Submitted:
28 February 2025
Posted:
28 February 2025
You are already at the latest version
Abstract
Plant glutathione peroxidase-like (GPXL) enzymes are thiol-based peroxidases that reduce H₂O₂ or hydroperoxides to water or alcohols using electrons principally from thioredoxin. Arabidopsis thaliana possesses eight isoenzymes located in different plant’s organelles and have various roles in redox-dependent processes. Determination of the redox potential of 6-day-old Atgpxl1-8 T-DNA insertional mutants using cytosolic redox sensitive fluorescent probe (roGFP2) uncovered more oxidized redox status in shoot and/or root of the untreated mutants except Atgpxl5. To investigate the involvement of AtGPXLs in growth and abiotic stress responses of seedlings, the 4-day-old Atgpxls were exposed to salt- and osmotic stresses for two weeks. Evaluation of the reactive oxygen species (ROS) levels of untreated 18-day-old plants using fluorescent microscopy revealed the elevated accumulation of total ROS in the shoots and, in some cases, the roots of the mutants. Regarding the growth of roots, both the length of primary roots and/or number of lateral roots were affected by mutation of AtGPXLs. Strong negative correlation was observed between the ROS level of wild type shoots and development of lateral roots, but it was altered in mutants: while in case of Atgpxl1, -5 and -7 seedlings it disappeared, in other mutants (Atgpxl4, -6 and -8) the correlation became stronger. Our analysis underpins the discrete role of AtGPXL enzymes in controlling the growth and development of plants by fine-tuning the ROS contents and redox status in an organ-specific way. Differences in root phenotype and metabolic activity between Atgpxl mutants and wild-type plants highlight the essential role of AtGPXLs in ROS processing to support growth, which is particularly evident when one GPXL isoenzyme is absent or its activity is reduced, both under normal and abiotic stress conditions.
Keywords:
1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. Analysis of the Redox Potential by Ratiometric Measurements of the roGFP2 Fluorescent Probe
2.3. Measurements of Root Parameters
2.4. Detection of the Vitality, Superoxide Radical and Total ROS Levels in Roots and Leaves
2.5. Correlation Analysis
2.6. Statistical Analysis
3. Results
3.1. The Redox Status of 6-Day-Old Atgpxl Seedlings Are More Oxidized Compared to the Wild Type
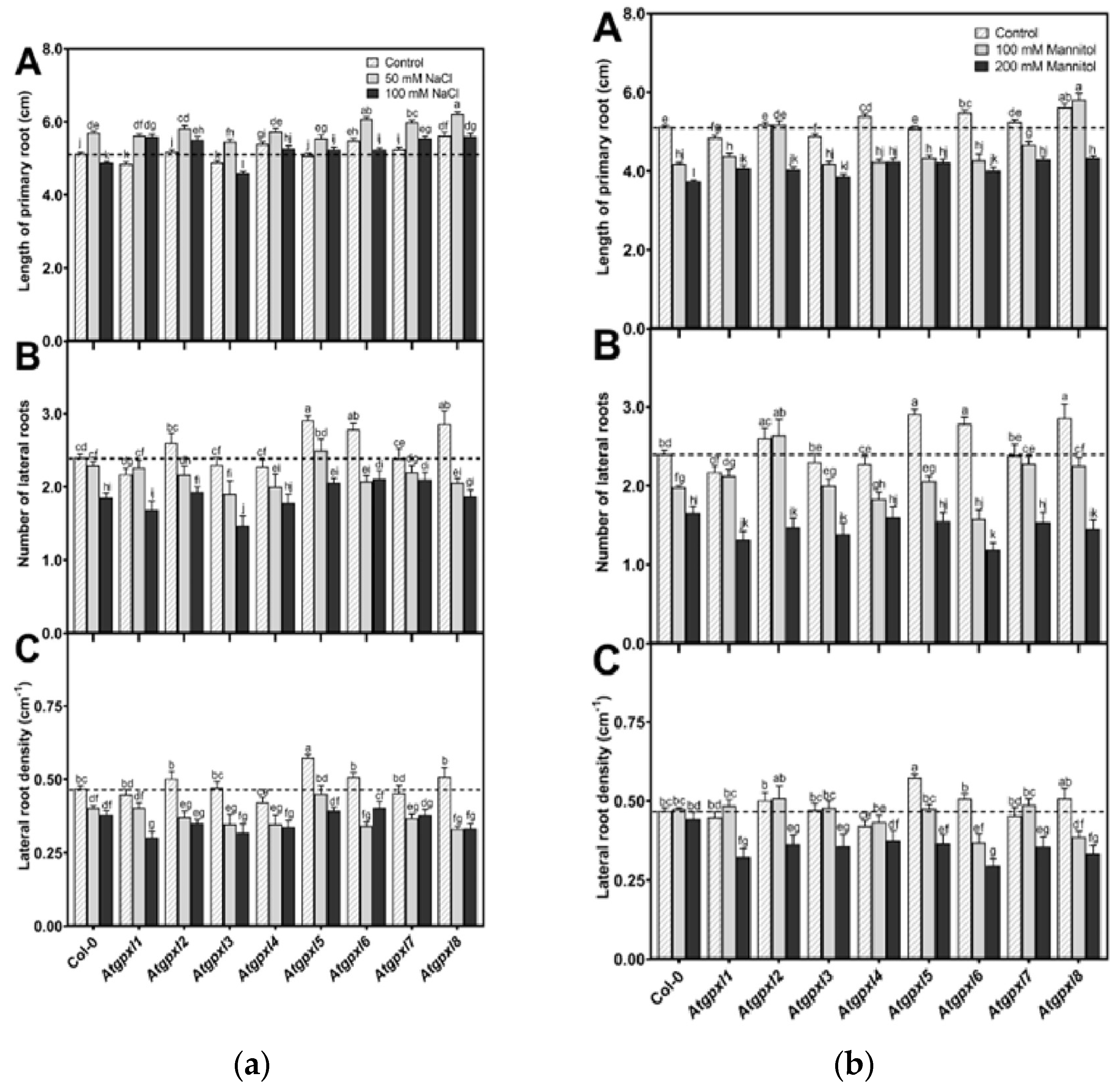
3.2. Several Atgpxl Mutants Had Longer Primary Roots Than the Wild Type Controls, and 50 mM NaCl Promoted the Growth of All Genotypes Compared to the Untreated Col-0 Primary Roots
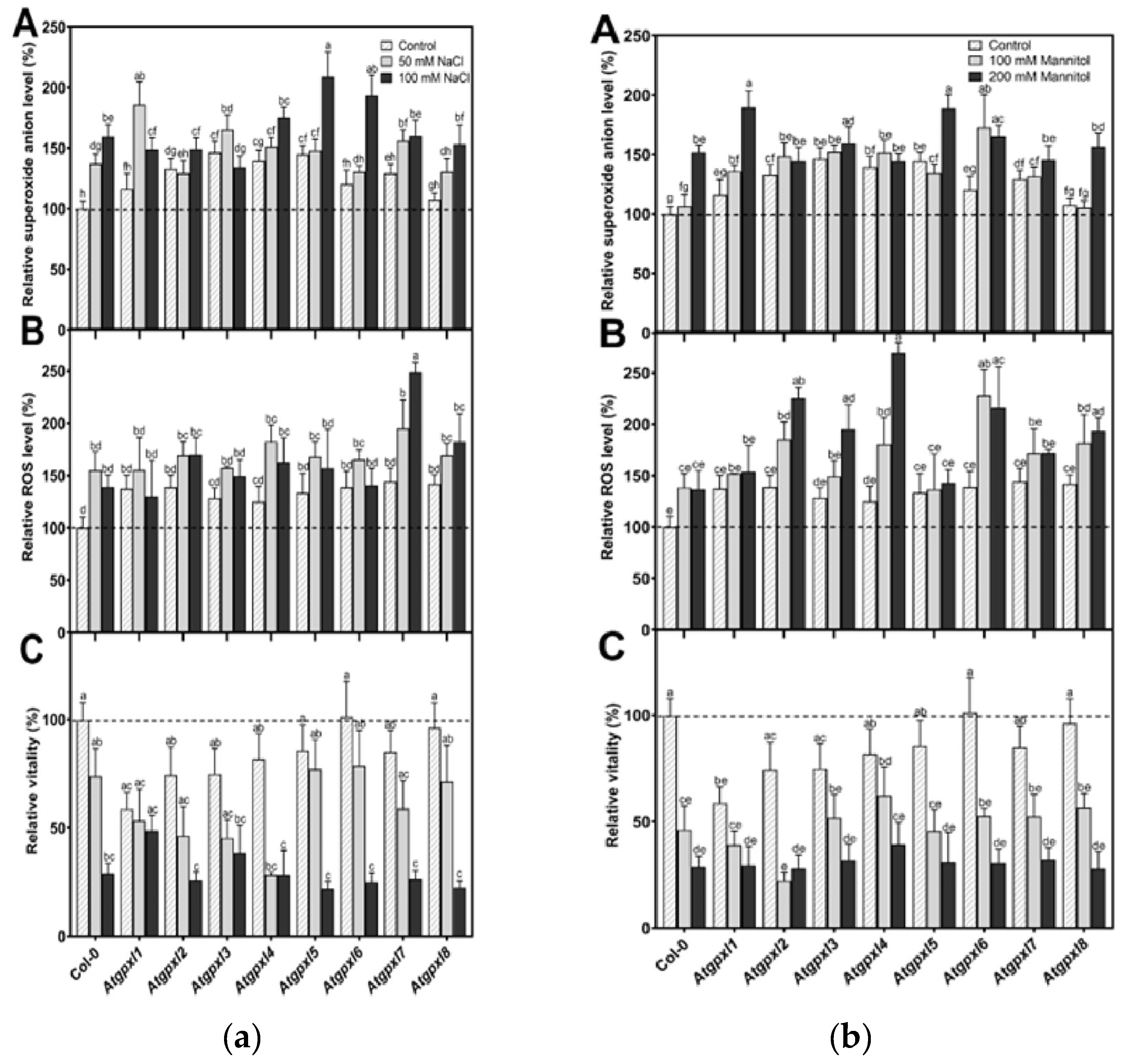
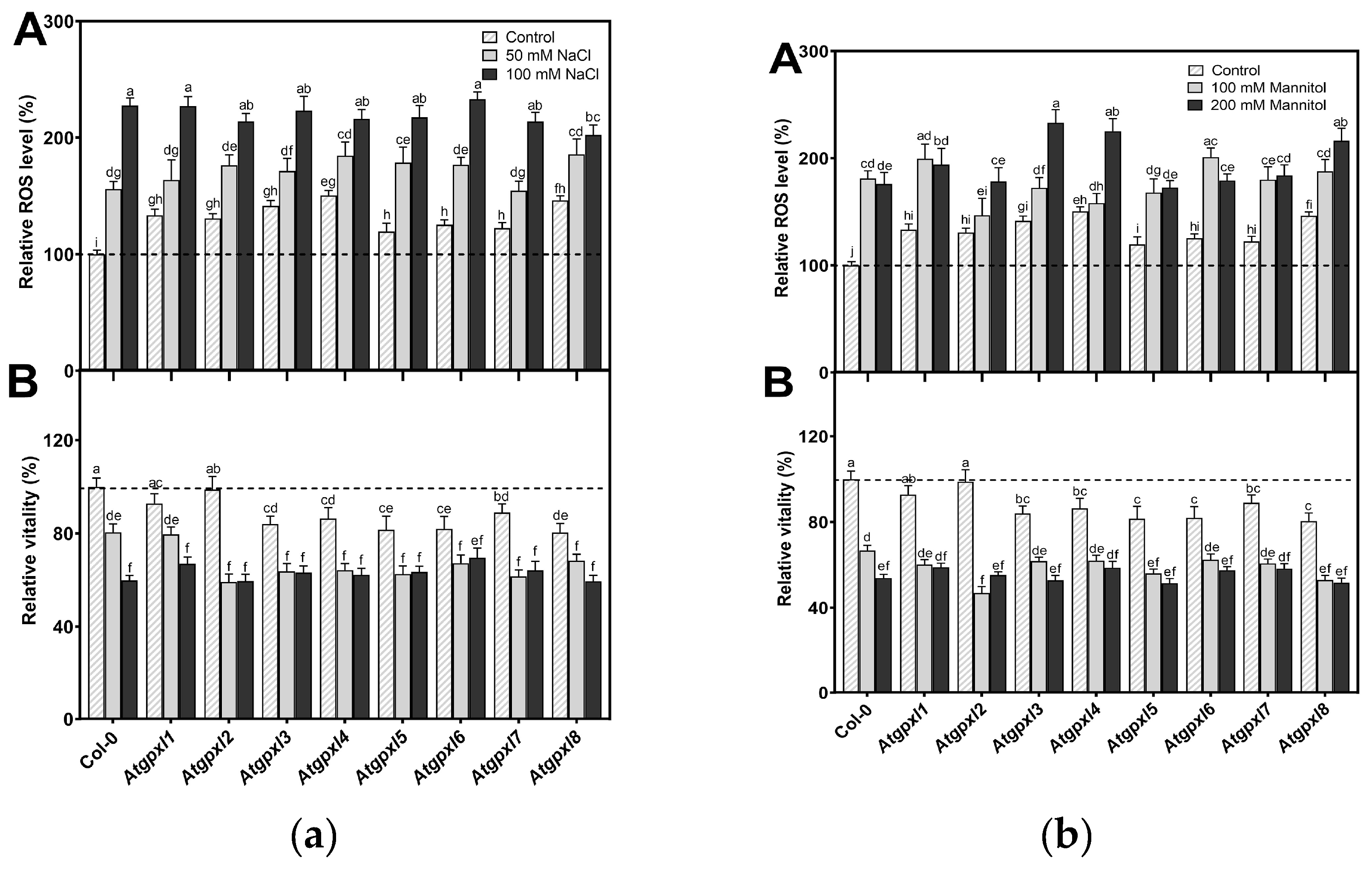
3.3. Slightly Elevated ROS Levels Decreased the Vitality of the Mutant Roots
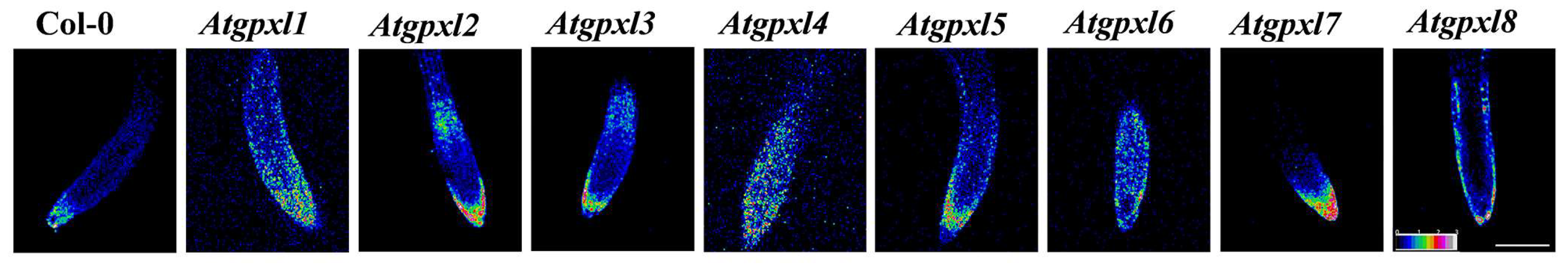
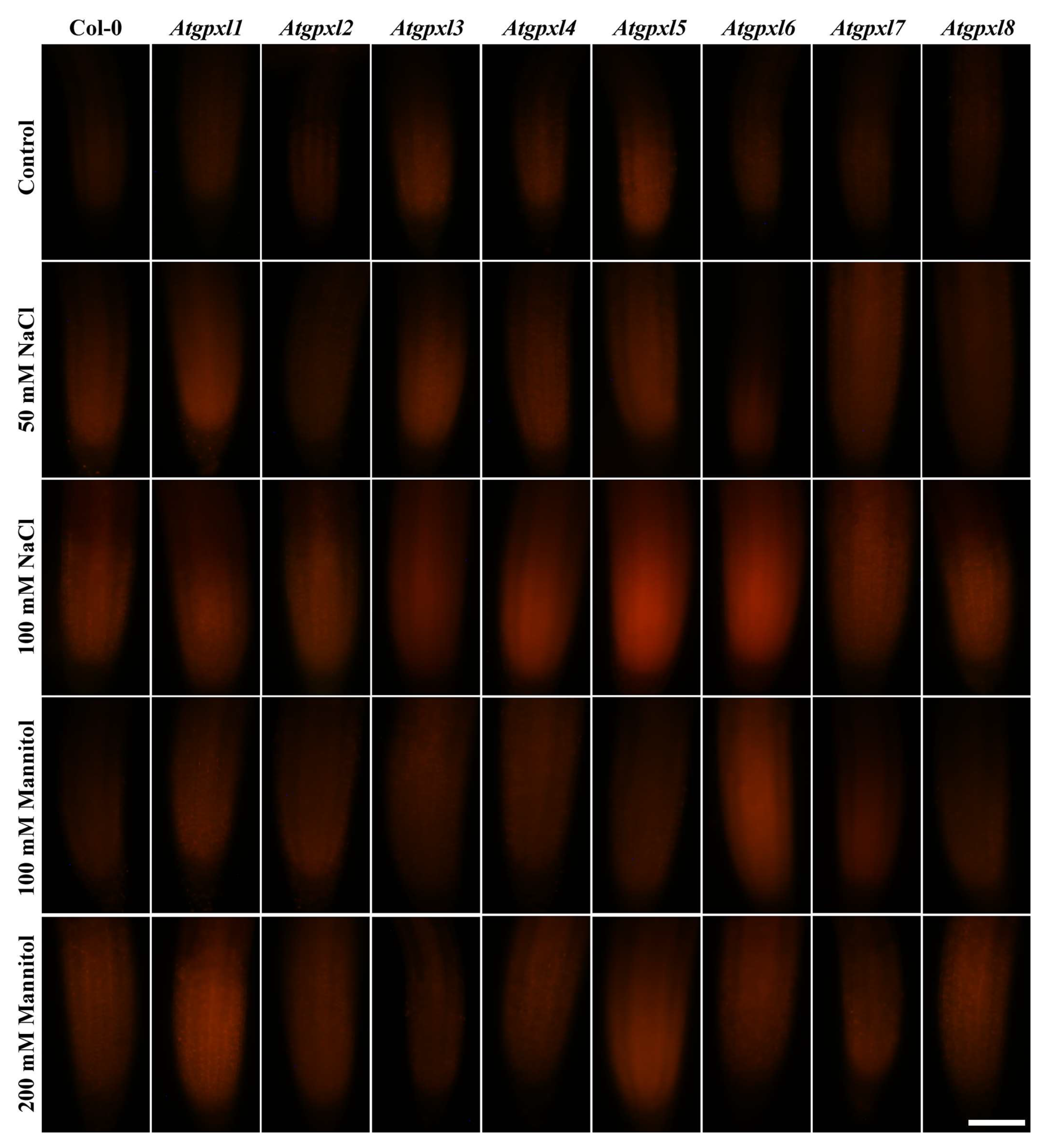
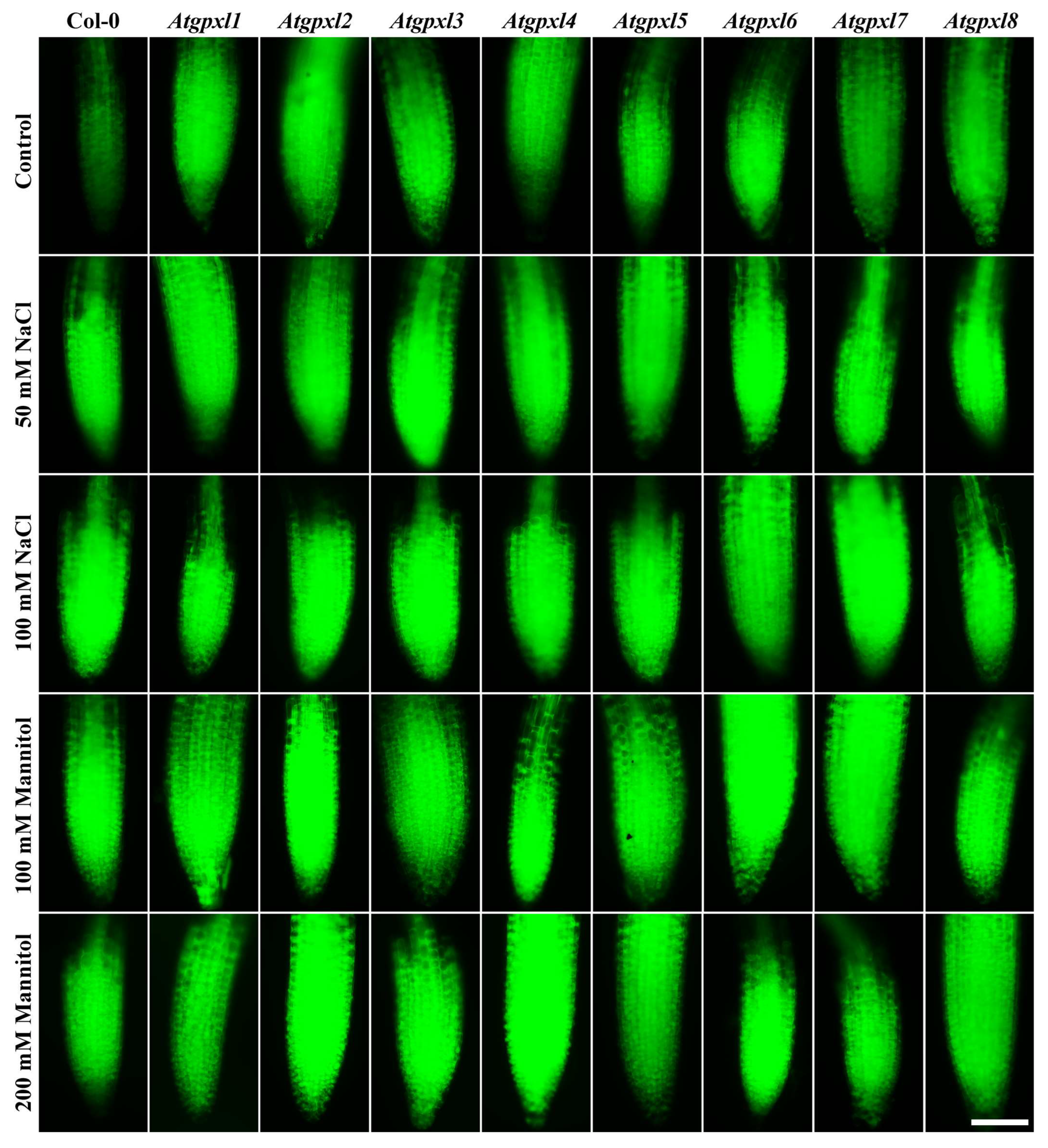
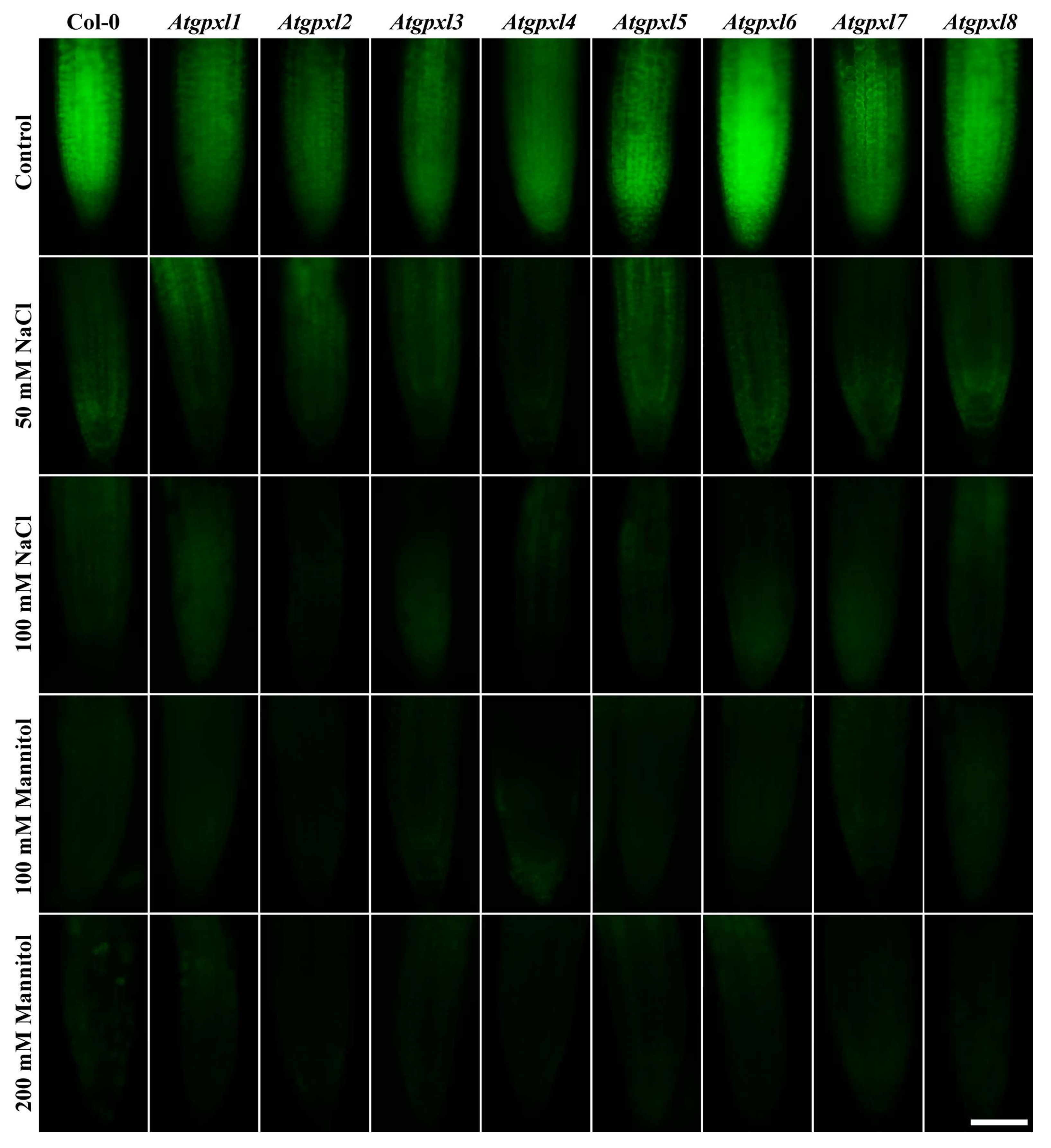
3.4. The Leaves of Untreated Mutant Seedlings Accumulated More Total ROS and Most of Them Showed Less Metabolic Activity
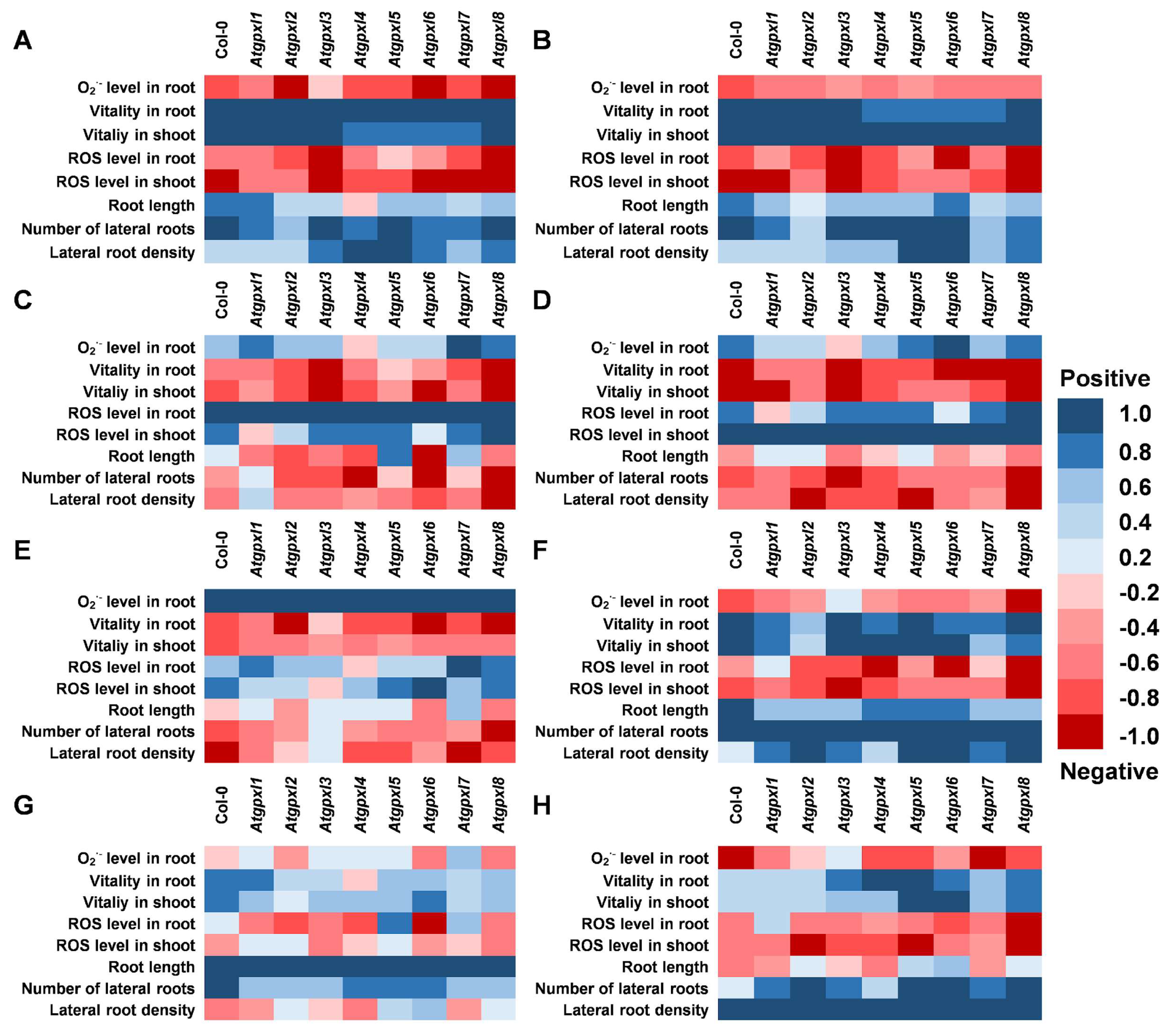
3.5. Correlations Between the Measured Parameters Strengthen the Involvement of AtGPXLs in ROS Homeostasis
4. Discussion
4.1. The ROS-Processing Roles of AtGPXLs in the Mutants Were Not Substituted Completely Either Under Control or Abiotic Stress Conditions
4.2. AtGPXLs Have Specific Functions in Tissues and Organs
4.3. AtGPXls Are Related to Redox Signaling
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mittler, R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Noctor, G. Redox homeostasis and antioxidant signaling: A metabolic interface between stress perception and physiological responses. Plant Cell 2005, 17, 1866–1875. [Google Scholar] [CrossRef] [PubMed]
- Boguszewska, D.; Zagdańska, B. ROS as signaling molecules and enzymes of plant response to unfavorable environmental conditions. In Oxidative stress–molecular mechanisms and biological effects; InTech: Rijeka, Croatia, 2012; pp. 341–362. [Google Scholar] [CrossRef]
- Baxter, A.; Mittler, R.; Suzuki, N. ROS as key players in plant stress signalling. J. Exp. Bot. 2014, 65, 1229–1240. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R. ROS Are Good. Trends Plant Sci. 2017, 22, 11–19. [Google Scholar] [CrossRef]
- Huang, H.; Ullah, F.; Zhou, D.X.; Yi, M.; Zhao, Y. Mechanisms of ROS regulation of plant development and stress responses. Front. Plant Sci. 2019, 10, 800. [Google Scholar] [CrossRef]
- Chapman, J.M.; Muhlemann, J.K.; Gayomba, S.R.; Muday, G.K. RBOH-Dependent ROS Synthesis and ROS Scavenging by Plant Specialized Metabolites To Modulate Plant Development and Stress Responses. Chem. Res. Toxicol. 2019, 32, 370–396. [Google Scholar] [CrossRef]
- Mase, K.; Tsukagoshi, H. Reactive Oxygen Species Link Gene Regulatory Networks During Arabidopsis Root Development. Front. Plant Sci. 2021, 12, 660274. [Google Scholar] [CrossRef] [PubMed]
- Imai, H.; Nakagawa, Y. Biological significance of phospholipid hydroperoxide glutathione peroxidase (PHGPx, GPx4) in mammalian cells. Free Radic. Biol. Med. 2003, 34, 145–169. [Google Scholar] [CrossRef]
- Margis, R.; Dunand, C.; Teixeira, F.K.; Margis-Pinheiro, M. Glutathione peroxidase family–an evolutionary overview. FEBS J. 2008, 275, 3959–3970. [Google Scholar] [CrossRef]
- Brigelius-Flohé, R.; Maiorino, M. Glutathione peroxidases. Biochim. Biophys. Acta (BBA)-General Subj. 2013, 1830, 3289–3303. [Google Scholar] [CrossRef]
- Luo, D.; Smith, S.W.; Anderson, B.D. Kinetics and mechanism of the reaction of cysteine and hydrogen peroxide in aqueous solution. J. Pharm. Sci. 2005, 94, 304–316. [Google Scholar] [CrossRef] [PubMed]
- Marinho, H.S.; Real, C.; Cyrne, L.; Soares, H.; Antunes, F. Hydrogen peroxide sensing, signaling and regulation of transcription factors. Redox Biol. 2014, 2, 535–562. [Google Scholar] [CrossRef]
- Conrad, M.; Moreno, S.G.; Sinowatz, F.; Ursini, F.; Kolle, S.; Roveri, A.; Brielmeier, M.; Wurst, W.; Maiorino, M.; Bornkamm, G.W. The nuclear form of phospholipid hydroperoxide glutathione peroxidase is a protein thiol peroxidase contributing to sperm chromatin stability. Mol. Cell. Biol. 2005, 25, 7637–7644. [Google Scholar] [CrossRef] [PubMed]
- Brigelius-Flohe, R.; Kipp, A. Glutathione peroxidases in different stages of carcinogenesis. Biochim. Biophys. Acta (BBA)-General Subj. 2009, 1790, 1555–1568. [Google Scholar] [CrossRef]
- Iqbal, A.; Yabuta, Y.; Takeda, T.; Nakano, Y.; Shigeoka, S. Hydroperoxide reduction by thioredoxin-specific glutathione peroxidase isoenzymes of Arabidopsis thaliana. FEBS J. 2006, 273, 5589–5597. [Google Scholar] [CrossRef]
- Navrot, N.; Collin, V.; Gualberto, J.; Gelhaye, E.; Hirasawa, M.; Rey, P.; Knaff, D.B.; Issakidis, E.; Jacquot, J.-P.; Rouhier, N. Plant glutathione peroxidases are functional peroxiredoxins distributed in several subcellular compartments and regulated during biotic and abiotic stresses. Plant Physiol. 2006, 142, 1364–1379. [Google Scholar] [CrossRef] [PubMed]
- Bela, K.; Riyazuddin, R.; Csiszár, J. Plant Glutathione Peroxidases: Non-Heme Peroxidases with Large Functional Flexibility as a Core Component of ROS-Processing Mechanisms and Signalling. Antioxidants 2022, 11, 1624. [Google Scholar] [CrossRef]
- Attacha, S.; Solbach, D.; Bela, K.; Moseler, A.; Wagner, S.; Schwarzländer, M.; Aller, I.; Müller, S.J.; Meyer, A.J. Glutathione peroxidase-like enzymes cover five distinct cell compartments and membrane surfaces in Arabidopsis thaliana. Plant. Cell Environ. 2017, 40, 1281–1295. [Google Scholar] [CrossRef]
- Miao, Y.; Lv, D.; Wang, P.; Wang, X.-C.; Chen, J.; Miao, C.; Song, C.-P. An Arabidopsis glutathione peroxidase functions as both a redox transducer and a scavenger in abscisic acid and drought stress responses. Plant Cell 2006, 18, 2749–2766. [Google Scholar] [CrossRef]
- Bela, K.; Horváth, E.; Gallé, Á.; Szabados, L.; Tari, I.; Csiszár, J. Plant glutathione peroxidases: Emerging role of the antioxidant enzymes in plant development and stress responses. J. Plant Physiol. 2015, 176, 192–201. [Google Scholar] [CrossRef]
- Haskirli, H.; Yilmaz, O.; Ozgur, R.; Uzilday, B.; Turkan, I. Melatonin mitigates UV-B stress via regulating oxidative stress response, cellular redox and alternative electron sinks in Arabidopsis thaliana. Phytochem. 2020, 182, 112592. [Google Scholar] [CrossRef]
- Madhu; Sharma, A.; Kaur, A.; Tyagi, S.; Upadhyay, S.K. Glutathione Peroxidases in Plants: Innumerable Role in Abiotic Stress Tolerance and Plant Development. J. Plant Growth Regul. 2023, 42, 598–613. [Google Scholar] [CrossRef]
- Herbette, S.; Menn, A.L.; Rousselle, P.; Ameglio, T.; Faltin, Z.; Branlard, G.; Eshdat, Y.; Julien, J.L.; Drevet, J.R.; Roeckel-Drevet, P. Modification of photosynthetic regulation in tomato overexpressing glutathione peroxidase. Biochim. Biophys. Acta 2005, 1724, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Herbette, S.; de Labrouhe, D.T.; Drevet, J.R.; Roeckel-Drevet, P. Transgenic tomatoes showing higher glutathione peroxydase antioxidant activity are more resistant to an abiotic stress but more susceptible to biotic stresses. Plant Sci. 2011, 180, 548–553. [Google Scholar] [CrossRef]
- Diao, Y.; Xu, H.; Li, G.; Yu, A.; Yu, X.; Hu, W.; Zheng, X.; Li, S.; Wang, Y.; Hu, Z. Cloning a glutathione peroxidase gene from Nelumbo nucifera and enhanced salt tolerance by overexpressing in rice. Mol. Biol. Rep. 2014, 41, 4919–4927. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, K.; Miyao, K.; Gaber, A.; Takeda, T.; Kanaboshi, H.; Miyasaka, H.; Shigeoka, S. Enhancement of stress tolerance in transgenic tobacco plants overexpressing Chlamydomonas glutathione peroxidase in chloroplasts or cytosol. Plant J. 2004, 37, 21–33. [Google Scholar] [CrossRef]
- Gaber, A.; Ogata, T.; Maruta, T.; Yoshimura, K.; Tamoi, M.; Shigeoka, S. The involvement of Arabidopsis glutathione peroxidase 8 in the suppression of oxidative damage in the nucleus and cytosol. Plant Cell Physiol. 2012, 53, 1596–1606. [Google Scholar] [CrossRef] [PubMed]
- Zhai, C.Z.; Zhao, L.; Yin, L.J.; Chen, M.; Wang, Q.Y.; Li, L.C.; Xu, Z.S.; Ma, Y.Z. Two wheat glutathione peroxidase genes whose products are located in chloroplasts improve salt and H2O2 tolerances in Arabidopsis. PLoS ONE 2013, 8, e73989. [Google Scholar] [CrossRef]
- Riyazuddin, R.; Bela, K.; Horváth, E.; Rigó, G.; Gallé, Á.; Szabados, L.; Fehér, A.; Csiszár, J. Overexpression of the Arabidopsis glutathione peroxidase-like 5 gene (AtGPXL5) resulted in altered plant development and redox status. Environ. Exp. Bot. 2019, 167, 103849. [Google Scholar] [CrossRef]
- Zhang, L.; Wu, M.; Teng, Y.; Jia, S.; Yu, D.; Wei, T.; Chen, C.; Song, W. Overexpression of the glutathione peroxidase 5 (RcGPX5) gene from Rhodiola crenulata increases drought tolerance in Salvia miltiorrhiza. Front. Plant Sci. 2019, 9, 1950. [Google Scholar] [CrossRef]
- Passaia, G.; Queval, G.; Bai, J.; Margis-Pinheiro, M.; Foyer, C.H. The Effects of Redox Controls Mediated by Glutathione Peroxidases on Root Architecture in Arabidopsis thaliana. J. Exp. Bot. 2014, 65, 1403–1413. [Google Scholar] [CrossRef]
- Benitez-Alfonso, Y.; Cilia, M.; San Roman, A.; Thomas, C.; Maule, A.; Hearn, S.; Jackson, D. Control of Arabidopsis meristem development by thioredoxin-dependent regulation of intercellular transport. Proc. Natl. Acad. Sci. 2009, 106, 3615–3620. [Google Scholar] [CrossRef] [PubMed]
- Schnaubelt, D.; Queval, G.; Dong, Y.; Diaz-Vivancos, P.; Makgopa, M.E.; Howell, G.; De Simone, A.; Bai, J.; Hannah, M.A.; Foyer, C.H. Low glutathione regulates gene expression and the redox potentials of the nucleus and cytosol in Arabidopsis thaliana. Plant Cell Environ. 2015, 38, 266–279. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Noctor, G. Redox signaling in plants. Antioxid. Redox Signal. 2013, 18, 2087–2090. [Google Scholar] [CrossRef] [PubMed]
- Gullner, G.; Zechmann, B.; Künstler, A.; Király, L. The signaling roles of glutathione in plant disease resistance. In Glutathione in Plant Growth, Development, and Stress Tolerance; Springer: Cham, Switzerland, 2017; pp. 331–357. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Nahar, K.; Anee, T.I.; Fujita, M. Glutathione in plants: Biosynthesis and physiological role in environmental stress tolerance. Physiol. Mol. Biol. Plants 2017, 23, 249–268. [Google Scholar] [CrossRef]
- Locato, V.; Cimini, S.; De Gara, L. Glutathione as a key player in plant abiotic stress responses and tolerance. In Glutathione in Plant Growth, Development, and Stress Tolerance; Springer: Cham, Switzerland, 2017; pp. 127–145. [Google Scholar] [CrossRef]
- Foyer, C.H.; Kunert, K. The ascorbate-glutathione cycle coming of age. J. Exp. Bot. 2024, 75, 2682–2699. [Google Scholar] [CrossRef]
- Foyer, C.H.; Noctor, G. Ascorbate and glutathione: The heart of the redox hub. Plant Physiol. 2011, 155, 2–18. [Google Scholar] [CrossRef]
- Potters, G.; Horemans, N.; Jansen, M.A.K. The cellular redox state in plant stress biology – a charging concept. Plant Physiol. Biochem. 2010, 48, 292–300. [Google Scholar] [CrossRef]
- Schafer, F.Q.; Buettner, G.R. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic. Biol. Med. 2001, 30, 1191–1212. [Google Scholar] [CrossRef]
- Meyer, A.J.; Brach, T.; Marty, L.; Kreye, S.; Rouhier, N.; Jacquot, J.P.; Hell, R. Redox-sensitive GFP in Arabidopsis thaliana is a quantitative biosensor for the redox potential of the cellular glutathione redox buffer. Plant J. 2007, 52, 973–986. [Google Scholar] [CrossRef]
- Szalai, G.; Kellős, T.; Galiba, G.; Kocsy, G. Glutathione as an Antioxidant and Regulatory Molecule in Plants Under Abiotic Stress Conditions. J Plant Growth Regul. 2009, 28, 66–80. [Google Scholar] [CrossRef]
- Foyer, C.H.; Noctor, G. Stress-triggered redox signalling: what’s in pROSpect? Plant Cell Environ. 2016, 39, 951–964. [Google Scholar] [CrossRef]
- Schwarzländer, M.; Fricker, M.D.; Müller, C.; Marty, L.; Brach, T.; Novak, J.; Sweetlove, L.J.; Hell, R.; Meyer, A.J. Confocal imaging of glutathione redox potential in living plant cells. J. Microsc. 2008, 231, 299–316. [Google Scholar] [CrossRef] [PubMed]
- Jiang, K.; Moe-Lange, J.; Hennet, L.; Feldman, L.J. Salt stress affects the redox status of Arabidopsis root meristems. Front. Plant Sci. 2016, 7, 81. [Google Scholar] [CrossRef]
- Bohle, F.; Klaus, A.; Ingelfinger, J.; Tegethof, H.; Safari, N.; Schwarzländer, M.; Hochholdinger, F.; Hahn, M.; Meyer, A.J.; Acosta, I.F.; Müller-Schüssele, S.J. Contrasting cytosolic glutathione redox dynamics under abiotic and biotic stress in barley as revealed by the biosensor Grx1-roGFP2. J. Exp. Bot. 2024, 75, 2299–2312. [Google Scholar] [CrossRef]
- Horváth, E.; Bela, K.; Holinka, B.; Riyazuddin, R.; Gallé, Á.; Hajnal, Á.; Hurton, Á.; Fehér, A.; Csiszár, J. The Arabidopsis glutathione transferases, ATGSTF8 and ATGSTU19 are involved in the maintenance of root redox homeostasis affecting meristem size and salt stress sensitivity. Plant Science 2019, 283, 366–374. [Google Scholar] [CrossRef] [PubMed]
- Ugalde, J.M.; Lamig, L.; Herrera-Vásquez, A.; Fuchs, P.; Homagk, M.; Kopriva, S.; Müller-Schüssele, S.J.; Holuigue, L.; Meyer, A.J. A dual role for glutathione transferase U7 in plant growth and protection from methyl viologen-induced oxidative stress. Plant Physiol. 2021, 187, 2451–2468. [Google Scholar] [CrossRef]
- Schwarzländer, M.; Fricker, M.D.; Sweetlove, L.J. Monitoring the in vivo redox state of plant mitochondria: Effect of respiratory inhibitors, abiotic stress and assessment of recovery from oxidative challenge. Biochim. Biophys. Acta 2009, 1787, 468–475. [Google Scholar] [CrossRef]
- Meyer, A.J.; Dick, T.P. Fluorescent protein-based redox probes. Antioxid. Redox Signal. 2010, 13, 621–650. [Google Scholar] [CrossRef]
- Dooley, C.T.; Dore, T.M.; Hanson, G.T.; Jackson, W.C.; Remington, S.J.; Tsien, R.Y. Imaging dynamic redox changes in mammalian cells with green fluorescent protein indicators. J. Biol. Chem. 2004, 279, 22284–22293. [Google Scholar] [CrossRef]
- Passaia G, Margis-Pinheiro M Glutathione peroxidases as redox sensor proteins in plant cells. Plant Sci. 2015, 234, 22–26. [CrossRef]
- Meyer, A.J.; Dreyer, A.; Ugalde, J.M.; Feitosa-Araujo, E.; Dietz, K.-J.; Schwarzländer, M. Shifting paradigms and novel players in Cys-based redox regulation and ROS signaling in plants - and where to go next. Biol. Chem. 2021, 402, 399–423. [Google Scholar] [CrossRef] [PubMed]
- Rattanawong, K.; Koiso, N.; Toda, E.; Kinoshita, A.; Tanaka, M.; Tsuji, H.; Okamoto, T. Regulatory functions of ROS dynamics via glutathione metabolism and glutathione peroxidase activity in developing rice zygote. Plant J. 2021, 108, 1097–1115. [Google Scholar] [CrossRef] [PubMed]
- Bela, K.; Riyazuddin, R.; Horváth, E.; Hurton, Á.; Gallé, Á.; Takács, Z.; Zsigmond, L.; Szabados, L.; Tari, I.; Csiszár, J. Comprehensive analysis of antioxidant mechanisms in Arabidopsis glutathione peroxidase-like mutants under salt-and osmotic stress reveals organ-specific significance of the AtGPXL’s activities. Environ. Exp. Bot. 2018, 150, 127–140. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Gutscher, M.; Pauleau, A.L.; Marty, L.; Brach, T.; Wabnitz, G.H.; Samstag, Y.; Meyer, A.J.; Dick, T.P. Real-time imaging of the intracellular glutathione redox potential, Nat. Methods 2008, 6, 553–559. [Google Scholar] [CrossRef]
- Horváth, E.; Bela, K.; Holinka, B.; Riyazuddin, R.; Gallé, Á.; Hajnal, Á.; Hurton, Á.; Fehér, A.; Csiszár, J. The Arabidopsis glutathione transferases, AtGSTF8 and AtGSTU19 are involved in the maintenance of root redox homeostasis affecting meristem size and salt stress sensitivity. Plant Sci. 2019, 283, 366–374. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Horváth, E.; Bela, K.; Papdi, C.; Gallé, Á.; Szabados, L.; Tari, I.; Csiszár, J. The role of Arabidopsis glutathione transferase F9 gene under oxidative stress in seedlings. Acta Biol. Hung. 2015, 66, 406–418. [Google Scholar] [CrossRef]
- Chen, S.; Vaghchhipawala, Z.; Li, W.; Asard, H.; Dickman, M.B. Tomato phospholipid hydroperoxide glutathione peroxidase inhibits cell death induced by Bax and oxidative stresses in yeast and plants. Plant Physiol. 2004, 135, 1630–1641. [Google Scholar] [CrossRef]
- Bela, K.; Bangash, S.A.K.; Csiszár, J. Plant Glutathione Peroxidases: Antioxidant enzymes in plant stress responses and tolerance. In Glutathione in Plant Growth, Development, and Stress Tolerance; Springer: Cham, Switzerland, 2017; pp. 113–126. [Google Scholar]
- Horváth, E.; Bela, K.; Gallé, Á.; Riyazuddin, R.; Csomor, G.; Csenki, D.; Csiszár, J. Compensation of mutation in Arabidopsis glutathione transferase (AtGSTU) genes under control or salt stress conditions. Int. J, Mol. Sci. 2020, 21, 2349. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez Milla, M.A.; Maurer, A.; Rodriguez Huete, A.; Gustafson, J.P. Glutathione peroxidase genes in Arabidopsis are ubiquitous and regulated by abiotic stresses through diverse signaling pathways. Plant J. 2003, 36, 602–615. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.C.; Slesak, I.; Jordá, L.; Sotnikov, A.; Melzer, M.; Miszalski, Z.; Mullineaux, P.M.; Parker, J.E.; Karpinska, B.; Karpinski, S. Arabidopsis chloroplastic glutathione peroxidases play a role in cross talk between photooxidative stress and immune responses. Plant Physiol. 2009, 150, 670–683. [Google Scholar] [CrossRef]
- Tsukagoshi, H. Control of root growth and development by reactive oxygen species. Curr. Opin. Plant Biol. 2016, 29, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.; Singh, M.; Singh, A.; Prasad, S.M. Generation mechanisms of reactive oxygen species in the plant cell: An Overview. In Revisiting the role of reactive oxygen species (ROS) in plants: ROS Boon or Bane for Plants; Wiley Publishing: Hoboken, New Jersey, USA, 2017; pp. 1–22. [Google Scholar] [CrossRef]
- Szőllősi, R. Superoxide dismutase (SOD) and abiotic stress tolerance in plants: An overview. In Oxidative Damage to Plants; Academic Press: Cambridge, Massachusetts, USA, 2014; pp. 89–129. [Google Scholar] [CrossRef]
- Xu, X.M.; Møller, S.G. ROS removal by DJ-1: Arabidopsis as a new model to understand Parkinson’s disease. Plant Signal. Behav. 2010, 5, 1034–1036. [Google Scholar] [CrossRef]
- Noctor, G.; Reichheld, J.P.; Foyer, C.H. ROS-related redox regulation and signaling in plants. Semin. Cell Biol. 2018, 80, 3–12. [Google Scholar] [CrossRef]
- He, H.; Van Breusegem, F.; Mhamdi, A. Redox-dependent control of nuclear transcription in plants. J. Exp. Bot. 2018, 69, 3359–3372. [Google Scholar] [CrossRef]
- Bashandy, T.; Guilleminot, J.; Vernoux, T.; Caparros-Ruiz, D.; Ljung, K.; Meyer, Y.; Reichheld, J.P. Interplay between the NADP-linked thioredoxin and glutathione systems in Arabidopsis auxin signaling. Plant Cell 2010, 22, 376–391. [Google Scholar] [CrossRef]
- De Gara, L.; Locato, V.; Dipierro, S.; de Pinto, M.C. Redox homeostasis in plants. The challenge of living with endogenous oxygen production. Respir. Physiol. Neurobiol. 2010, 173, S13–S19. [Google Scholar] [CrossRef]
- Lu, J.; Holmgren, A. The thioredoxin antioxidant system. Free Rad. Biol. Med. 2014, 66, 75–87. [Google Scholar] [CrossRef]
- Martins, L.; Trujillo-Hernandez, J.A.; Reichheld, J.P. Thiol Based Redox Signaling in Plant Nucleus. Front. Plant Sci. 2018, 9, 705. [Google Scholar] [CrossRef]
- Vernoux, T.; Wilson, R.C.; Seeley, K.A.; Reichheld, J.P.; Muroy, S.; Brown, S.; Maughan, S.C.; Cobbett, C.S.; Van Montagu, M.; Inzé, D.; May, M.J. The ROOT MERISTEMLESS1/CADMIUM SENSITIVE2 gene defines a glutathione-dependent pathway involved in initiation and maintenance of cell division during postembryonic root development. Plant Cell 2000, 12, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Diaz Vivancos, P.; Wolff, T.; Markovic, J.; Pallardó, F.V.; Foyer, C.H. A nuclear glutathione cycle within the cell cycle. Biochem. J. 2010, 431, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Dietz, K.J. Redox regulation of transcription factors in plant stress acclimation and development. Antiox. Redox. Signal. 2014, 21, 1356–1372. [Google Scholar] [CrossRef]
- Diaz-Vivancos, P.; de Simone, A.; Kiddle, G.; Foyer, C.H. Glutathione – linking cell proliferation to oxidative stress. Free Rad. Biol. Med. 2015, 89, 1154–1164. [Google Scholar] [CrossRef]
- De Tullio, M.C.; Jiang, K.; Feldman, L.J. Redox regulation of root apical meristem organization: Connecting root development to its environment. Plant Physiol. Biochem. 2010, 48, 328–336. [Google Scholar] [CrossRef]
- Zolla, G.; Heimer, Y.M.; Barak, S. Mild salinity stimulates a stress-induced morphogenic response in Arabidopsis thaliana roots. J. Exp. Bot. 2010, 61, 211–224. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Duan, X.; Luo, L.; Dai, S.; Ding, Z.; Xia, G. How Plant Hormones Mediate Salt Stress Responses. Trends Plant Sci. 2020, 25, 1117–1130. [Google Scholar] [CrossRef]
- Balasubramaniam, T.; Shen, G.; Esmaeili, N.; Zhang, H. Plants’ Response Mechanisms to Salinity Stress. Plants 2023, 12, 2253. [Google Scholar] [CrossRef]
- Chen, C.; Miller, G. ASCORBATE PEROXIDASE 6 protects Arabidopsis desiccating and germinating seeds from stress and mediates cross talk between reactive oxygen species, abscisic acid, and auxin. Plant Physiol. 2014, 166, 370–383. [Google Scholar] [CrossRef]
- Fu, Y.; Yang, Y.; Chen, S.; Ning, N.; Hu, H. Arabidopsis IAR4 modulates primary root growth under salt stress through ROS-mediated modulation of auxin distribution. Front. Plant Sci. 2019, 10, 522. [Google Scholar] [CrossRef] [PubMed]
- Correaaragunde, N.; Foresi, N.; Delledonne, M.; Lamattina, L. Auxin induces redox regulation of ascorbate peroxidase 1 activity by S-nitrosylation/denitrosylation balance resulting in changes of root growth pattern in Arabidopsis. J. Exp. Bot. 2013, 64, 3339–3349. [Google Scholar] [CrossRef] [PubMed]
- Iglesias, M.J.; Terrile, M.C.; Bartoli, C.G.; D’Ippólito, S.; Casalongué, C.A. Auxin signaling participates in the adaptative response against oxidative stress and salinity by interacting with redox metabolism in Arabidopsis. Plant Mol. Biol. 2010, 74, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Riyazuddin, R.; Bela, K.; Poór, P.; Szepesi, Á.; Horváth, E.; Rigó, G.; Szabados, L.; Fehér, A.; Csiszár, J. Crosstalk between the Arabidopsis glutathione peroxidase-like 5 isoenzyme (AtGPXL5) and ethylene. Int. J. Mol. Sci. 2022, 23, 5749. [Google Scholar] [CrossRef]
| Genotype | Cotyledon | Root |
|---|---|---|
| Col-0 | -302.27±4.66d | -302.48±5.88c |
| gpxl1 | -274.25±11.92a | -269.30±1.02a |
| gpxl2 | -294.59±2.76c | -289.84±4.01b |
| gpxl3 | -285.03±3.06b | -296.69±1.76c |
| gpxl4 | -298.93±6.89cd | -271.86±9.25a |
| gpxl5 | -298.46±4.96cd | -298.77±4.30c |
| gpxl6 | -287.90±6.88b | -278.71±8.39a |
| gpxl7 | -277.97±3.36a | -300.50±0.89c |
| gpxl8 | -284.01±8.28ab | -281.32±6.43ab |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





