Submitted:
17 February 2025
Posted:
18 February 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Generation of an Updated miRNA Annotation for the SL4.0 Tomato Genome Assembly
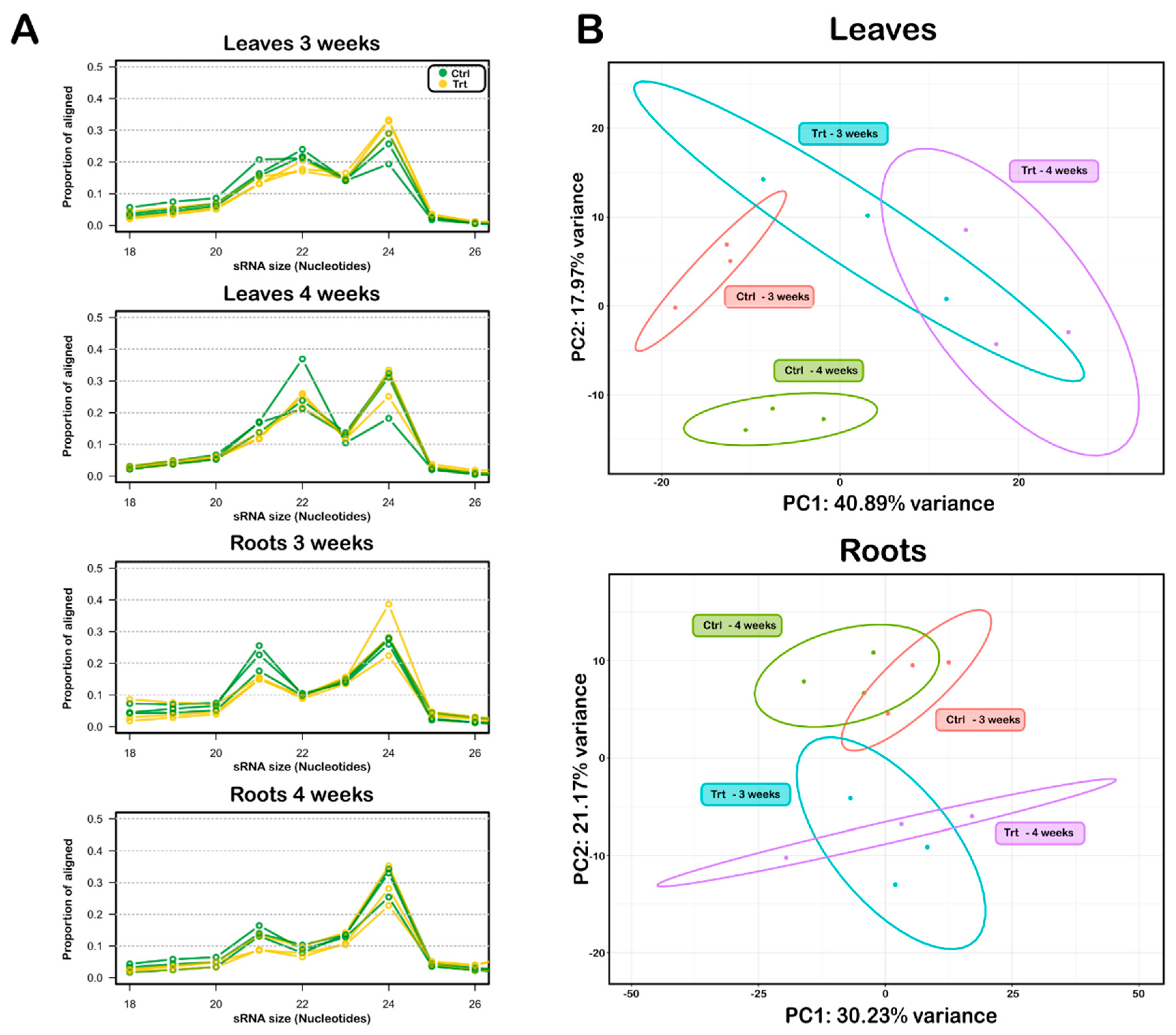
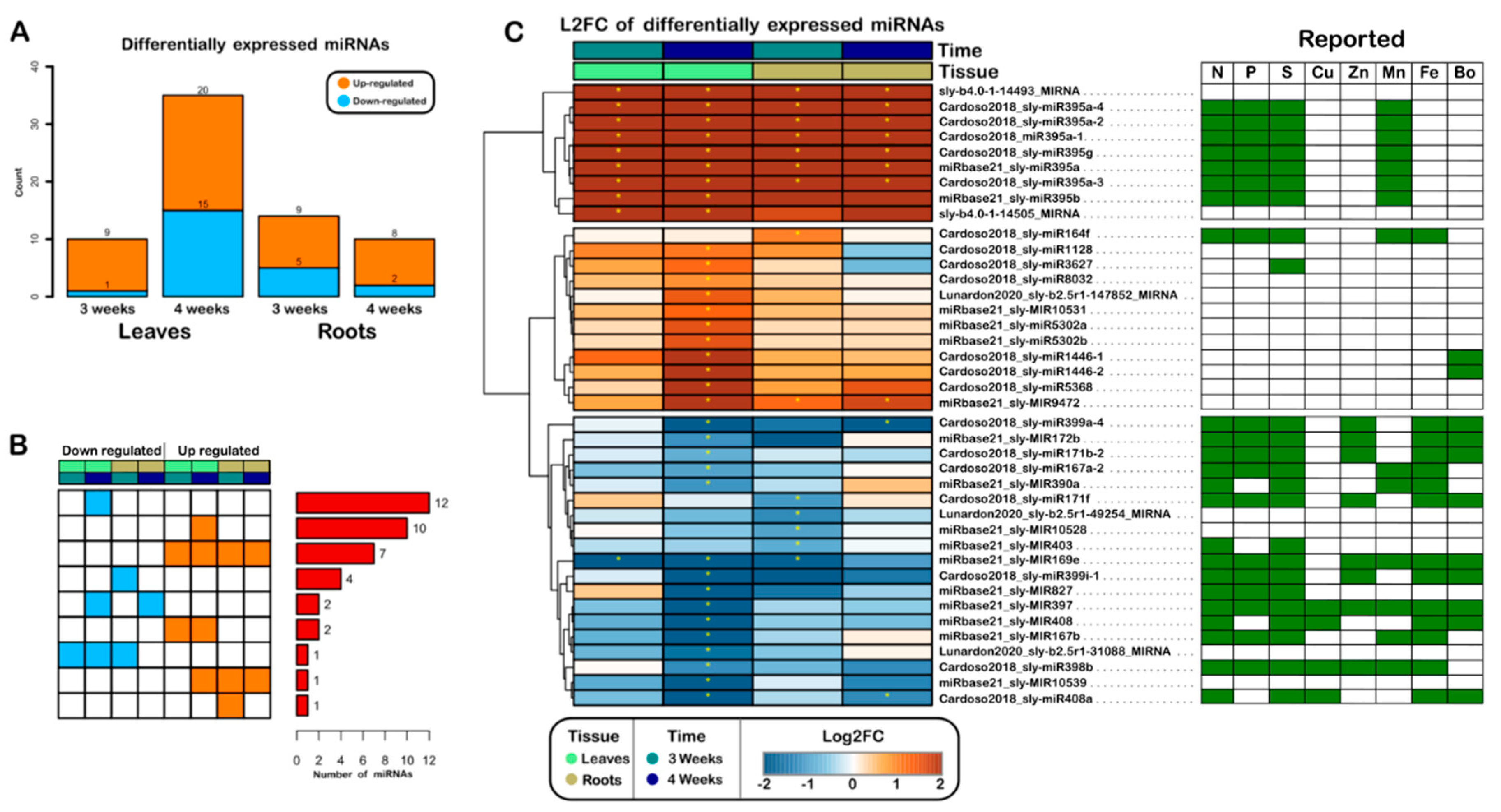
2.2. miRNAs Are Regulated in a Tissue- and Time-Specific Manner in the Post-Transcriptional Response of Tomato to S Deficiency
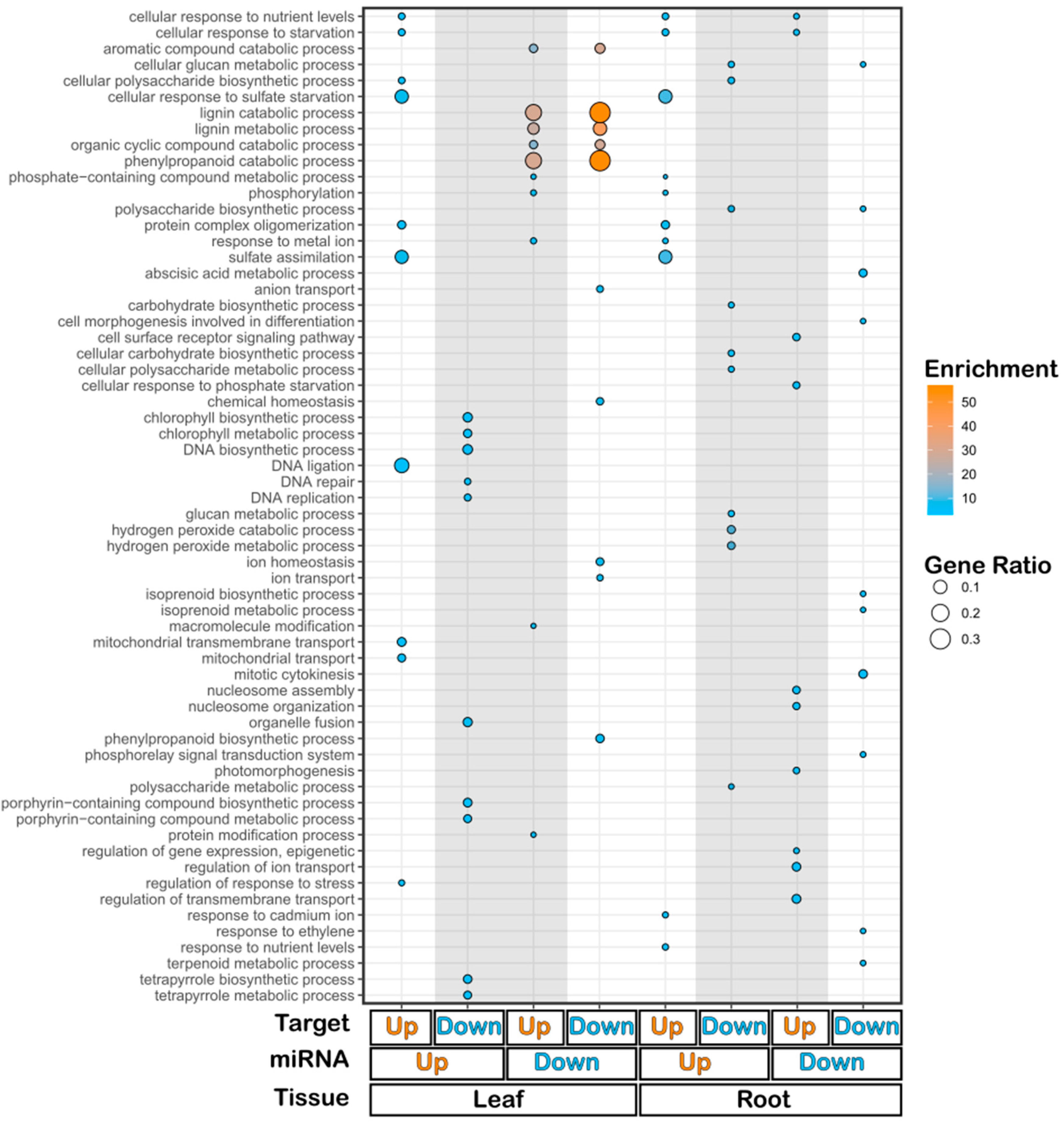
2.3. Target Prediction of Differentially Expressed miRNAs Identifies Novel Pathways Involved in Tomato's Response to Sulfate Deficiency
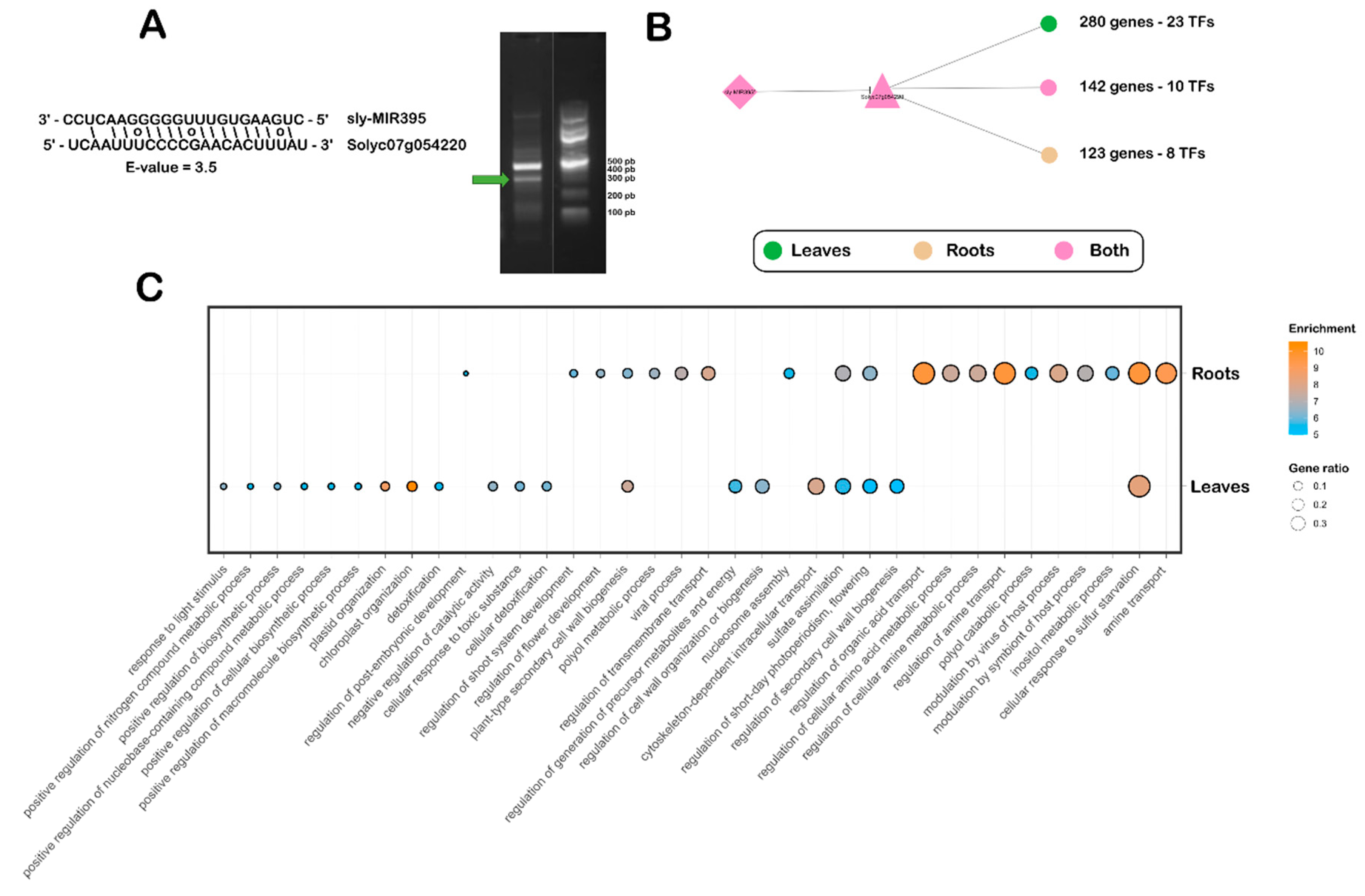
2.4. Ethylene-Responsive Transcription Factor SlERF2a as a Novel Target of miRNA395 in the Tomato Sulfate Deficiency Response
3. Discussion
3.1. A Comprehensive miRNA Annotation for Tomato in the SL4.0 Genome
3.2. Spatiotemporal miRNA Regulation Reveals Specific Post-Transcriptional Control of Sulfur Deficiency Adaptation in Tomato
3.3. Prediction of Differentially Expressed miRNA Targets Uncovers Novel Regulatory Pathways in Tomato Sulfate Deficiency Response
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. sRNA Library Preparation and Sequencing
4.3. sRNA-Seq Read Alignment and Differential Expression Analysis
4.4. miRNA Annotation in the SL4.0 Genome Assembly
4.5. RNA-Seq Read Alignment and Differential Expression Analysis
4.6. Identification of mRNA Targets of miRNAs
4.7. Gene Ontology Analysis
4.8. Real-Time qPCR Analysis
4.9. Stem-Loop qPCR Analysis
4.10. Target Validation by RNA Ligase-Mediated Rapid Amplification of cDNA Ends (RLM-RACE)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| S | Sulfur |
| SULTR | Sulfate transporter |
| ATPS | ATP sulfurylase |
| OAS | O-acetyl serine |
| SLIM1 | Sulfur limitation 1 |
| TF | Transcription factor |
| sRNA | Small RNA |
| miRNA | Micro RNA |
| AGO | Argonaute |
| RISC | RNA-induced silencing complex |
| MS | Murashige and Skoog |
| GO | Gene ontology |
| DE | Differentially expressed |
| DEGs | Differentially expressed genes |
| ERF | Ethylene-responsive factor |
| SAM | S-adenosyl methionine |
References
- Maruyama-Nakashita A. Metabolic changes sustain the plant life in low-sulfur environments. Curr Opin Plant Biol. 2017 Oct 1;39:144–51. [CrossRef]
- 2Kopriva S, Malagoli M, Takahashi H. Sulfur nutrition: impacts on plant development, metabolism, and stress responses. J Exp Bot. 2019 Aug 19;70(16):4069–73. [CrossRef]
- Takahashi H. Sulfate transport systems in plants: functional diversity and molecular mechanisms underlying regulatory coordination. J Exp Bot. 2019 Aug 19;70(16):4075–87. [CrossRef]
- Kataoka T, Hayashi N, Yamaya T, Takahashi H. Root-to-shoot transport of sulfate in Arabidopsis. Evidence for the role of SULTR3;5 as a component of low-affinity sulfate transport system in the root vasculature. Plant Physiol. 2004 Dec;136(4):4198–204. [CrossRef]
- Saito K. Sulfur Assimilatory Metabolism. The Long and Smelling Road. Plant Physiol. 2004 Sep;136(1):2443–50.
- Nakai Y, Maruyama-Nakashita A. Biosynthesis of Sulfur-Containing Small Biomolecules in Plants. Int J Mol Sci. 2020 May 14;21(10):3470. [CrossRef]
- Aula L, Dhillon JS, Omara P, Wehmeyer GB, Freeman KW, Raun WR. World Sulfur Use Efficiency for Cereal Crops. Agron J. 2019;111(5):2485–92. [CrossRef]
- Hawkesford MJ. Plant responses to sulphur deficiency and the genetic manipulation of sulphate transporters to improve S-utilization efficiency. J Exp Bot. 2000 Jan 1;51(342):131–8.
- McNeill A m., Eriksen J, Bergström L, Smith K a., Marstorp H, Kirchmann H, et al. Nitrogen and sulphur management: challenges for organic sources in temperate agricultural systems. Soil Use Manag. 2005;21(1):82–93.
- Zhang L, Kawaguchi R, Morikawa-Ichinose T, Allahham A, Kim SJ, Maruyama-Nakashita A. Sulfur Deficiency-Induced Glucosinolate Catabolism Attributed to Two β-Glucosidases, BGLU28 and BGLU30, is Required for Plant Growth Maintenance under Sulfur Deficiency. Plant Cell Physiol. 2020 Apr 1;61(4):803–13.
- Zuchi S, Cesco S, Varanini Z, Pinton R, Astolfi S. Sulphur deprivation limits Fe-deficiency responses in tomato plants. Planta. 2009 Jun 1;230(1):85–94. [CrossRef]
- Yu X, Gong H, Cao L, Hou Y, Qu S. MicroRNA397b negatively regulates resistance of Malus hupehensis to Botryosphaeria dothidea by modulating MhLAC7 involved in lignin biosynthesis. Plant Sci. 2020 Mar 1;292:110390. [CrossRef]
- Zörb C, Steinfurth D, Seling S, LangenkÄmper G, Koehler P, Wieser H, et al. Quantitative Protein Composition and Baking Quality of Winter Wheat as Affected by Late Sulfur Fertilization. J Agric Food Chem. 2009 May 13;57(9):3877–85. [CrossRef]
- Bielecka M, Watanabe M, Morcuende R, Scheible WR, Hawkesford MJ, Hesse H, et al. Transcriptome and metabolome analysis of plant sulfate starvation and resupply provides novel information on transcriptional regulation of metabolism associated with sulfur, nitrogen and phosphorus nutritional responses in Arabidopsis. Front Plant Sci. 2015 Jan 28;5:805. [CrossRef]
- Henríquez-Valencia C, Arenas-M A, Medina J, Canales J. Integrative Transcriptomic Analysis Uncovers Novel Gene Modules That Underlie the Sulfate Response in Arabidopsis thaliana. Front Plant Sci [Internet]. 2018 Apr 10 [cited 2024 Jul 10];9. Available from: https://www.frontiersin.org/journals/plant-science/articles/10.3389/fpls.2018.00470/full.
- Maruyama-Nakashita A, Inoue E, Watanabe-Takahashi A, Yamaya T, Takahashi H. Transcriptome Profiling of Sulfur-Responsive Genes in Arabidopsis Reveals Global Effects of Sulfur Nutrition on Multiple Metabolic Pathways. Plant Physiol. 2003 Jun;132(2):597–605. [CrossRef]
- Maruyama-Nakashita A, Nakamura Y, Tohge T, Saito K, Takahashi H. Arabidopsis SLIM1 Is a Central Transcriptional Regulator of Plant Sulfur Response and Metabolism. Plant Cell. 2006 Nov;18(11):3235–51. [CrossRef]
- Iyer-Pascuzzi AS, Jackson T, Cui H, Petricka JJ, Busch W, Tsukagoshi H, et al. Cell identity regulators link development and stress responses in the Arabidopsis root. Dev Cell. 2011 Oct 18;21(4):770–82. [CrossRef]
- Fernández JD, Miño I, Canales J, Vidal EA. Gene regulatory networks underlying sulfate deficiency responses in plants. J Exp Bot. 2024 May 20;75(10):2781–98. [CrossRef]
- Zhang B, Pan X, Cobb GP, Anderson TA. Plant microRNA: A small regulatory molecule with big impact. Dev Biol. 2006 Jan 1;289(1):3–16. [CrossRef]
- Axtell MJ. Classification and Comparison of Small RNAs from Plants. Annu Rev Plant Biol. 2013 Apr 29;64(Volume 64, 2013):137–59.
- Song X, Li Y, Cao X, Qi Y. MicroRNAs and Their Regulatory Roles in Plant–Environment Interactions. Annu Rev Plant Biol. 2019 Apr 29;70(1):489–525. [CrossRef]
- Liang G, Yang F, Yu D. MicroRNA395 mediates regulation of sulfate accumulation and allocation in Arabidopsis thaliana. Plant J. 2010;62(6):1046–57. [CrossRef]
- Liu HH, Tian X, Li YJ, Wu CA, Zheng CC. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA. 2008 May;14(5):836–43. [CrossRef]
- Achkar NP, Cambiagno DA, Manavella PA. miRNA Biogenesis: A Dynamic Pathway. Trends Plant Sci. 2016 Dec 1;21(12):1034–44.
- Voinnet O. Origin, Biogenesis, and Activity of Plant MicroRNAs. Cell. 2009 Feb 20;136(4):669–87.
- Kurihara Y, Watanabe Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc Natl Acad Sci U S A. 2004 Aug 24;101(34):12753–8. [CrossRef]
- Denli AM, Tops BBJ, Plasterk RHA, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004 Nov;432(7014):231–5. [CrossRef]
- Okada C, Yamashita E, Lee SJ, Shibata S, Katahira J, Nakagawa A, et al. A High-Resolution Structure of the Pre-microRNA Nuclear Export Machinery. Science. 2009 Nov 27;326(5957):1275–9.
- Iwakawa H oki, Tomari Y. Molecular insights into microRNA-mediated translational repression in plants. Mol Cell. 2013 Nov 21;52(4):591–601.
- Yang G, Li Y, Wu B, Zhang K, Gao L, Zheng C. MicroRNAs transcriptionally regulate promoter activity in Arabidopsis thaliana. J Integr Plant Biol. 2019;61(11):1128–33. [CrossRef]
- Dolata J, Bajczyk M, Bielewicz D, Niedojadlo K, Niedojadlo J, Pietrykowska H, et al. Salt Stress Reveals a New Role for ARGONAUTE1 in miRNA Biogenesis at the Transcriptional and Posttranscriptional Levels1[OPEN]. Plant Physiol. 2016 Sep;172(1):297–312.
- Baumberger N, Baulcombe DC. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci. 2005 Aug 16;102(33):11928–33.
- Li L, Yi H, Xue M, Yi M. miR398 and miR395 are involved in response to SO2 stress in Arabidopsis thaliana. Ecotoxicol Lond Engl. 2017 Nov;26(9):1181–7. [CrossRef]
- Huang SQ, Xiang AL, Che LL, Chen S, Li H, Song JB, et al. A set of miRNAs from Brassica napus in response to sulphate deficiency and cadmium stress. Plant Biotechnol J. 2010;8(8):887–99. [CrossRef]
- Iqrar S, Ashrafi K, Khan S, Saifi M, Nasrullah N, Abdin MZ. Set of miRNAs Involved in Sulfur Uptake and the Assimilation Pathway of Indian Mustard (B. juncea) in Response to Sulfur Treatments. ACS Omega. 2022 Apr 19;7(15):13228–42.
- Shu L, Hu Z. Characterization and differential expression of microRNAs elicited by sulfur deprivation in Chlamydomonas reinhardtii. BMC Genomics. 2012 Mar 22;13(1):108. [CrossRef]
- Liang G, Li Y, He H, Wang F, Yu D. Identification of miRNAs and miRNA-mediated regulatory pathways in Carica papaya. Planta. 2013 Oct 1;238(4):739–52. [CrossRef]
- Liang G, Ai Q, Yu D. Uncovering miRNAs involved in crosstalk between nutrient deficiencies in Arabidopsis. Sci Rep. 2015 Jul 2;5(1):11813.
- Kawashima CG, Matthewman CA, Huang S, Lee BR, Yoshimoto N, Koprivova A, et al. Interplay of SLIM1 and miR395 in the regulation of sulfate assimilation in Arabidopsis. Plant J. 2011;66(5):863–76. [CrossRef]
- Shahzad R, Harlina P, Ayaad M, Nishawy E, Fahad S, Subthain H, et al. Dynamic roles of microRNAs in nutrient acquisition and plant adaptation under nutrient stress: A review. Plant OMICS. 2018 Feb 1;11:58–79. [CrossRef]
- Barciszewska-Pacak M, Milanowska K, Knop K, Bielewicz D, Nuc P, Plewka P, et al. Arabidopsis microRNA expression regulation in a wide range of abiotic stress responses. Front Plant Sci. 2015;6:410. [CrossRef]
- Our World in Data [Internet]. [cited 2025 Feb 16]. Tomato production. Available from: https://ourworldindata.org/grapher/tomato-production.
- Kimura S, Sinha N. Tomato (Solanum lycopersicum): A Model Fruit-Bearing Crop. Cold Spring Harb Protoc. 2008 Nov 1;2008(11):pdb.emo105. [CrossRef]
- Gascuel Q, Diretto G, Monforte AJ, Fortes AM, Granell A. Use of Natural Diversity and Biotechnology to Increase the Quality and Nutritional Content of Tomato and Grape. Front Plant Sci [Internet]. 2017 May 12 [cited 2024 Jul 10];8. Available from: https://www.frontiersin.org/journals/plant-science/articles/10.3389/fpls.2017.00652/full.
- Cerdá A, Martínez V, Caro M, Fernández FG. Effect of sulfur deficiency and excess on yield and sulfur accumulation in tomato plants. J Plant Nutr. 1984 Nov 1;7(11):1529–43.
- Siddiqui MH, Alamri S, Alsubaie QD, Ali HM, Khan MN, Al-Ghamdi A, et al. Exogenous nitric oxide alleviates sulfur deficiency-induced oxidative damage in tomato seedlings. Nitric Oxide. 2020 Jan 1;94:95–107.
- Hasan MK, Liu CX, Pan YT, Ahammed GJ, Qi ZY, Zhou J. Melatonin alleviates low-sulfur stress by promoting sulfur homeostasis in tomato plants. Sci Rep. 2018 Jul 5;8(1):10182. [CrossRef]
- Lopez J, Tremblay N, Voogt W, Dubé S, Gosselin A. Effects of varying sulphate concentrations on growth, physiology and yield of the greenhouse tomato. Sci Hortic. 1996 Dec 1;67(3):207–17. [CrossRef]
- Canales J, Uribe F, Henríquez-Valencia C, Lovazzano C, Medina J, Vidal EA. Transcriptomic analysis at organ and time scale reveals gene regulatory networks controlling the sulfate starvation response of Solanum lycopersicum. BMC Plant Biol. 2020 Aug 24;20(1):385. [CrossRef]
- Moxon S, Jing R, Szittya G, Schwach F, Rusholme Pilcher RL, Moulton V, et al. Deep sequencing of tomato short RNAs identifies microRNAs targeting genes involved in fruit ripening. Genome Res. 2008 Oct;18(10):1602–9. [CrossRef]
- Zhao G, Yu H, Liu M, Lu Y, Ouyang B. Identification of salt-stress responsive microRNAs from Solanum lycopersicum and Solanum pimpinellifolium. Plant Growth Regul. 2017 Sep 1;83(1):129–40. [CrossRef]
- Jin W, Wu F. Characterization of miRNAs associated with Botrytis cinerea infection of tomato leaves. BMC Plant Biol. 2015 Jan 16;15(1):1.
- Yogindran S, Rajam MV. Host-derived artificial miRNA-mediated silencing of ecdysone receptor gene provides enhanced resistance to Helicoverpa armigera in tomato. Genomics. 2021 Jan 1;113(1, Part 2):736–47.
- Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019 Jan 8;47(D1):D155–62.
- Lunardon A, Johnson NR, Hagerott E, Phifer T, Polydore S, Coruh C, et al. Integrated annotations and analyses of small RNA–producing loci from 47 diverse plants. Genome Res. 2020 Mar;30(3):497–513. [CrossRef]
- Baldrich P, Bélanger S, Kong S, Pokhrel S, Tamim S, Teng C, et al. The evolutionary history of small RNAs in Solanaceae. [cited 2025 Jan 15]; Available from: https://dx.doi.org/10.1093/plphys/kiac089.
- Liu J, Liu X, Zhang S, Liang S, Luan W, Ma X. TarDB: an online database for plant miRNA targets and miRNA-triggered phased siRNAs. BMC Genomics. 2021 May 13;22(1):348. [CrossRef]
- Chang L, Zhou G, Soufan O, Xia J. miRNet 2.0: network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res. 2020 Jul 2;48(W1):W244–51.
- Hosmani PS, Flores-Gonzalez M, van de Geest H, Maumus F, Bakker LV, Schijlen E, et al. An improved de novo assembly and annotation of the tomato reference genome using single-molecule sequencing, Hi-C proximity ligation and optical maps. bioRxiv. 2019 Jan 1;767764.
- Murashige T, Skoog F. A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol Plant. 1962;15(3):473–97. [CrossRef]
- Johnson NR, Yeoh JM, Coruh C, Axtell MJ. Improved Placement of Multi-mapping Small RNAs. G3 GenesGenomesGenetics. 2016 Jul 1;6(7):2103–11. [CrossRef]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014 Dec 5;15(12):550.
- Cardoso TC de S, Alves TC, Caneschi CM, Santana D dos RG, Fernandes-Brum CN, Reis GLD, et al. New insights into tomato microRNAs. Sci Rep. 2018 Oct 30;8(1):16069.
- McGinnis S, Madden TL. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004 Jul 1;32(Web Server issue):W20–5.
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019 Aug;37(8):907–15. [CrossRef]
- Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014 Apr 1;30(7):923–30. [CrossRef]
- Dai X, Zhuang Z, Zhao PX. psRNATarget: a plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018 Jul 2;46(W1):W49–54.
- Bioconductor [Internet]. [cited 2025 Feb 16]. topGO. Available from: http://bioconductor.org/packages/topGO/.
- Tian T, Liu Y, Yan H, You Q, Yi X, Du Z, et al. agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017 Jul 3;45(W1):W122–9. [CrossRef]
- Zhao S, Fernald RD. Comprehensive Algorithm for Quantitative Real-Time Polymerase Chain Reaction. J Comput Biol J Comput Mol Cell Biol. 2005 Oct;12(8):1047–64. [CrossRef]
- Su X, Wang B, Geng X, Du Y, Yang Q, Liang B, et al. A high-continuity and annotated tomato reference genome. BMC Genomics. 2021 Dec 15;22(1):898. [CrossRef]
- Fernandez-Pozo N, Menda N, Edwards JD, Saha S, Tecle IY, Strickler SR, et al. The Sol Genomics Network (SGN)—from genotype to phenotype to breeding. Nucleic Acids Res. 2015 Jan 28;43(Database issue):D1036–41. [CrossRef]
- Bizouerne E, Ly Vu B, Ly Vu J, Verdier J, Buitink J, Leprince O. Dataset for transcriptome and physiological response of mature tomato seed tissues to light and heat during fruit ripening. Data Brief. 2021 Feb 1;34:106671. [CrossRef]
- Wang Q, Xu X, Cao X, Hu T, Xia D, Zhu J, et al. Identification, Classification, and Expression Analysis of the Triacylglycerol Lipase (TGL) Gene Family Related to Abiotic Stresses in Tomato. Int J Mol Sci. 2021 Jan;22(3):1387.
- Wang J, Feng Y, Ding X, Huo J, Nie WF. Identification of Long Non-Coding RNAs Associated with Tomato Fruit Expansion and Ripening by Strand-Specific Paired-End RNA Sequencing. Horticulturae. 2021 Dec;7(12):522. [CrossRef]
- Axtell MJ. ShortStack: Comprehensive annotation and quantification of small RNA genes. RNA. 2013 Jun;19(6):740–51. [CrossRef]
- Shahid S, Axtell MJ. Identification and annotation of small RNA genes using ShortStack. Methods San Diego Calif. 2014 May 1;67(1):20–7. [CrossRef]
- Khatabi B, Arikit S, Xia R, Winter S, Oumar D, Mongomake K, et al. High-resolution identification and abundance profiling of cassava (Manihot esculenta Crantz) microRNAs. BMC Genomics. 2016 Jan 28;17(1):85. [CrossRef]
- Wan LC, Wang F, Guo X, Lu S, Qiu Z, Zhao Y, et al. Identification and characterization of small non-coding RNAs from Chinese fir by high throughput sequencing. BMC Plant Biol. 2012 Aug 15;12(1):146. [CrossRef]
- Won SY, Yumul RE, Chen X. Small RNAs in Plants. In: Howell SH, editor. Molecular Biology [Internet]. New York, NY: Springer; 2014 [cited 2024 Jul 10]. p. 95–127. Available from: . [CrossRef]
- You C, Cui J, Wang H, Qi X, Kuo LY, Ma H, et al. Conservation and divergence of small RNA pathways and microRNAs in land plants. Genome Biol. 2017 Aug 23;18(1):158. [CrossRef]
- Hsieh LC, Lin SI, Shih ACC, Chen JW, Lin WY, Tseng CY, et al. Uncovering Small RNA-Mediated Responses to Phosphate Deficiency in Arabidopsis by Deep Sequencing. Plant Physiol. 2009 Dec 1;151(4):2120–32.
- Dong F, Wang C, Dong Y, Hao S, Wang L, Sun X, et al. Differential expression of microRNAs in tomato leaves treated with different light qualities. BMC Genomics. 2020 Jan 13;21(1):37.
- Wang Z, Hardcastle TJ, Canto Pastor A, Yip WH, Tang S, Baulcombe DC. A novel DCL2-dependent miRNA pathway in tomato affects susceptibility to RNA viruses. Genes Dev. 2018 Sep 1;32(17–18):1155–60. [CrossRef]
- Matthewman CA, Kawashima CG, Húska D, Csorba T, Dalmay T, Kopriva S. miR395 is a general component of the sulfate assimilation regulatory network in Arabidopsis. FEBS Lett. 2012 Sep 21;586(19):3242–8. [CrossRef]
- Apodiakou A, Hoefgen R. New insights into the regulation of plant metabolism by O-acetylserine: sulfate and beyond. J Exp Bot. 2023 Jun 6;74(11):3361–78. [CrossRef]
- Jin J, Tian F, Yang DC, Meng YQ, Kong L, Luo J, et al. PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017 Jan 4;45(D1):D1040–5.
- Zhang Q, Li Y, Zhang Y, Wu C, Wang S, Hao L, et al. Md-miR156ab and Md-miR395 Target WRKY Transcription Factors to Influence Apple Resistance to Leaf Spot Disease. Front Plant Sci. 2017 Apr 19;8:526.
- Kawashima CG, Yoshimoto N, Maruyama-Nakashita A, Tsuchiya YN, Saito K, Takahashi H, et al. Sulphur starvation induces the expression of microRNA-395 and one of its target genes but in different cell types. Plant J. 2009;57(2):313–21. [CrossRef]
- Pokoo R, Ren S, Wang Q, Motes CM, Hernandez TD, Ahmadi S, et al. Genotype- and tissue-specific miRNA profiles and their targets in three alfalfa (Medicago sativa L) genotypes. BMC Genomics. 2018 Dec 31;19(10):913. [CrossRef]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009 Dec 15;10:421.
- Olmo R, Quijada NM, Morán-Diez ME, Hermosa R, Monte E. Identification of Tomato microRNAs in Late Response to Trichoderma atroviride. Int J Mol Sci. 2024 Jan;25(3):1617. [CrossRef]
- Zhou R, Yu X, Ottosen CO, Zhang T, Wu Z, Zhao T. Unique miRNAs and their targets in tomato leaf responding to combined drought and heat stress. BMC Plant Biol. 2020 Mar 6;20(1):107. [CrossRef]
- Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS. The Sequential Action of miR156 and miR172 Regulates Developmental Timing in Arabidopsis. Cell. 2009 Aug 21;138(4):750–9.
- Chiou TJ, Aung K, Lin SI, Wu CC, Chiang SF, Su C lin. Regulation of Phosphate Homeostasis by MicroRNA in Arabidopsis. Plant Cell. 2006 Feb 1;18(2):412–21. [CrossRef]
- Krieg DR. Photosynthetic activity during stress. Agric Water Manag. 1983 Sep 1;7(1):249–63.
- Pedrosa L de F, Fabi JP. Polysaccharides from Medicinal Plants: Bridging Ancestral Knowledge with Contemporary Science. Plants. 2024 Jan;13(13):1721.
- Kaashyap M, Ford R, Kudapa H, Jain M, Edwards D, Varshney R, et al. Differential Regulation of Genes Involved in Root Morphogenesis and Cell Wall Modification is Associated with Salinity Tolerance in Chickpea. Sci Rep. 2018 Mar 19;8(1):4855. [CrossRef]
- Yang Z, Hui S, Lv Y, Zhang M, Chen D, Tian J, et al. miR395-regulated sulfate metabolism exploits pathogen sensitivity to sulfate to boost immunity in rice. Mol Plant. 2022 Apr 4;15(4):671–88. [CrossRef]
- Yuan N, Yuan S, Li Z, Li D, Hu Q, Luo H. Heterologous expression of a rice miR395 gene in Nicotiana tabacum impairs sulfate homeostasis. Sci Rep. 2016 Jun 28;6(1):28791.
- Liu M, Gomes BL, Mila I, Purgatto E, Peres LEP, Frasse P, et al. Comprehensive Profiling of Ethylene Response Factor Expression Identifies Ripening-Associated ERF Genes and Their Link to Key Regulators of Fruit Ripening in Tomato. Plant Physiol. 2016 Mar 1;170(3):1732–44. [CrossRef]
- Xie X lan, Yin X ren, Chen K song. Roles of APETALA2/Ethylene-Response Factors in Regulation of Fruit Quality. Crit Rev Plant Sci. 2016 Mar 3;35(2):120–30.
- Huang Z, Zhang Z, Zhang X, Zhang H, Huang D, Huang R. Tomato TERF1 modulates ethylene response and enhances osmotic stress tolerance by activating expression of downstream genes. FEBS Lett. 2004 Aug 27;573(1):110–6. [CrossRef]
- Zhang G, Chen M, Li L, Xu Z, Chen X, Guo J, et al. Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J Exp Bot. 2009 Sep 1;60(13):3781–96.
- Fukao T, Yeung E, Bailey-Serres J. The Submergence Tolerance Regulator SUB1A Mediates Crosstalk between Submergence and Drought Tolerance in Rice. Plant Cell. 2011 Jan 1;23(1):412–27.
- Gao Y, Fan ZQ, Zhang Q, Li HL, Liu GS, Jing Y, et al. A tomato NAC transcription factor, SlNAM1, positively regulates ethylene biosynthesis and the onset of tomato fruit ripening. Plant J. 2021;108(5):1317–31. [CrossRef]
- Roeder S, Dreschler K, Wirtz M, Cristescu SM, van Harren FJM, Hell R, et al. SAM levels, gene expression of SAM synthetase, methionine synthase and ACC oxidase, and ethylene emission from N. suaveolens flowers. Plant Mol Biol. 2009 Jul 1;70(5):535–46. [CrossRef]
- Moniuszko G, Skoneczny M, Zientara-Rytter K, Wawrzyńska A, Głów D, Cristescu SM, et al. Tobacco LSU-like protein couples sulphur-deficiency response with ethylene signalling pathway. J Exp Bot. 2013 Nov;64(16):5173–82. [CrossRef]
- Lewandowska M, Wawrzyńska A, Moniuszko G, Łukomska J, Zientara K, Piecho M, et al. A Contribution to Identification of Novel Regulators of Plant Response to Sulfur Deficiency: Characteristics of a Tobacco Gene UP9C, Its Protein Product and the Effects of UP9C Silencing. Mol Plant. 2010 Mar;3(2):347–60. [CrossRef]
- Iqbal N, Khan NA, Nazar R, Silva JAT da. Ethylene-stimulated photosynthesis results from increased nitrogen and sulfur assimilation in mustard types that differ in photosynthetic capacity. Environ Exp Bot. 2012 May 1;78:84–90. [CrossRef]
- Liu C, Ma D, Wang Z, Chen N, Ma X, He XQ. MiR395c Regulates Secondary Xylem Development Through Sulfate Metabolism in Poplar. Front Plant Sci. 2022 Jun 9;13:897376. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




