 2020, 196, 105507. [Google Scholar]
- Parween, S.; Fernandez-Cancio, M.; Benito-Sanz, S.; Camats, N.; Rojas Velazquez, M. N.; Lopez-Siguero, J. P.; Udhane, S. S.; Kagawa, N.; Fluck, C. E.; Audi, L.; Pandey, A. V. , Molecular Basis of CYP19A1 Deficiency in a 46,XX Patient With R550W Mutation in POR: Expanding the PORD Phenotype. J Clin Endocrinol Metab 2020, 105. [Google Scholar] [CrossRef]
- Alatzoglou, K. S.; Webb, E. A.; Le Tissier, P.; Dattani, M. T. , Isolated growth hormone deficiency (GHD) in childhood and adolescence: recent advances. Endocr Rev 2014, 35, 376–432. [Google Scholar]
- Pandey, A. V. , Bioinformatics tools and databases for the study of human growth hormone. Endocr Dev 2012, 23, 71–85. [Google Scholar]
- Procter, A. M.; Phillips, J. A., 3rd; Cooper, D. N. , The molecular genetics of growth hormone deficiency. Human genetics 1998, 103, 255–72. [Google Scholar]
- Hirt, H.; Kimelman, J.; Birnbaum, M. J.; Chen, E. Y.; Seeburg, P. H.; Eberhardt, N. L.; Barta, A. , The human growth hormone gene locus: structure, evolution, and allelic variations. DNA 1987, 6, 59–70. [Google Scholar] [CrossRef]
- Mullis, P. E. , Genetic control of growth. Eur J Endocrinol 2005, 152, 11–31. [Google Scholar]
- Phillips, J. A., 3rd; Cogan, J. D. , Genetic basis of endocrine disease. 6. Molecular basis of familial human growth hormone deficiency. J Clin Endocrinol Metab 1994, 78, 11–6. [Google Scholar] [PubMed]
- de Vos, A. M.; Ultsch, M.; Kossiakoff, A. A. , Human growth hormone and extracellular domain of its receptor: crystal structure of the complex. Science 1992, 255, 306–12. [Google Scholar] [CrossRef] [PubMed]
- Bluet-Pajot, M. T.; Epelbaum, J.; Gourdji, D.; Hammond, C.; Kordon, C. , Hypothalamic and hypophyseal regulation of growth hormone secretion. Cellular and molecular neurobiology 1998, 18, 101–23. [Google Scholar] [PubMed]
- Kojima, M.; Hosoda, H.; Date, Y.; Nakazato, M.; Matsuo, H.; Kangawa, K. , Ghrelin is a growth-hormone-releasing acylated peptide from stomach. Nature 1999, 402, 656–60. [Google Scholar]
- Mullis, P. E.; Deladoey, J.; Dannies, P. S. , Molecular and cellular basis of isolated dominant-negative growth hormone deficiency, IGHD type II: insights on the secretory pathway of peptide hormones. Horm Res 2002, 58, 53–66. [Google Scholar]
- Petkovic, V.; Miletta, M. C.; Eble, A.; Iliev, D. I.; Binder, G.; Fluck, C. E.; Mullis, P. E. , Effect of zinc binding residues in growth hormone (GH) and altered intracellular zinc content on regulated GH secretion. Endocrinology.
- Norrelund, H. , The metabolic role of growth hormone in humans with particular reference to fasting. Growth Horm IGF Res 2005, 15, 95–122. [Google Scholar]
- Meinhardt, U. J.; Ho, K. K. , Modulation of growth hormone action by sex steroids. Clin Endocrinol (Oxf) 2006, 65, 413–22. [Google Scholar] [CrossRef]
- Van Cauter, E.; Latta, F.; Nedeltcheva, A.; Spiegel, K.; Leproult, R.; Vandenbril, C.; Weiss, R.; Mockel, J.; Legros, J. J.; Copinschi, G. , Reciprocal interactions between the GH axis and sleep. Growth Horm IGF Res 2004, 14, S10–7. [Google Scholar] [CrossRef]
- Widdowson, W. M.; Healy, M. L.; Sonksen, P. H.; Gibney, J. , The physiology of growth hormone and sport. Growth Horm IGF Res 2009, 19, 308–19. [Google Scholar]
- Walenkamp, M. J.; Wit, J. M. , Genetic disorders in the GH IGF-I axis in mouse and man. Eur J Endocrinol 2007, 157, S15–26. [Google Scholar]
- Clark, R. G.; Mortensen, D. L.; Carlsson, L. M.; Spencer, S. A.; McKay, P.; Mulkerrin, M.; Moore, J.; Cunningham, B. C. , Recombinant human growth hormone (GH)-binding protein enhances the growth-promoting activity of human GH in the rat. Endocrinology 1996, 137, 4308–15. [Google Scholar] [PubMed]
- Leung, D. W.; Spencer, S. A.; Cachianes, G.; Hammonds, R. G.; Collins, C.; Henzel, W. J.; Barnard, R.; Waters, M. J.; Wood, W. I. , Growth hormone receptor and serum binding protein: purification, cloning and expression. Nature 1987, 330, 537–43. [Google Scholar]
- Niall, H. D. , Revised primary structure for human growth hormone. Nat New Biol 1971, 230, 90–1. [Google Scholar]
- Hartman, M. L.; Faria, A. C.; Vance, M. L.; Johnson, M. L.; Thorner, M. O.; Veldhuis, J. D. , Temporal structure of in vivo growth hormone secretory events in humans. The American journal of physiology 1991, 260, E101–10. [Google Scholar]
- Brown, R. J.; Adams, J. J.; Pelekanos, R. A.; Wan, Y.; McKinstry, W. J.; Palethorpe, K.; Seeber, R. M.; Monks, T. A.; Eidne, K. A.; Parker, M. W.; Waters, M. J. , Model for growth hormone receptor activation based on subunit rotation within a receptor dimer. Nat Struct Mol Biol 2005, 12, 814–21. [Google Scholar]
- Gent, J.; Van Den Eijnden, M.; Van Kerkhof, P.; Strous, G. J. , Dimerization and signal transduction of the growth hormone receptor. Mol Endocrinol 2003, 17, 967–75. [Google Scholar]
- Herrington, J.; Carter-Su, C. , Signaling pathways activated by the growth hormone receptor. Trends Endocrinol Metab 2001, 12, 252–7. [Google Scholar]
- Chesover, A. D.; Dattani, M. T. , Evaluation of growth hormone stimulation testing in children. Clin Endocrinol (Oxf) 2016, 84, 708–14. [Google Scholar]
- Cerbone, M.; Dattani, M. T. , Progression from isolated growth hormone deficiency to combined pituitary hormone deficiency. Growth Horm IGF Res 2017, 37, 19–25. [Google Scholar]
- Murray, P. G.; Dattani, M. T.; Clayton, P. E. , Controversies in the diagnosis and management of growth hormone deficiency in childhood and adolescence. Arch Dis Child 2016, 101, 96–100. [Google Scholar]
- Petkovic, V.; Godi, M.; Pandey, A. V.; Lochmatter, D.; Buchanan, C. R.; Dattani, M. T.; Eblé, A.; Flück, C. E.; Mullis, P. E. , Growth hormone (GH) deficiency type II: a novel GH-1 gene mutation (GH-R178H) affecting secretion and action. J Clin Endocrinol Metab 2010, 95, 731–9. [Google Scholar] [PubMed]
- Petkovic, V.; Eblé, A.; Pandey, A. V.; Betta, M.; Mella, P.; Flück, C. E.; Buzi, F.; Mullis, P. E. , A novel GH-1 gene mutation (GH-P59L) causes partial GH deficiency type II combined with bioinactive GH syndrome. Growth Horm IGF Res 2011, 21, 160–6. [Google Scholar] [PubMed]
- Petkovic, V.; Miletta, M. C.; Boot, A. M.; Losekoot, M.; Flück, C. E.; Pandey, A. V.; Eblé, A.; Wit, J. M.; Mullis, P. E. , Short stature in two siblings heterozygous for a novel bioinactive GH mutant (GH-P59S) suggesting that the mutant also affects secretion of the wild-type GH. Eur J Endocrinol 2013, 168, K35–43. [Google Scholar] [PubMed]
- Miletta, M. C.; Eblé, A.; Janner, M.; Parween, S.; Pandey, A. V.; Flück, C. E.; Mullis, P. E. , IGHD II: A Novel GH-1 Gene Mutation (GH-L76P) Severely Affects GH Folding, Stability, and Secretion. J Clin Endocrinol Metab 2015, 100, E1575–83. [Google Scholar]
- Webb, E. A.; Dattani, M. T. , Diagnosis of growth hormone deficiency. Endocr Dev 2010, 18, 55–66. [Google Scholar]
- Franca, M. M.; Jorge, A. A.; Alatzoglou, K. S.; Carvalho, L. R.; Mendonca, B. B.; Audi, L.; Carrascosa, A.; Dattani, M. T.; Arnhold, I. J. , Absence of GH-releasing hormone (GHRH) mutations in selected patients with isolated GH deficiency. J Clin Endocrinol Metab 2011, 96, E1457–60. [Google Scholar]
- Alatzoglou, K. S.; Kular, D.; Dattani, M. T. , Autosomal Dominant Growth Hormone Deficiency (Type II). Pediatr Endocrinol Rev 2015, 12, 347–55. [Google Scholar]
- Altschul, S. F.; Madden, T. L.; Schaffer, A. A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D. J. , Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997, 25, 3389–402. [Google Scholar]
- Ashkenazy, H.; Erez, E.; Martz, E.; Pupko, T.; Ben-Tal, N. , ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res 2010, 38, W529–33. [Google Scholar]
- Krieger, E.; Darden, T.; Nabuurs, S. B.; Finkelstein, A.; Vriend, G. , Making optimal use of empirical energy functions: force-field parameterization in crystal space. Proteins 2004, 57, 678–83. [Google Scholar]
- Magrane, M. , UniProt Knowledgebase: a hub of integrated protein data. Database (Oxford) 2011, 2011, bar009. [Google Scholar] [PubMed]
- Worth, C. L.; Preissner, R.; Blundell, T. L. , SDM--a server for predicting effects of mutations on protein stability and malfunction. Nucleic Acids Res 2011, 39, W215–22. [Google Scholar] [PubMed]
- Pandurangan, A. P.; Ochoa-Montano, B.; Ascher, D. B.; Blundell, T. L. , SDM: a server for predicting effects of mutations on protein stability. Nucleic Acids Res 2017, 45, W229–W235. [Google Scholar]
- Topham, C. M.; Srinivasan, N.; Blundell, T. L. , Prediction of the stability of protein mutants based on structural environment-dependent amino acid substitution and propensity tables. Protein Eng 1997, 10, 7–21. [Google Scholar]
- Reumers, J.; Schymkowitz, J.; Rousseau, F. , Using structural bioinformatics to investigate the impact of non synonymous SNPs and disease mutations: scope and limitations. BMC Bioinformatics 2009, 10, S9. [Google Scholar]
- Karczewski, K. J.; Weisburd, B.; Thomas, B.; Solomonson, M.; Ruderfer, D. M.; Kavanagh, D.; Hamamsy, T.; Lek, M.; Samocha, K. E.; Cummings, B. B.; Birnbaum, D.; Daly, M. J.; MacArthur, D. G. , The ExAC browser: displaying reference data information from over 60 000 exomes. Nucleic Acids Res 2017, 45, D840–D845. [Google Scholar]
- Auton, A.; Brooks, L. D.; Durbin, R. M.; Garrison, E. P.; Kang, H. M.; Korbel, J. O.; Marchini, J. L.; McCarthy, S.; McVean, G. A.; Abecasis, G. R. , A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar]
- Millar, D. S.; Lewis, M. D.; Horan, M.; Newsway, V.; Easter, T. E.; Gregory, J. W.; Fryklund, L.; Norin, M.; Crowne, E. C.; Davies, S. J.; Edwards, P.; Kirk, J.; Waldron, K.; Smith, P. J.; Phillips, J. A., 3rd; Scanlon, M. F.; Krawczak, M.; Cooper, D. N.; Procter, A. M. , Novel mutations of the growth hormone 1 (GH1) gene disclosed by modulation of the clinical selection criteria for individuals with short stature. Human mutation 2003, 21, 424–40. [Google Scholar]
- Miyata, I.; Cogan, J. D.; Prince, M. A.; Kamijo, T.; Ogawa, M.; Phillips, J. A. , Detection of Growth Hormone Gene Defects by Dideoxy Fingerprinting (ddF). Endocr J 1997, 44, 149–154. [Google Scholar]
- Deladoey, J.; Stocker, P.; Mullis, P. E. , Autosomal dominant GH deficiency due to an Arg183His GH-1 gene mutation: clinical and molecular evidence of impaired regulated GH secretion. J Clin Endocrinol Metab 2001, 86, 3941–7. [Google Scholar]
- Takahashi, Y.; Kaji, H.; Okimura, Y.; Goji, K.; Abe, H.; Chihara, K. , Brief report: short stature caused by a mutant growth hormone. N Engl J Med 1996, 334, 432–6. [Google Scholar] [PubMed]
- Petkovic, V.; Besson, A.; Thevis, M.; Lochmatter, D.; Eble, A.; Fluck, C. E.; Mullis, P. E. , Evaluation of the biological activity of a growth hormone (GH) mutant (R77C) and its impact on GH responsiveness and stature. J Clin Endocrinol Metab 2007, 92, 2893–901. [Google Scholar] [PubMed]
- Takahashi, Y.; Shirono, H.; Arisaka, O.; Takahashi, K.; Yagi, T.; Koga, J.; Kaji, H.; Okimura, Y.; Abe, H.; Tanaka, T.; Chihara, K. , Biologically inactive growth hormone caused by an amino acid substitution. J Clin Invest 1997, 100, 1159–65. [Google Scholar] [PubMed]
- Rojas Velazquez, M. N.; Therkelsen, S.; Pandey, A. V. , Exploring Novel Variants of the Cytochrome P450 Reductase Gene (POR) from the Genome Aggregation Database by Integrating Bioinformatic Tools and Functional Assays. Biomolecules 2023, 13, 1728. [Google Scholar] [CrossRef]
- Prado, M. J.; Ligabue-Braun, R.; Zaha, A.; Rossetti, M. L. R.; Pandey, A. V. , Variant predictions in congenital adrenal hyperplasia caused by mutations in CYP21A2. Front Pharmacol 2022, 13, 931089. [Google Scholar]

| Organism | NCBI Seq ID | Uniprot Seq. ID | Uniprot Seq. Name | Seq length | Seq Identity % | Seq Similarity % | Signal Pep |
|---|---|---|---|---|---|---|---|
| Human | NP_000506 | P01241 | SOMA_HUMAN | 217 | 100 | 100 | 26 |
| Rhesus macaque | NP_001036203 | P33093 | SOMA_MACMU | 217 | 96 | 97 | 26 |
| Rat | NP_001030020 | P01244 | SOMA_RAT | 216 | 65 | 76 | 26 |
| Mouse | NP_032143 | P06880 | SOMA_MOUSE | 216 | 67 | 77 | 26 |
| Horse | NP_001075417 | P01245 | SOMA_HORSE | 216 | 67 | 79 | 26 |
| Pig | NP_999034 | P01248 | SOMA_PIG | 216 | 68 | 78 | 26 |
| Bovine | NP_851339 | P01246 | SOMA_BOVIN | 217 | 67 | 77 | 26 |
| Sheep | NP_001009315 | P67930 | SOMA_SHEEP | 217 | 67 | 76 | 26 |
| Guinea pig | NP_001166330 | Q9JKM4 | SOMA_CAVPO | 216 | 65 | 77 | 26 |
| Common turkey | XP_010722827 | P22077 | SOMA_MELGA | 216 | 55 | 73 | 25 |
| Chicken | NP_989690 | P08998 | SOMA_CHICK | 214 | 57 | 74 | 25 |
| Common ostrich | BAA82959 | Q9PWG3 | SOMA_STRCA | 215 | 54 | 72 | 25 |
| Japanese eel | AAA48535 | P08899 | SOMA_ANGJA | 207 | 44 | 61 | 19 |
| Goldfish | AAC19389 | O93359 | SOMA1_CARAU | 210 | 38 | 58 | 22 |
| Atlantic salmon | AAU11454 | Q5SDS1 | Q5SDS1_SALSA | 208 | 36 | 52 | 22 |
| Growth hormone deficiency, isolated, 1B (IGHD1B) | |||||
| Natural Variant | Sequence Position | PDB No. | Effect of mutation | dbSNP | Publication |
| L → P | 16 | - | suppresses secretion | [49] Millar | |
| D → N | 37 | 11 | - | [49] Millar | |
| R → C | 42 | 16 | reduced secretion | rs71640273 | [49] Millar |
| T → I | 53 | 27 | reduced ability to activate the JAK/STAT pathway | [49] Millar | |
| K → R | 67 | 41 | reduced ability to activate the JAK/STAT pathway | [49] Millar | |
| N → D | 73 | 47 | reduced ability to activate the JAK/STAT pathway | rs71640276 | [49] Millar |
| S → F | 97 | 71 | reduced ability to activate the JAK/STAT pathway | [49] Millar | |
| E → K | 100 | 74 | - | [49] Millar | |
| Q → L | 117 | 91 | reduced secretion | Q→R | [49] Millar |
| S → C | 134 | 108 | [49] Millar | ||
| S → R | 134 | 108 | reduced ability to activate the JAK/STAT pathway | [49] Millar | |
| T → A | 201 | 175 | reduced ability to activate the JAK/STAT pathway | [49] Millar | |
| Growth hormone deficiency, isolated, 2 (IGHD2) | |||||
| R → H | 209 | 183 | rs137853223 | [50] Miyata[51] Deladoey | |
| Kowarski syndrome (KWKS) | |||||
| R → C | 103 | 77 | No effect on GHR signaling pathway; does not affect interaction with GHR; results in a stronger interaction with GHBP; does not affect the subcellular location. | rs137853220 | [52] Takahashi[53] Petkovic |
| D → G | 138 | 112 | Loss of activity | rs137853221 | [54] Takahashi |
| ClinicalSignificance | Protein Residue | AA Pos | Ref Prot Res | PDBRes | ConSurf Conservation | Contact R 1 | Contact R 2 | UniProt Disease Variant |
|---|---|---|---|---|---|---|---|---|
| Gln [Q] | 28 | Pro [P] | 2 | e | Yes (2) | |||
| Thr [T] | 30 | Ile [I] | 4 | b | Yes (4) | |||
| Lys [K] | 34 | Arg [R] | 8 | e | Yes (8) | |||
| His [H] | 38 | Asn [N] | 12 | e | Yes (12) | |||
| Phe [F] | 41 | Leu [L] | 15 | b | Yes (15) | |||
| His [H] | 42 | Arg [R] | 16 | e, f | Yes (16) | Yes (R>C) | ||
| Thr [T] | 43 | Ala [A] | 17 | b, s | ||||
| Arg [R] | 44 | His [H] | 18 | e | Yes (18) | |||
| Tyr [Y] | 47 | His [H] | 21 | e, f | Yes (21) | |||
| Thr [T] | 50 | Ala [A] | 24 | b, s | ||||
| B | Tyr [Y] | 51 | Phe [F] | 25 | e | Yes (25) | ||
| Pro [P] | 71 | Leu [L] | 45 | e | Yes (45) | |||
| Lys [K] | 73 | Asn [N] | 47 | e | Yes (N>D) | |||
| Thr [T] | 74 | Pro [P] | 48 | e | Yes (48) | |||
| P | Ser [S] | 79 | Cys [C] | 53 | b, s | |||
| Cys [C] | 88 | Ser [S] | 62 | e | Yes (62) | |||
| Lys [K] | 89 | Asn [N] | 63 | e | Yes (63) | |||
| His [H] | 103 | Arg [R] | 77 | e | Yes (R>C) | |||
| P | Cys [C] | 103 | 77 | |||||
| Cys [C] | 105 | Ser [S] | 79 | b, s | ||||
| Glu [E] | 110 | Gln [Q] | 84 | e, f | ||||
| Arg [R] | 117 | Gln [Q] | 91 | e | Yes (Q>L) | |||
| P | Gly [G] | 138 | Asp [D] | 112 | e | Yes (D>G) | ||
| Glu [E] | 142 | Asp [D] | 116 | e | Yes (116) | |||
| Asp [D] | 145 | Glu [E] | 119 | e | Yes (119) | |||
| Ser [S] | 146 | Gly [G] | 120 | b | Yes (120) | |||
| Met [M] | 149 | Thr [T] | 123 | b | Yes (123) | |||
| Pro [P] | 188 | Leu [L] | 162 | b, s | ||||
| His [H] | 190 | Tyr [Y] | 164 | b | Yes (164) | |||
| Glu [E] | 195 | Asp [D] | 169 | e, f | ||||
| Asn [N] | 198 | Lys [K] | 172 | e, f | Yes (172) | |||
| Lys [K] | 200 | Glu [E] | 174 | e | Yes (174) | |||
| Met [M] | 205 | Ile [I] | 179 | b | Yes (179) | |||
| Arg [R] | 208 | Cys [C] | 182 | b, s | Yes (182) | |||
| P | His [H] | 209 | Arg [R] | 183 | e | Yes (R>H) | ||
| Tyr [Y] | 215 | Cys [C] | 189 | e, f | Yes (189) | |||
| Ser [S] | 216 | Gly [G] | 190 | e | Yes (190) |
| SNV | Conservation score | Residue variety across species |
|---|---|---|
| P2Q | 8 | P,Y,V |
| I4T | 5 | A,F,T,P,E,V,M,I,L |
| R8K | 4 | S,W,N,K,E,H,Q,D,R,G |
| N12H | 7 | S,T,N,K,E,H,M,C,I,R,L |
| L15F | 1 | S,F,T,N,K,E,V,H,Q,M,R,I,G,L |
| R16H, R16L, R16C | 7 | H,Q,R,Y,L,V |
| A17T | 8 | S,A,T,I,L,V |
| H18R | 7 | S,W,T,N,E,H,Q,D |
| H21Y | 7 | F,S,H,K,R,Y,V |
| A24T | 8 | S,A,T,N,Y,V |
| F25Y, F25I | 6 | S,A,F,T,K,E,Y,Q,M,D,R,I,G,L |
| L45P | 6 | L |
| N47K, N47D | 3 | S,A,T,N,P,K,V,H,M,D,I,G |
| C53S, C53F | 9 | C |
| S62C | 5 | A,S,T,N,K,E,V,H,Q,M,I,G |
| N63K | 7 | S,D,N,P,G,E |
| R77H, R77C | 5 | S,N,K,H,Q,D,R,G,L |
| S79C | 7 | A,S,M,T,I,G,V |
| Q84F | 6 | S,W,P,Y,E,V,H,Q,M,D,R,I,L |
| Q91R | 3 | S,F,A,N,K,E,Y,V,H,Q,D,R,I,G,L |
| D112G, G112H | 1 | A,S,T,N,K,P,E,H,Q,D,R,G,L |
| D116E, D116N | 4 | A,S,N,K,E,Y,V,Q,D,R,I,G |
| E119D | 4 | S,A,T,N,K,E,V,Q,M,D,R,L |
| G120S, G120C | 9 | A,F,T,G,Y |
| T123M | 2 | S,A,T,N,K,E,V,M,R,I,L |
| L162P | 7 | F,T,M,N,K,I,L,V |
| Y164H | 6 | A,S,T,N,Y,H,M,C,R |
| D169E | 9 | D,E |
| K172N | 9 | H,M,N,R,K |
| E174K | 9 | S,Q,D,Y,E |
| I179V, I179M, I179S | 7 | F,T,M,I,L,V |
| C182R | 9 | C |
| R183H, R183C | 9 | Q,K,R |
| C189Y | 9 | C |
| G190S | 4 | S,A,T,G |
| Mutation | WT_SSE | WT_RSA (%) | WT_DEPTH (Å) | WT_OSP | WT_SS | WT_SN | WT_SO | MT_SSE | MT_RSA (%) | MT_DEPTH (Å) | MT_OSP | MT_SS | MT_SN | MT_SO | Predicted ΔΔG | Stability |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P2Q | p | 89 | 3.2 | 0.11 | - | - | - | p | 99 | 3.3 | 0.08 | - | - | - | -0.8 | - |
| I4T | b | 55.1 | 3.5 | 0.33 | - | - | - | b | 70.7 | 3.4 | 0.26 | - | - | - | -1.09 | - |
| I4V | b | 55.1 | 3.5 | 0.33 | - | - | - | b | 55.1 | 3.3 | 0.33 | - | - | - | -0.07 | - |
| R8K | H | 59.6 | 3.4 | 0.31 | + | - | - | H | 66.7 | 3.5 | 0.27 | - | - | - | -0.43 | - |
| N12H | H | 57.8 | 3.5 | 0.33 | + | - | + | H | 58.6 | 3.5 | 0.26 | + | - | - | 0.68 | + |
| L15F | H | 74.6 | 3.2 | 0.23 | - | - | - | H | 79.7 | 3.3 | 0.19 | - | - | - | -0.63 | - |
| R16H | H | 44.2 | 3.8 | 0.34 | + | - | - | H | 31.9 | 3.9 | 0.4 | + | - | + | 0.19 | + |
| R16L | H | 44.2 | 3.8 | 0.34 | + | - | - | H | 20.9 | 4.1 | 0.44 | - | - | - | 0.39 | + |
| R16C | H | 44.2 | 3.8 | 0.34 | + | - | - | H | 22.1 | 4.2 | 0.44 | + | - | + | -0.76 | - |
| A17T | H | 1.4 | 6.7 | 0.54 | - | - | - | H | 0.3 | 6.9 | 0.59 | - | + | - | -1.88 | - |
| H18R | H | 69.2 | 3.4 | 0.26 | + | - | - | H | 70.1 | 3.4 | 0.2 | - | - | - | 0.06 | + |
| H21Y | H | 18.8 | 4.3 | 0.47 | - | - | - | H | 25.8 | 4.5 | 0.41 | - | - | - | 0.65 | + |
| A24T | H | 0 | 8.3 | 0.57 | - | - | - | H | 0 | 8.4 | 0.65 | - | - | + | -3.21 | - |
| F25Y | H | 57.4 | 3.6 | 0.27 | - | - | - | H | 57.6 | 3.6 | 0.27 | - | - | - | 0.47 | + |
| F25I | H | 57.4 | 3.6 | 0.27 | - | - | - | H | 47.8 | 3.6 | 0.34 | - | - | - | 0.31 | + |
| L45P | H | 40.9 | 3.6 | 0.33 | - | - | - | H | 34 | 3.7 | 0.33 | - | - | - | -2.23 | - |
| P48T | H | 78 | 3.1 | 0.2 | - | - | - | H | 95 | 3.2 | 0.16 | - | - | - | 0.34 | + |
| C53S | b | 3.7 | 5.9 | 0.43 | + | - | + | b | 4.5 | 5.8 | 0.42 | + | - | - | -1.11 | - |
| N47K | b | 62.6 | 3.3 | 0.39 | + | + | + | b | 82.8 | 3.3 | 0.2 | - | - | - | -0.32 | - |
| N47D | b | 62.6 | 3.3 | 0.39 | + | + | + | b | 67.5 | 3.3 | 0.33 | - | - | + | -0.44 | - |
| C53F | b | 3.7 | 5.9 | 0.43 | + | - | + | b | 3.6 | 5.2 | 0.51 | - | - | - | -0.62 | - |
| S62C | a | 84.6 | 3.1 | 0.14 | + | - | - | a | 90.8 | 3.2 | 0.11 | - | - | - | 0.62 | + |
| N63K | b | 71.2 | 3.5 | 0.3 | + | + | - | b | 83.5 | 3.3 | 0.16 | - | - | - | -0.18 | - |
| R77H | H | 19.4 | 4.9 | 0.45 | - | - | + | H | 17 | 4.7 | 0.54 | + | - | + | -0.07 | - |
| R77C | H | 19.4 | 4.9 | 0.45 | - | - | + | H | 12.1 | 5 | 0.57 | - | - | + | -0.71 | - |
| S79C | H | 0 | 10.9 | 0.57 | - | - | + | H | 0 | 10.8 | 0.61 | - | + | + | 1.52 | + |
| Q84E | H | 21.4 | 4 | 0.43 | + | - | - | H | 12.1 | 4.4 | 0.45 | + | - | - | 0.4 | + |
| Q91R | H | 64.4 | 3.4 | 0.24 | - | - | - | H | 78.7 | 3.4 | 0.16 | - | - | - | -0.15 | - |
| Q91K | H | 64.4 | 3.4 | 0.24 | - | - | - | H | 57.2 | 3.4 | 0.21 | - | - | - | -0.44 | - |
| Q91L | H | 64.4 | 3.4 | 0.24 | - | - | - | H | 62.9 | 3.4 | 0.23 | - | - | - | 0.29 | + |
| D112G | H | 78.3 | 3.4 | 0.27 | - | - | + | H | 82.6 | 3.7 | 0.34 | - | - | - | -0.16 | - |
| D112H | H | 78.3 | 3.4 | 0.27 | - | - | + | H | 76.7 | 3.4 | 0.25 | - | - | + | 0.88 | + |
| D116E | H | 54.7 | 3.6 | 0.31 | + | - | - | H | 57.1 | 3.8 | 0.26 | - | - | - | 1.25 | + |
| D116N | H | 54.7 | 3.6 | 0.31 | + | - | - | H | 61.9 | 3.7 | 0.29 | + | - | - | -0.35 | - |
| E119D | H | 85.5 | 3.3 | 0.2 | - | - | - | H | 80.4 | 3.3 | 0.25 | - | - | - | -1.48 | - |
| G120S | H | 64.2 | 4.6 | 0.46 | - | - | - | H | 30.8 | 4.2 | 0.42 | - | - | + | 0.18 | + |
| G120C | H | 64.2 | 4.6 | 0.46 | - | - | - | H | 30.1 | 4.2 | 0.4 | - | - | + | 0.7 | + |
| T123M | H | 47.5 | 3.8 | 0.29 | - | - | + | H | 52.2 | 3.6 | 0.24 | - | - | - | 1.19 | + |
| L162P | H | 3.5 | 6 | 0.48 | - | - | - | H | 16.4 | 5.4 | 0.39 | - | - | - | -4.31 | - |
| Y164H | H | 13.3 | 5.2 | 0.48 | - | - | - | H | 10.8 | 4.9 | 0.48 | - | - | - | -1.27 | - |
| D169E | H | 1.4 | 7.7 | 0.53 | + | - | + | H | 3 | 9.3 | 0.6 | + | - | + | -0.01 | - |
| K172N | H | 27.5 | 3.9 | 0.4 | - | - | - | H | 35 | 4.3 | 0.42 | - | - | + | -0.69 | - |
| E174K | H | 24.3 | 3.7 | 0.4 | + | - | - | H | 32.8 | 4.2 | 0.33 | - | - | - | -1.01 | - |
| I179M | H | 23 | 4.2 | 0.4 | - | - | - | H | 32.8 | 4.1 | 0.32 | + | - | - | -0.02 | - |
| I179S | H | 23 | 4.2 | 0.4 | - | - | - | H | 19.5 | 4.5 | 0.39 | - | - | + | -0.8 | - |
| I179V | H | 23 | 4.2 | 0.4 | - | - | - | H | 20.4 | 4.3 | 0.43 | - | - | - | -0.35 | - |
| C182R | H | 28.7 | 3.5 | 0.4 | + | - | + | H | 68.5 | 3.6 | 0.21 | - | - | - | 1.04 | + |
| R183H | H | 31.8 | 3.7 | 0.3 | + | - | + | H | 77.5 | 3.4 | 0.16 | - | - | - | -1.1 | - |
| R183C | H | 31.8 | 3.7 | 0.3 | + | - | + | H | 72.9 | 3.2 | 0.2 | - | - | - | -0.65 | - |
| C189Y | a | 22.9 | 3.9 | 0.29 | + | - | - | a | 75.2 | 3.4 | 0.11 | - | - | - | 0.78 | + |
| G190S | b | 199.3 | 3.5 | 0.07 | - | - | - | p | 107.2 | 3.1 | 0.08 | - | - | - | 0 | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





