1. Introduction
Oxidative stress plays a crucial role in the pathophysiology of various reproductive disorders in dairy cows, particularly in cases of postpartum uterine infections, such as endometritis. These infections are primarily caused by bacterial contaminants such as
Escherichia coli and
Staphylococcus aureus, which disrupt the normal function of the endometrium and create an unsuitable environment for embryo implantation, ultimately reducing the reproductive performance[
1,
2] The accumulation of reactive oxygen species(ROS) can lead to cellular damage and impaired lipid peroxidation health; therefore, studying the effects of oxidative stress on the health of the dairy cow endometrium is meaningful. [
3].
Acute endometritis in dairy cows may be treated with potassium permanganate (KMnO
4), recognized for its antibacterial and anti-inflammatory properties. However, KMnO
4 is a strong oxidizing agent, and incomplete uterine washing can result in residual potassium permanganate within the body, potentially creating a prolonged oxidative environment that may lead to oxidative damage of the endometrium[
4,
5,
6,
7,
8]. In the context of postpartum infections in dairy cows, KMnO
4 is utilized to cleanse the uterus, disinfect wounds, and treat hoof conditions. Additionally, it is employed in other animal species for wound debridement and the management of uterine adenomyosis in canines and felines. Nonetheless, potassium permanganate's potent oxidative properties raise concerns, as it has been shown to exhibit acute toxicity in rodent models. This study aims to investigate the potential for potassium permanganate to induce oxidative stress in the endometrium during therapeutic application[
9,
10,
11].
IGF1R (Insulin-like Growth Factor 1 Receptor) is essential in oxidative stress [
12]. It belongs to the receptor tyrosine kinase family. It can be activated by IGF1 or IGF2 [
13], activating its downstream PI3K/AKT signaling pathway, which regulates cell growth, differentiation, and responses to oxidative stress[
14]. IGF1R is significantly related to oxidative stress responses, particularly in the context of ischemia/reperfusion injury and mental disorders. Studies have shown that low concentrations of H
2O
2 during myocardial ischemia-reperfusion activate the IGF1R/PI3K/AKT signaling pathway to resist oxidative stress damage and prevent apoptosis.
MicroRNAs (miRNAs) are a class of small non-coding RNAs that regulate gene expression by binding to the 3' untranslated regions of mRNAs, leading to mRNA degradation or inhibition of translation[
15]. The let-7 family is a group of conserved microRNAs that play roles in various biological processes, including cell proliferation, differentiation, and apoptosis. The let-7 family is highly expressed in mitochondria and acts by regulating antioxidant-related genes [
16]. For example, let-7b and let-7c can indirectly regulate HMOX1 to alleviate oxidative damage in human hepatocytes [
17]. Studies have shown that let-7 family member let-7a inhibits oxidative stress and cellular damage by targeting arginase 2 (ARG2)[
18]. The let-7 family also plays a significant role in regulating cancer stem cells, participating in the maintenance of stem cell properties, promoting the proliferation of cancer cells, and resisting apoptosis [
19].
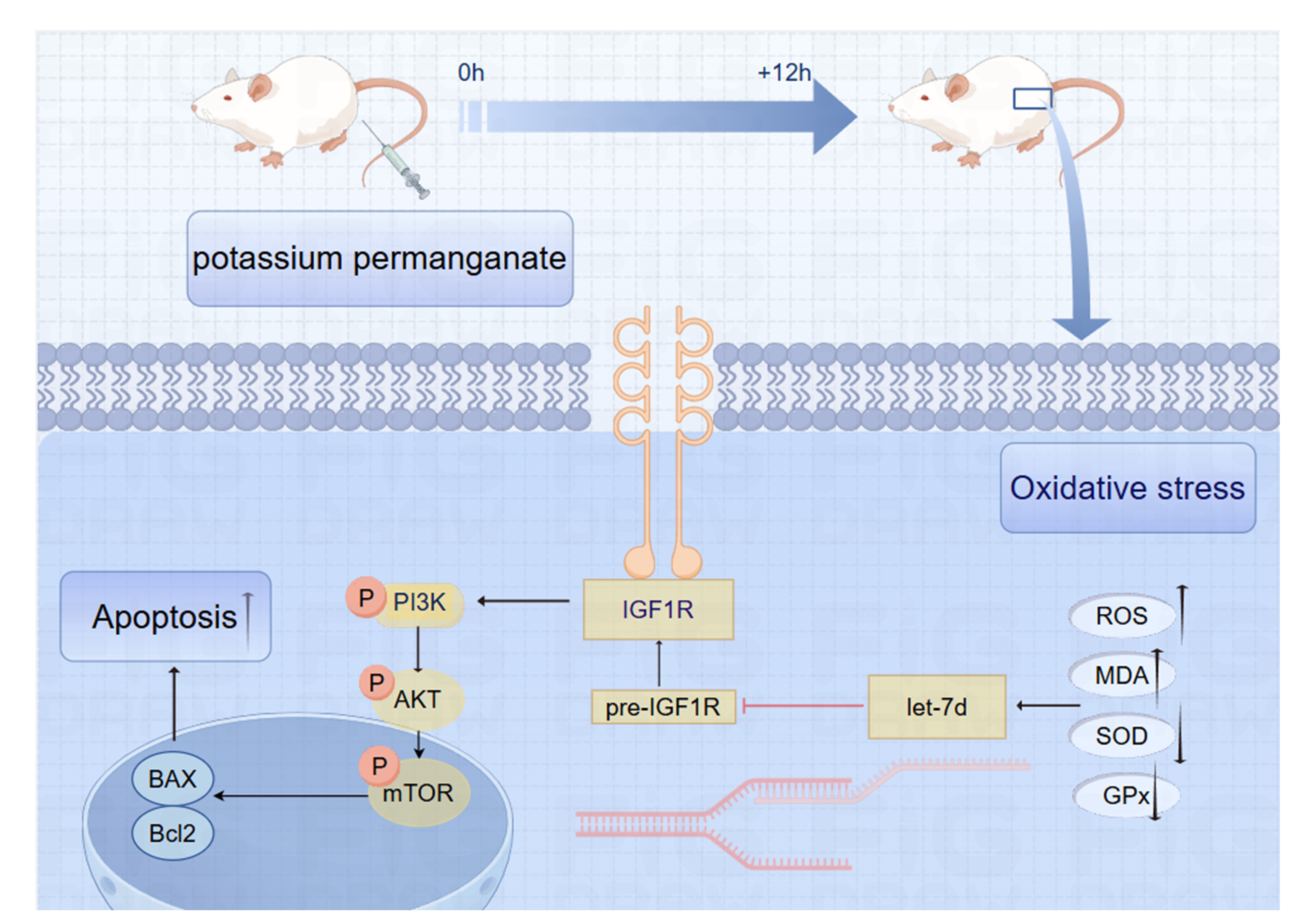
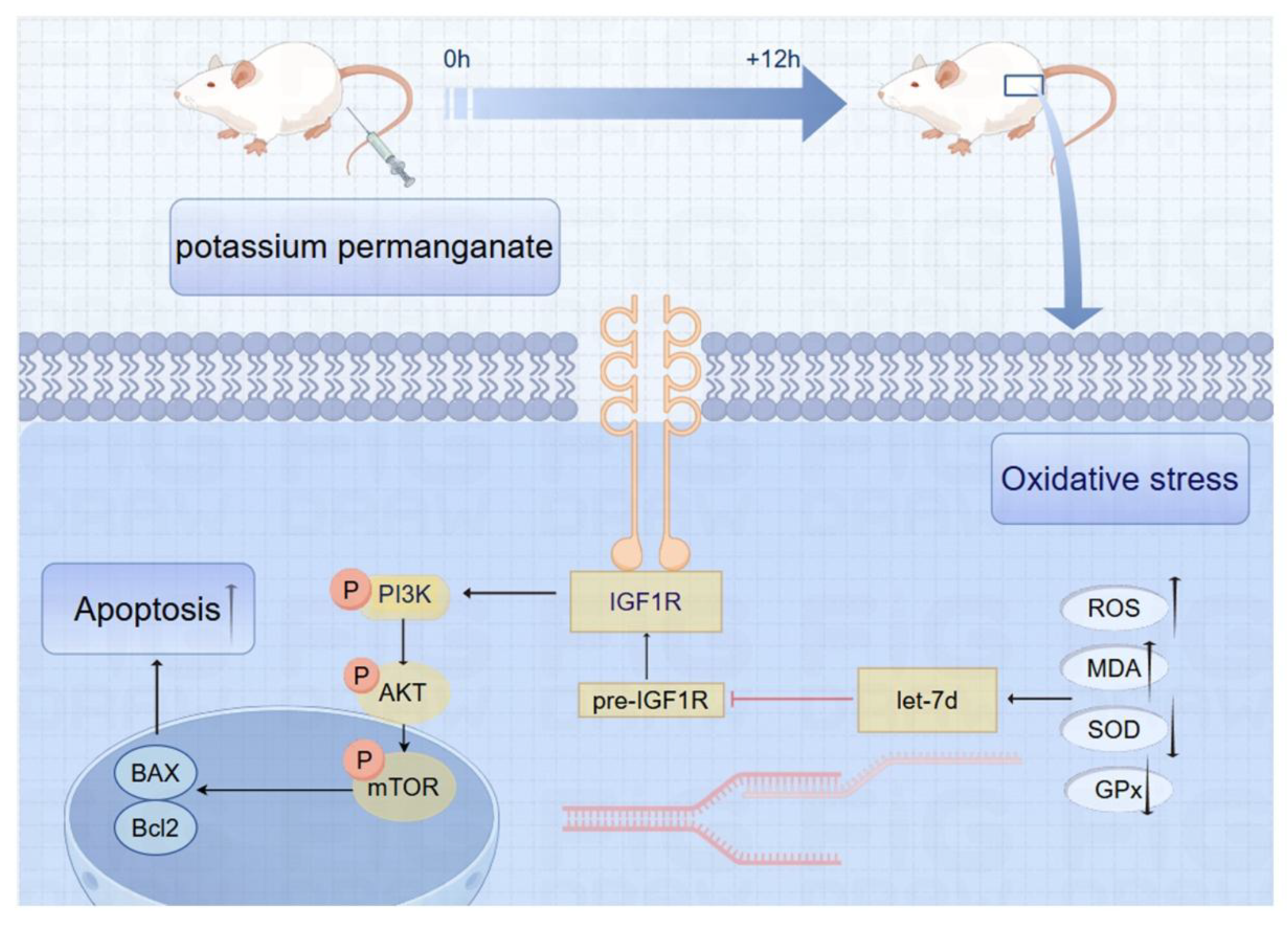
This research aims to investigate the implications of potassium permanganate treatment on oxidative stress in the endometrium of dairy cows. By examining the interactions between oxidative stress, IGF1R signaling, and miRNA regulation in the context of reproductive health, this study seeks to provide insights into potential therapeutic strategies for managing oxidative stress-related reproductive disorders in dairy cows. Understanding these mechanisms may pave the way for improved reproductive outcomes and economic viability in the dairy industry.
2. Material and methods
2.1. Collection of Bovine Uterine Samples and SD Rat Model
Bovine uterine tissue samples were obtained from a slaughterhouse in Wuhan, China. Following collection, the tissues were immediately immersed in liquid nitrogen and transported to the laboratory within 30 minutes. All animal procedures adhered to protocols approved by the Hubei Province Laboratory Animal Research Center and the Animal Research Ethics Committee of Huazhong Agricultural University (HZAUMO-2015-12). Additionally, 4 to 6-week-old Sprague-Dawley (SD) rats were procured from the Experimental Animal Center. Both uterine horns of the rats were exposed and flushed with varying concentrations of potassium permanganate (KMnO₄) (C1: 0.01%, C2: 0.025%, C3: 0.05%, C4: 0.1%), while the control group received an equivalent volume of phosphate-buffered saline (PBS).
2.2. Histological Analysis
Tissue samples were fixed in 10% formalin, followed by washing with buffer (e.g., PBS), dehydration through a graded ethanol series, clearing with xylene, and embedding in molten paraffin. The embedded tissues were placed in molds and allowed to solidify at room temperature or in cold storage. The solidified blocks were sectioned into thin slices (4-6 μm) using a microtome, deparaffinized with xylene, and stained with hematoxylin for nuclei and eosin for cytoplasmic visualization.
2.3. Bovine Endometrial Epithelial Cell Culture
Bovine endometrial epithelial cells (BEECs) were cultured in a medium supplemented with 10% fetal bovine serum (from the United States) and 1% penicillin-streptomycin (Biosharp). The incubation environment was maintained at 5% CO₂ and 37°C, with medium changes every 6 hours. Upon reaching approximately 80% confluence, the cells were digested with trypsin, subcultured, and stored at -80°C or in liquid nitrogen. To investigate the role of let-7d and IGF1R in oxidative stress within the bovine endometrium, an in vitro oxidative stress model was established by treating the cells with 0.01% KMnO₄.
2.4. bEECs Cell Transfection
When cell cultures reached approximately 60% confluence, transfections were performed using the jetPRIME reagent with bta-let-7d mimics (5’-3': S: AGAGGUAGUAGGUUGCAUAGUU, AS: CUAUGCAACCUACUACCUCUUU), bta-let-7d inhibitor (5’-3': S: AACUAUGCAACCUACUACCUCU), and IGF1R siRNA (si-IGF1R, 5’-3': S: GCACAACUACUGCUCCAAATT, AS: UUUGGAGCAGUAGUUGUGCTT). RNA and protein were extracted 24 hours post-transfection.
2.5. RNA Extraction and RT-qPCR Analysis
Total RNA from tissues or cells was extracted using Trizol reagent according to the manufacturer's instructions. Complementary DNA (cDNA) synthesis was conducted using the Yeasen one-step reverse transcription kit, with miRNA reverse transcription achieved through tailing. Gene expression was quantified via real-time PCR, employing β-actin and U6 as reference genes. The relative gene expression levels were analyzed using the 2^(-
ΔΔCT) method. Primer sequences are provided in
Table 1.
2.6. Western Blot Analysis
Following protein extraction from tissues and cells using RIPA buffer, protein concentrations in each group were quantified using a BCA assay. Samples underwent SDS-PAGE under constant voltage, followed by transfer to PVDF membranes using constant current. Membranes were blocked with ECL rapid blocking buffer for one hour and incubated overnight with primary antibodies at 4°C (16–18 hours). After three washes with TBST, membranes were incubated with secondary antibodies for two hours, rewashed thrice with TBST, and visualized. Results were analyzed using ImageJ software, with β-actin serving as an internal control.
2.7. miRNA Target Prediction
Potential miRNA targets for IGF1R were predicted using the TargetScan website, focusing on the top five candidates. These miRNAs were further assessed based on their expression in bovine endometrial tissue and previous studies regarding their functions. Ultimately, let-7d was selected due to its high expression in endometrial tissue and its significant role in mitochondrial function. Additional target predictions were conducted using miRDB (
http://mirdb.org/), PicTar (
https://pictar.mdc-berlin.de/), and miRcode (
http://www.mircode.org/).
2.8. Dual-Luciferase Reporter Assay
Wild-type and mutant plasmids for IGF1R were constructed using the primGLO backbone based on predicted binding sites. These plasmids, along with bta-let-7d mimics, were co-transfected into 293T cells. Fluorescence intensity was measured using the Yeasen Dual-Luciferase Assay Kit (Cat. No. 11402ES60).
2.9. Detection of MDA, ROS, T-AOC, GPx, and SOD
After stimulation, cells from each group were washed with PBS and collected using a scraper in 500 μL PBS. Supernatants were obtained following centrifugation for 20 minutes, and protein concentrations were determined via BCA assay. Assays for MDA (Cat. No. S0131S), ROS (Cat. No. S0033S), total antioxidant capacity (T-AOC) (Cat. No. S0119), glutathione peroxidase (GPx) (Cat. No. S0056) from Beyotime, and total superoxide dismutase (T-SOD) (Cat. No. A001-1-2) from Nanjing Jiancheng were performed according to the respective kit instructions.
2.10. Immunofluorescence Staining
Following stimulation, cells were washed twice with PBS and fixed with 4% paraformaldehyde for 20 minutes. Cells were then permeabilized with Triton for 15 minutes, washed three times, and blocked with blocking buffer at room temperature for three hours. Primary antibodies were added and incubated overnight at 4°C in the dark. After three additional washes with PBS, secondary antibodies were added and incubated at room temperature for three hours. Nuclei were stained with DAPI at room temperature for 15 minutes. Cells were rewashed three times and visualized using a fluorescence microscope.
2.11. Statistical Analysis
All data were processed using GraphPad 9.4, with each experiment performed in triplicate. Data are presented as mean ± standard error of the mean (SEM). T-tests were employed for comparisons between two groups, while one-way ANOVA was utilized for comparisons among multiple groups. Statistical significance was defined as P < 0.05, with highly significant values indicated at P < 0.01.
4. Discussion
The health status of the endometrium in dairy cows is crucial for embryo implantation and pregnancy maintenance[
20,
21] . The endometrium serves as a vital site for embryo implantation and development, and its health directly impacts successful implantation and pregnancy maintenance [
22]. Oxidative stress is widely recognized as a key factor affecting endometrial function, where excessive reactive oxygen species (ROS) can lead to cellular damage, thereby impairing reproductive performance [
23]. This study is the first to elucidate the mechanism by which bta-let-7d regulates oxidative stress in the bovine endometrium through targeting IGF1R, providing new insights into reproductive health.
In this study, potassium permanganate (KMnO₄) was utilized to induce oxidative stress in bovine endometrial epithelial cells (BEECs) [
24]. The findings showed significant increases in ROS and MDA levels, while the activities of GPX and SOD decreased, alongside a decline in total antioxidant capacity. These results indicate that oxidative stress inhibits antioxidant enzyme activity, leading to ROS accumulation and increased lipid peroxidation, contributing to cellular damage. This underscores the importance of maintaining intracellular ROS balance and highlights the protective role of antioxidant enzymes during oxidative stress. Given the critical importance of endometrial health for embryo implantation and pregnancy maintenance in dairy cows, understanding and regulating the oxidative stress response is vital for enhancing reproductive performance[
22]. Future investigations may explore modulating antioxidant enzyme activities to mitigate oxidative stress and improve reproductive efficiency in dairy herds.
Our study identified that the upregulation of bta-let-7d led to a significant decrease in IGF1R expression. IGF1R, a key receptor in the PI3K/AKT signaling pathway, is essential for regulating cellular proliferation, survival, and metabolism [
25]. Previous research has associated abnormal IGF1R expression with various reproductive disorders[
26,
27]. This receptor plays a significant role in preparing the uterus for embryo implantation. Studies indicate that stromal IGF1 and embryonic IGF2 activate IGF1R, which is vital for epithelial differentiation and receptivity during implantation [
28]. Therefore, by suppressing IGF1R expression, bta-let-7d may influence these physiological processes. IGF1R activation promotes cell proliferation and survival, while bta-let-7d inhibits this pathway, exacerbating oxidative stress-induced damage in endometrial cells.
Increased oxidative stress correlates with various diseases, including reproductive disorders[
29] [
30]. Our results demonstrated that ROS levels in endometrial cells significantly decreased following the downregulation of bta-let-7d, likely due to the subsequent increase in IGF1R levels. IGF1R activation can enhance the PI3K/AKT pathway, promoting the expression of antioxidant enzymes such as SOD and GPX. SOD scavenges superoxide radicals, and GPX removes hydrogen peroxide, working together to reduce MDA formation and alleviate oxidative stress while enhancing overall antioxidant capacity.
As understanding of miRNA functions in biology deepens, the potential of bta-let-7d as a regulatory factor becomes increasingly apparent[
31]. Studies show that the let-7 family plays critical roles in various biological processes, including cell cycle regulation and stress response[
32]. Thus, the regulatory mechanisms of bta-let-7d may extend beyond IGF1R to include other signaling pathways, such as MAPK and Wnt. influence endometrial cell responses to oxidative stress by regulating these pathways.
Thus, the Mechanism by which bta-let-7d regulates oxidative stress through IGF1R could provide a new strategy for enhancing reproductive efficiency in dairy cows. This finding provides a novel perspective on using molecular biology approaches to intervene in the reproductive health of dairy cows. Although this study provides new insights into the role of bta-let-7d and IGF1R in oxidative stress regulation, further research is needed on the following aspects: Mechanisms of bta-let-7d: Future studies could explore whether bta-let-7d influences oxidative stress through other target genes or pathways. Clinical application: Evaluating the practical application of bta-let-7d as a biomarker or therapeutic target in dairy cow reproductive management.
Overall, these findings provide valuable insights into the molecular mechanisms underlying oxidative stress in bovine endometrial tissues. The interplay between bta-let-7d and IGF1R is critical, and further exploration of these pathways may lead to novel strategies for improving reproductive health in dairy cows. Future research should investigate potential interventions to restore IGF1R levels and assess the therapeutic potential of modulating bta-let-7d expression to combat oxidative stress.
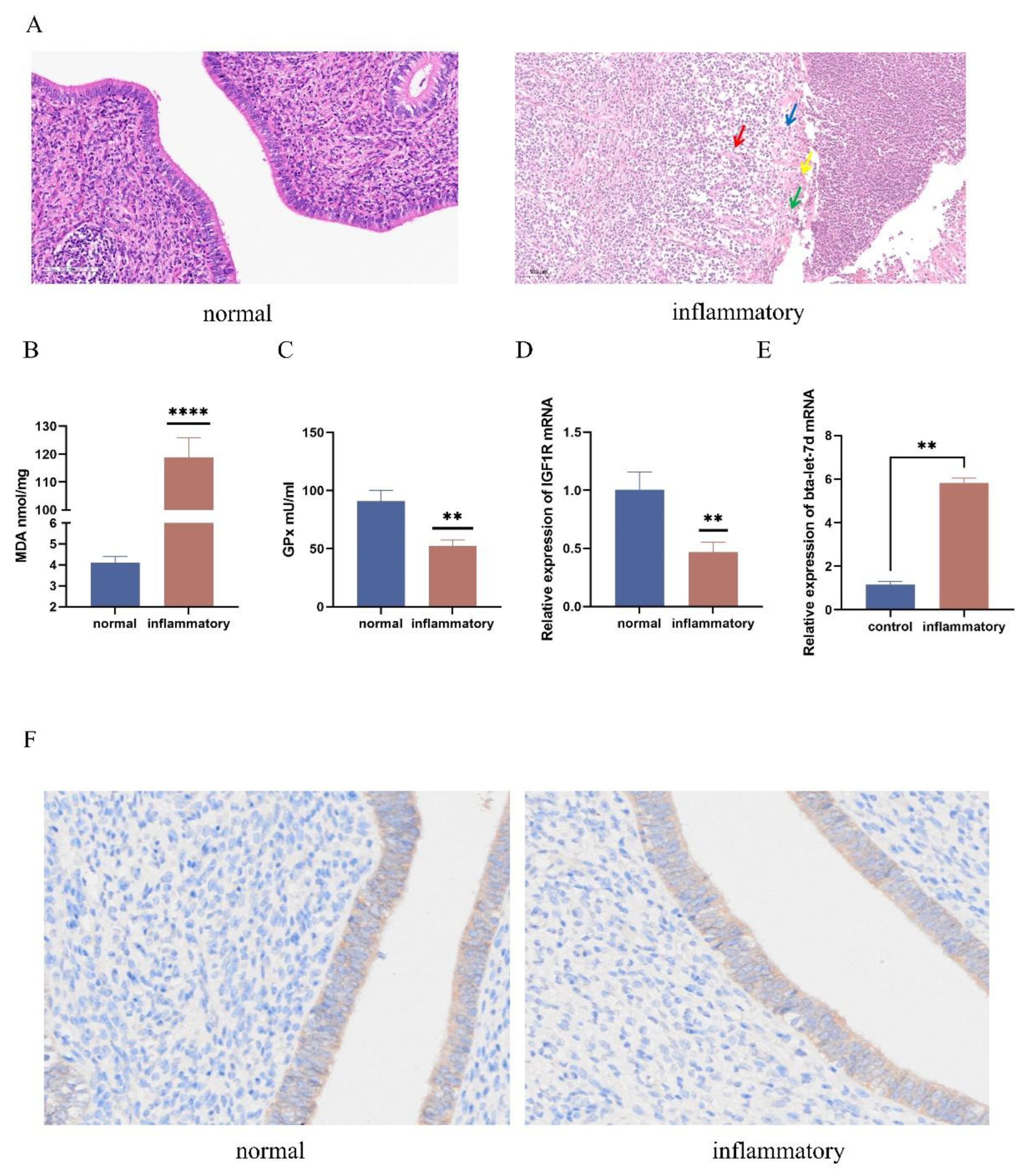
Figure 1.
Differential Expression of IGF1R and let-7d in Normal and Oxidative Stress-Affected Tissues. (A) Histopathological analysis of bovine uterine tissues stained with H&E, scale bar = 100 μm. (B) The malondialdehyde (MDA) levels in normal and inflammatory bovine uterine tissues were detected using a commercial kit. (C) Glutathione peroxidase (GPx) levels in normal and inflammatory bovine uterine tissues were detected using a commercial kit. (D) qPCR was used to measure the expression levels of IGF1R mRNA in the tissues. (E) qPCR was used to detect the expression levels of bta-let-7d in the tissues. (F) Immunohistochemical staining showing nuclear localization (blue) and IGF1R expression in normal and inflammatory bovine uterine tissues. Data are expressed as the mean ± SEM of three independent experiments. *p < 0.05, **p < 0.01.
Figure 1.
Differential Expression of IGF1R and let-7d in Normal and Oxidative Stress-Affected Tissues. (A) Histopathological analysis of bovine uterine tissues stained with H&E, scale bar = 100 μm. (B) The malondialdehyde (MDA) levels in normal and inflammatory bovine uterine tissues were detected using a commercial kit. (C) Glutathione peroxidase (GPx) levels in normal and inflammatory bovine uterine tissues were detected using a commercial kit. (D) qPCR was used to measure the expression levels of IGF1R mRNA in the tissues. (E) qPCR was used to detect the expression levels of bta-let-7d in the tissues. (F) Immunohistochemical staining showing nuclear localization (blue) and IGF1R expression in normal and inflammatory bovine uterine tissues. Data are expressed as the mean ± SEM of three independent experiments. *p < 0.05, **p < 0.01.
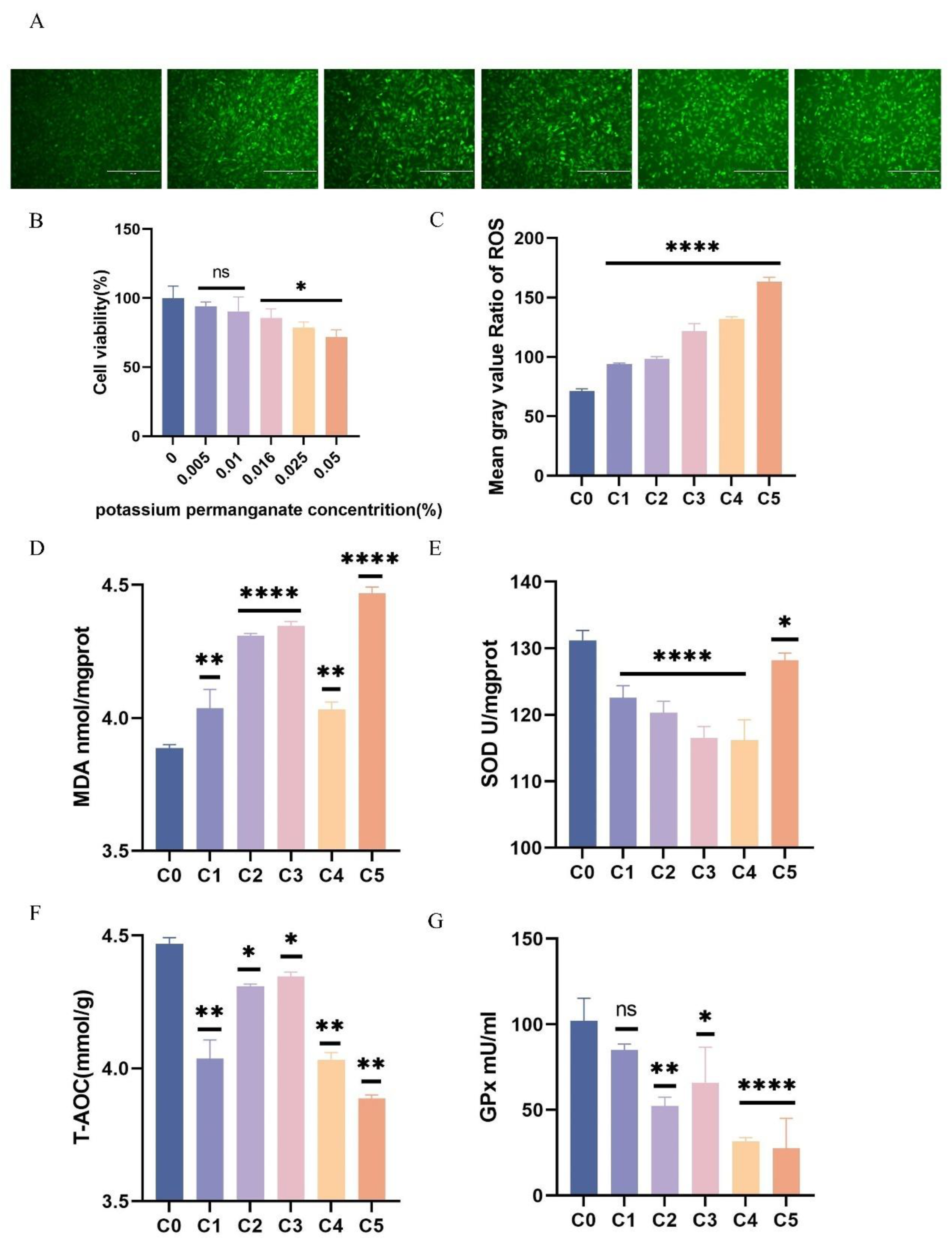
Figure 2.
Determination of potassium permanganate for modelling oxidative stress in BEECs. Fluorescence intensity of ROS in BEECs under different concentrations of potassium permanganate. (B) CCK-8 assay detecting cell viability in BEECs exposed to five different concentrations of potassium permanganate for 12 hours. (D) Lipid peroxidation (MDA) levels in BEECs under five concentrations of potassium permanganate. (E) Total superoxide dismutase (SOD) levels in BEECs under five concentrations of potassium permanganate. (F) Total antioxidant capacity (T-AOC) in BEECs under five concentrations of potassium permanganate. (G) Glutathione peroxidase (GPx) levels in BEECs under five concentrations of potassium permanganate. Data are expressed as the mean ± SEM of three independent experiments. *p < 0.05, **p < 0.01.
Figure 2.
Determination of potassium permanganate for modelling oxidative stress in BEECs. Fluorescence intensity of ROS in BEECs under different concentrations of potassium permanganate. (B) CCK-8 assay detecting cell viability in BEECs exposed to five different concentrations of potassium permanganate for 12 hours. (D) Lipid peroxidation (MDA) levels in BEECs under five concentrations of potassium permanganate. (E) Total superoxide dismutase (SOD) levels in BEECs under five concentrations of potassium permanganate. (F) Total antioxidant capacity (T-AOC) in BEECs under five concentrations of potassium permanganate. (G) Glutathione peroxidase (GPx) levels in BEECs under five concentrations of potassium permanganate. Data are expressed as the mean ± SEM of three independent experiments. *p < 0.05, **p < 0.01.
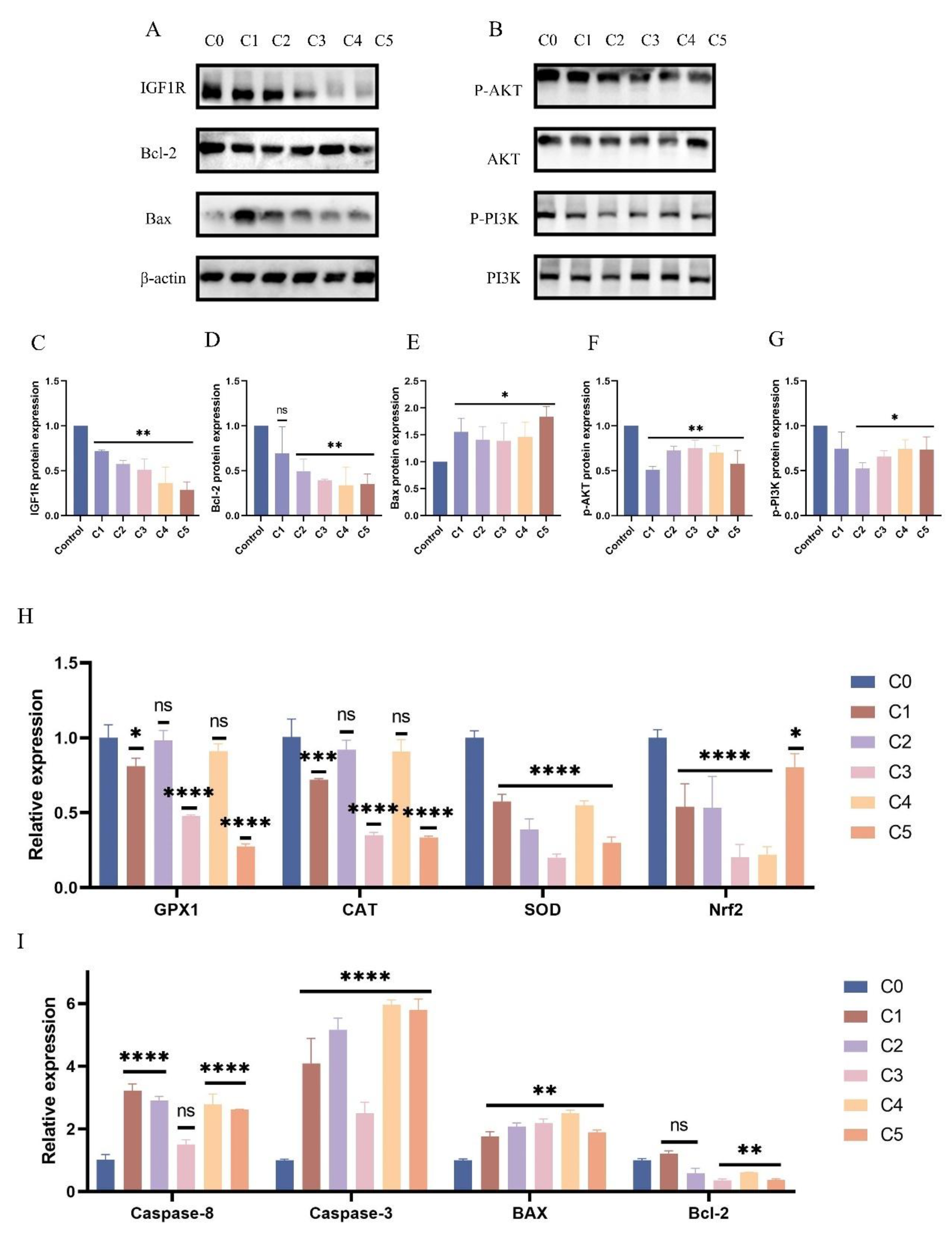
Figure 3.
Response Patterns of IGF1R and let-7d in Cellular Models. (A-G) BEECs were subjected to the same conditions as the previous step. Western blotting was used to detect the protein levels of IGF1R, Bcl-2, Bax, phosphorylated AKT, total AKT, phosphorylated PI3K, and total PI3K, with β-actin as an internal control. The protein bands were quantified using ImageJ software. (H-I) RT-qPCR was used to measure the relative expression levels of GPx1, CAT, SOD, Nrf2, Caspase-6, Caspase-3, Bax, and Bcl-2, with β-actin as an internal control. (J-K) IGF1R and p-mTOR fluorescence intensities were detected in BEECs stimulated by C1. Gray values of the indicated protein were detected using Image J gel analysis software. Data are presented as the mean ± SEM of three independent experiments. *P < 0.05; **P < 0.01,***P < 0.001,****P < 0.0001.
Figure 3.
Response Patterns of IGF1R and let-7d in Cellular Models. (A-G) BEECs were subjected to the same conditions as the previous step. Western blotting was used to detect the protein levels of IGF1R, Bcl-2, Bax, phosphorylated AKT, total AKT, phosphorylated PI3K, and total PI3K, with β-actin as an internal control. The protein bands were quantified using ImageJ software. (H-I) RT-qPCR was used to measure the relative expression levels of GPx1, CAT, SOD, Nrf2, Caspase-6, Caspase-3, Bax, and Bcl-2, with β-actin as an internal control. (J-K) IGF1R and p-mTOR fluorescence intensities were detected in BEECs stimulated by C1. Gray values of the indicated protein were detected using Image J gel analysis software. Data are presented as the mean ± SEM of three independent experiments. *P < 0.05; **P < 0.01,***P < 0.001,****P < 0.0001.
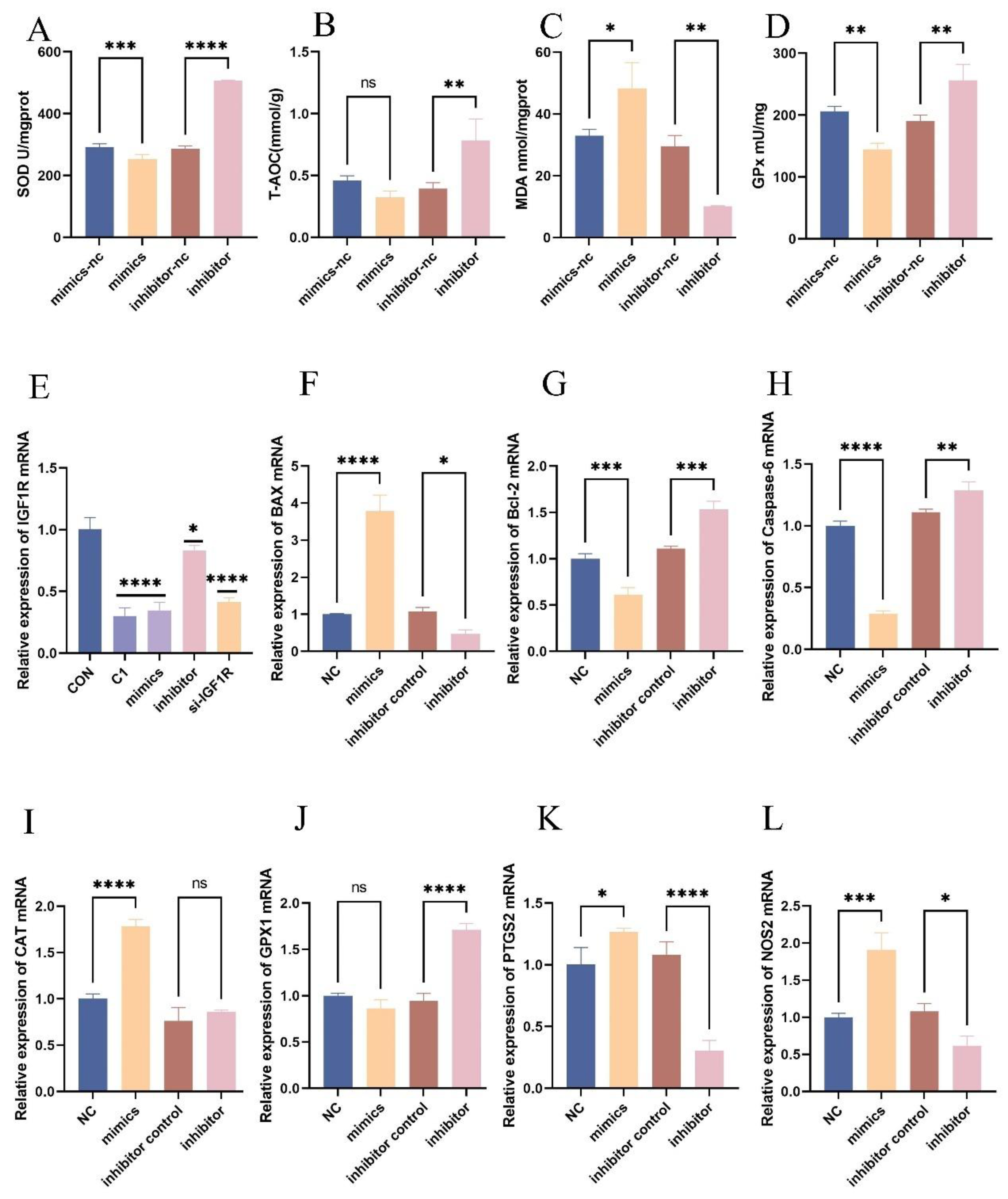
Figure 4.
Regulatory Role of let-7d on the Expression of Oxidative Stress Markers. BEECs were transfected with bta-let-7d mimics, inhibitors, and their respective negative controls. The expression levels of total superoxide dismutase (SOD), total antioxidant capacity (T-AOC), lipid peroxidation (MDA), and glutathione peroxidase (GPx) were measured using commercial kits. (E) RT-qPCR was used to measure the relative expression levels of IGF1R in BEECs treated with 0.01% potassium permanganate and transfected with bta-let-7d mimics, inhibitors, and si-IGF1R. (F-L) RT-qPCR was used to measure the relative expression levels of Bax, Bcl-2, Caspase-6, CAT, GPx1, PTGS2, and NOS2, with β-actin as an internal control. Data are presented as the mean ± SEM of three independent experiments. *P < 0.05; **P < 0.01,***P < 0.001,****P < 0.0001.
Figure 4.
Regulatory Role of let-7d on the Expression of Oxidative Stress Markers. BEECs were transfected with bta-let-7d mimics, inhibitors, and their respective negative controls. The expression levels of total superoxide dismutase (SOD), total antioxidant capacity (T-AOC), lipid peroxidation (MDA), and glutathione peroxidase (GPx) were measured using commercial kits. (E) RT-qPCR was used to measure the relative expression levels of IGF1R in BEECs treated with 0.01% potassium permanganate and transfected with bta-let-7d mimics, inhibitors, and si-IGF1R. (F-L) RT-qPCR was used to measure the relative expression levels of Bax, Bcl-2, Caspase-6, CAT, GPx1, PTGS2, and NOS2, with β-actin as an internal control. Data are presented as the mean ± SEM of three independent experiments. *P < 0.05; **P < 0.01,***P < 0.001,****P < 0.0001.
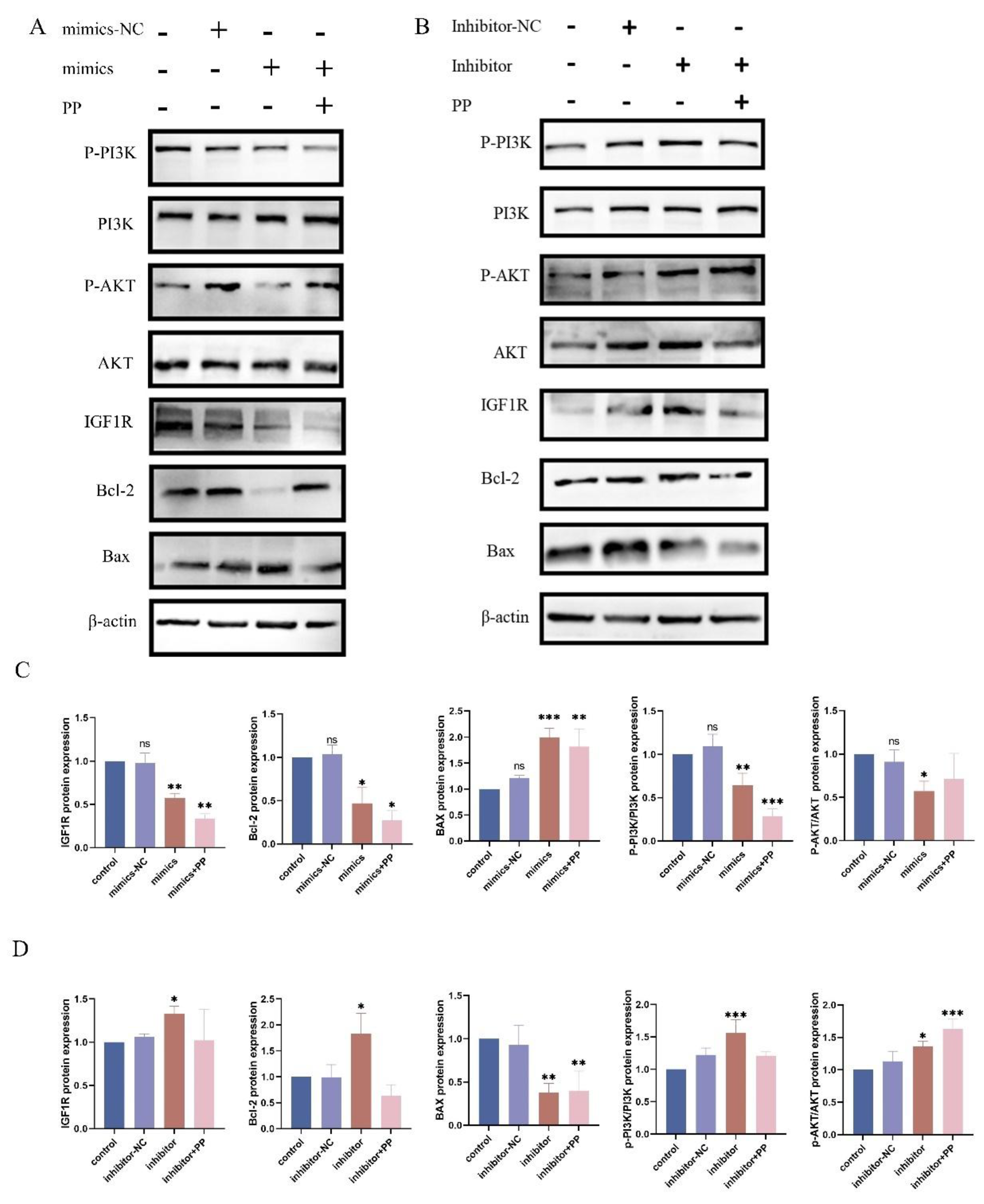
Figure 5.
Interaction between IGF1R and let-7d in the Signaling Pathway. (I) BEECs were transfected with bta-let-7d mimics and negative controls, and the expression levels of PI3K pathway-related proteins were detected using Western blotting. (K) The grey values of the specified proteins were quantified using ImageJ gel analysis software. (C) BEECs were transfected with bta-let-7d inhibitors and their negative controls, and PI3K pathway-related proteins were detected by Western blotting. (L) The grey values of the specified proteins were quantified using ImageJ software. Data are presented as the mean ± SEM of three independent experiments. *P < 0.05; **P < 0.01,***P < 0.001,****P < 0.0001.
Figure 5.
Interaction between IGF1R and let-7d in the Signaling Pathway. (I) BEECs were transfected with bta-let-7d mimics and negative controls, and the expression levels of PI3K pathway-related proteins were detected using Western blotting. (K) The grey values of the specified proteins were quantified using ImageJ gel analysis software. (C) BEECs were transfected with bta-let-7d inhibitors and their negative controls, and PI3K pathway-related proteins were detected by Western blotting. (L) The grey values of the specified proteins were quantified using ImageJ software. Data are presented as the mean ± SEM of three independent experiments. *P < 0.05; **P < 0.01,***P < 0.001,****P < 0.0001.
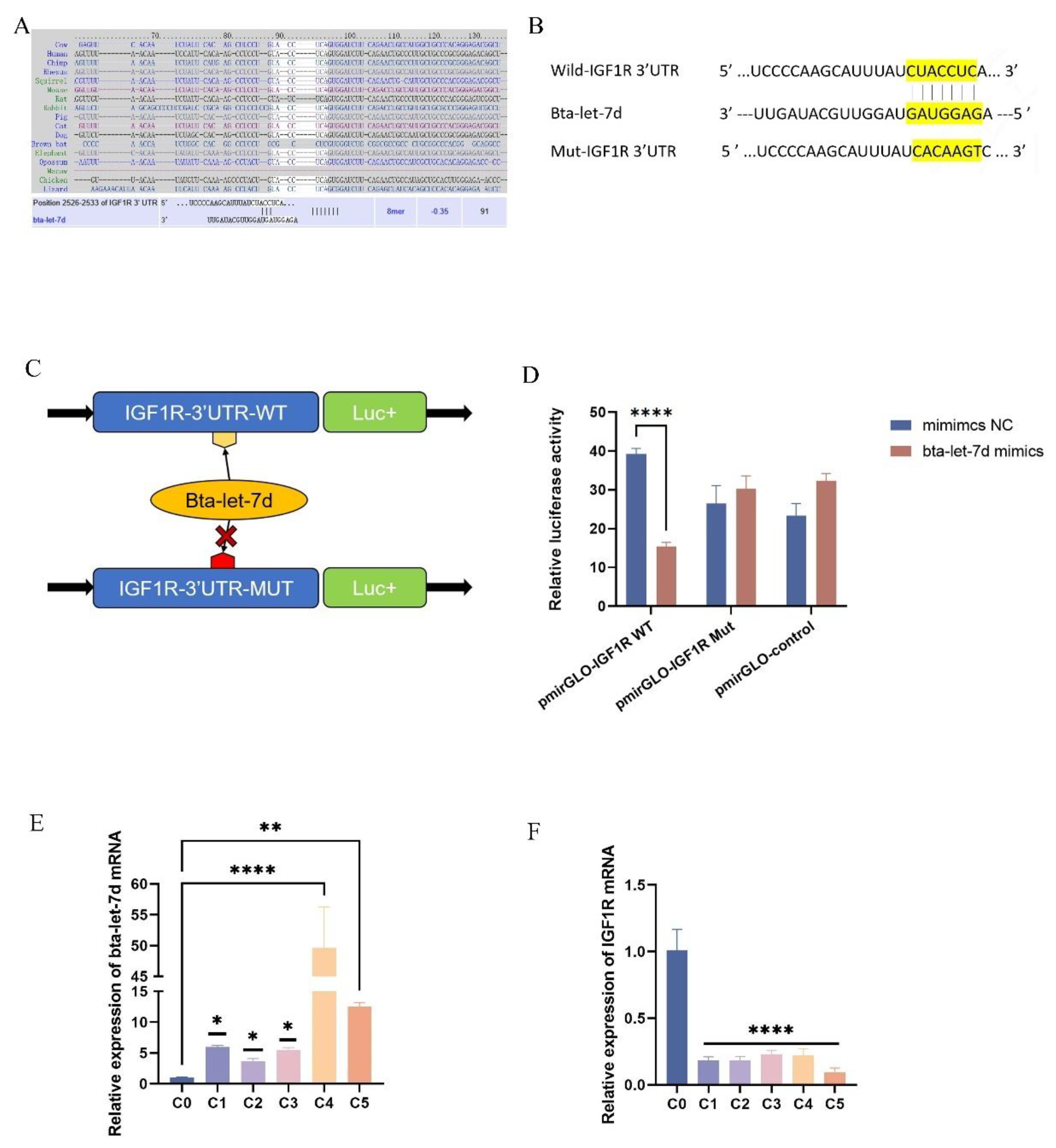
Figure 6.
IGF1R is a Target Gene of let-7d. (A) Conservation of the let-7d target sequence in IGF1R across different species and the predicted alignment between bta-let-7d and IGF1R 3’-UTR calculated by TargetScan and miRDB (bottom). (B-C) Schematic showing the construction of wild-type and mutant plasmids containing the 3’-UTR of IGF1R with or without the predicted bta-let-7d binding site for dual-luciferase reporter assays. (D) 293T cells were transfected with bta-let-7d mimics and their negative controls, and luciferase activity was measured using a dual-luciferase reporter assay. Luciferase activity is expressed as the ratio of firefly to Renilla luciferase activity. (E-F) RT-qPCR was used to detect the relative expression levels of IGF1R and bta-let-7d, with β-actin and U6 as internal controls, respectively. Data are presented as the mean ± SEM of three independent experiments. *P < 0.05; **P < 0.01,***P < 0.001,****P < 0.0001.
Figure 6.
IGF1R is a Target Gene of let-7d. (A) Conservation of the let-7d target sequence in IGF1R across different species and the predicted alignment between bta-let-7d and IGF1R 3’-UTR calculated by TargetScan and miRDB (bottom). (B-C) Schematic showing the construction of wild-type and mutant plasmids containing the 3’-UTR of IGF1R with or without the predicted bta-let-7d binding site for dual-luciferase reporter assays. (D) 293T cells were transfected with bta-let-7d mimics and their negative controls, and luciferase activity was measured using a dual-luciferase reporter assay. Luciferase activity is expressed as the ratio of firefly to Renilla luciferase activity. (E-F) RT-qPCR was used to detect the relative expression levels of IGF1R and bta-let-7d, with β-actin and U6 as internal controls, respectively. Data are presented as the mean ± SEM of three independent experiments. *P < 0.05; **P < 0.01,***P < 0.001,****P < 0.0001.
Figure 7.
Negative Regulation of Oxidative Stress by IGF1R. (A, C-F) Western blotting was used to detect the protein expression levels of IGF1R, PI3K, phosphorylated PI3K, AKT, and phosphorylated AKT after IGF1R knockdown, with β-actin as an internal control. The grey values of the specified proteins were quantified using ImageJ software. (B) RT-qPCR measured IGF1R mRNA levels in BEECs transfected with si-NC or si-IGF1R. (G) Total superoxide dismutase (SOD), total antioxidant capacity (T-AOC), lipid peroxidation (MDA), and glutathione peroxidase (GPx) levels were measured in BEECs 24 hours after transfection with si-NC or si-IGF1R. (H) RT-qPCR was used to measure the relative expression levels of GPx1, CAT, SOD, and Nrf2 in BEECs 24 hours after transfection with si-NC or si-IGF1R, with β-actin as an internal control. (I) RT-qPCR was used to measure the relative expression levels of Bax, Bcl-2, Caspase-3, and Caspase-8 in BEECs 24 hours after transfection with si-NC or si-IGF1R, with β-actin as an internal control. (G-J) BEECs were subjected to the same conditions as the previous step. Protein levels were measured using Western blotting, with β-actin as an internal control. The protein bands were quantified using ImageJ. Data are presented as the mean ± SEM of three independent experiments. *P < 0.05; **P < 0.01,***P < 0.001,****P < 0.0001.
Figure 7.
Negative Regulation of Oxidative Stress by IGF1R. (A, C-F) Western blotting was used to detect the protein expression levels of IGF1R, PI3K, phosphorylated PI3K, AKT, and phosphorylated AKT after IGF1R knockdown, with β-actin as an internal control. The grey values of the specified proteins were quantified using ImageJ software. (B) RT-qPCR measured IGF1R mRNA levels in BEECs transfected with si-NC or si-IGF1R. (G) Total superoxide dismutase (SOD), total antioxidant capacity (T-AOC), lipid peroxidation (MDA), and glutathione peroxidase (GPx) levels were measured in BEECs 24 hours after transfection with si-NC or si-IGF1R. (H) RT-qPCR was used to measure the relative expression levels of GPx1, CAT, SOD, and Nrf2 in BEECs 24 hours after transfection with si-NC or si-IGF1R, with β-actin as an internal control. (I) RT-qPCR was used to measure the relative expression levels of Bax, Bcl-2, Caspase-3, and Caspase-8 in BEECs 24 hours after transfection with si-NC or si-IGF1R, with β-actin as an internal control. (G-J) BEECs were subjected to the same conditions as the previous step. Protein levels were measured using Western blotting, with β-actin as an internal control. The protein bands were quantified using ImageJ. Data are presented as the mean ± SEM of three independent experiments. *P < 0.05; **P < 0.01,***P < 0.001,****P < 0.0001.
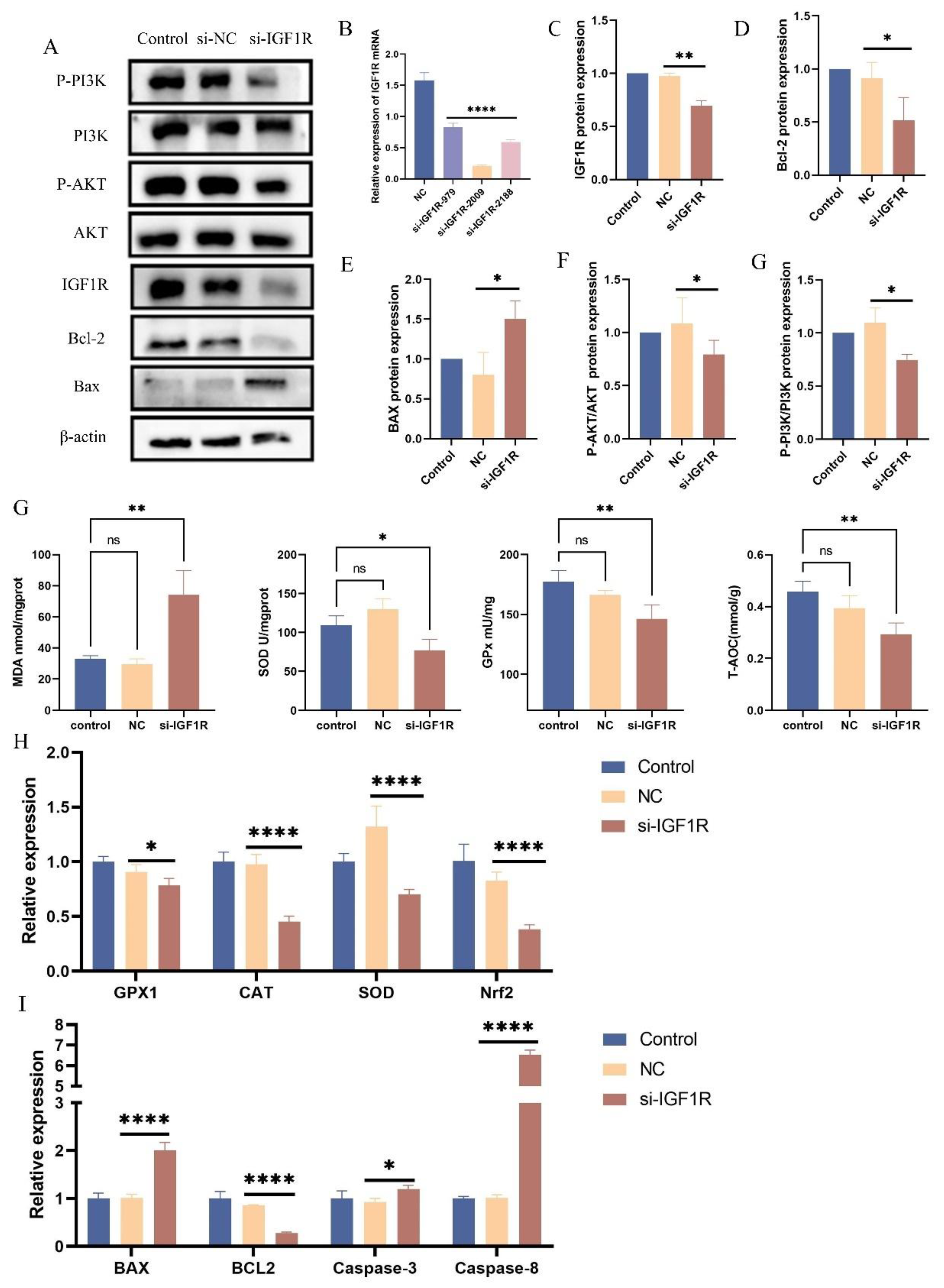
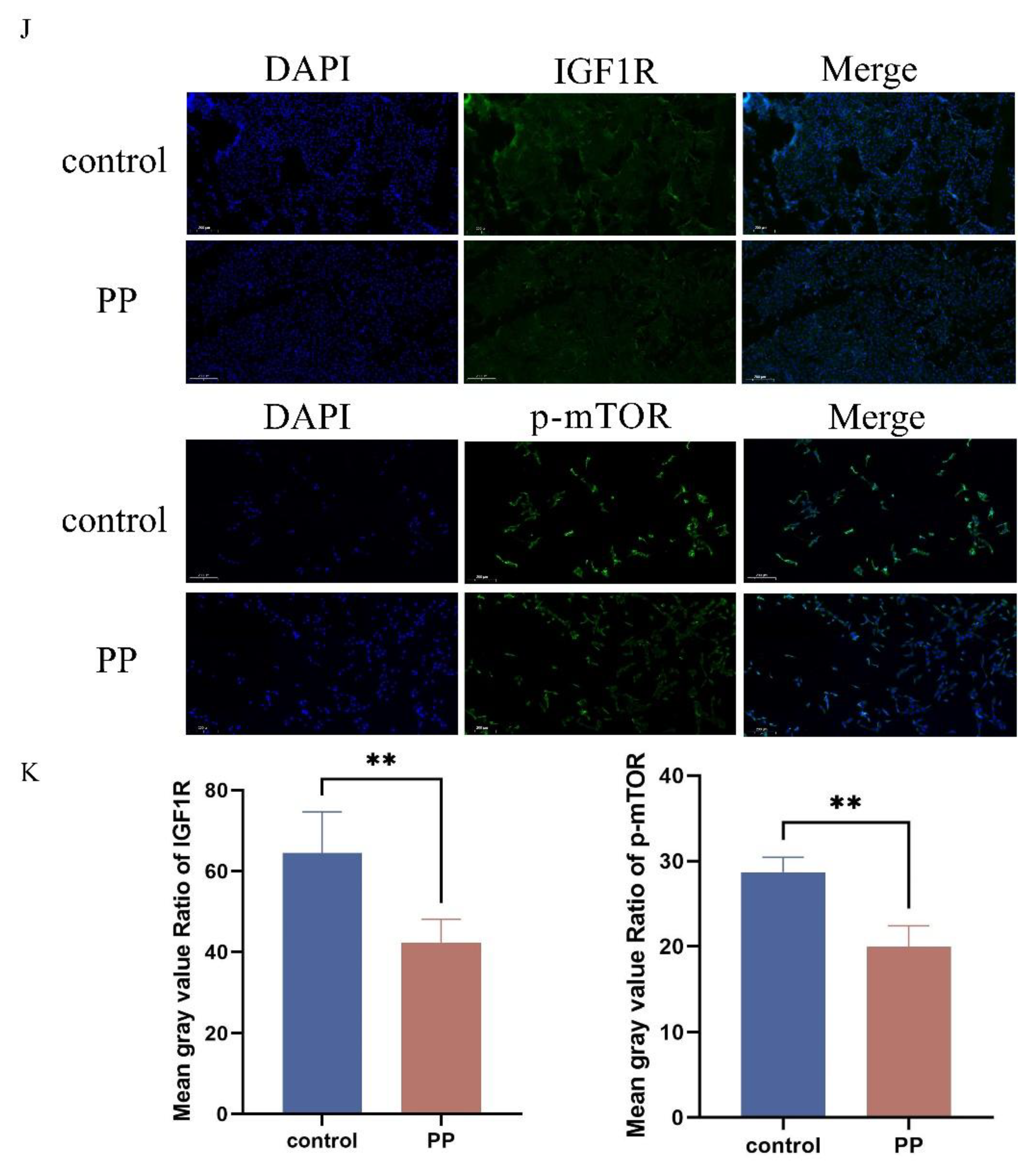
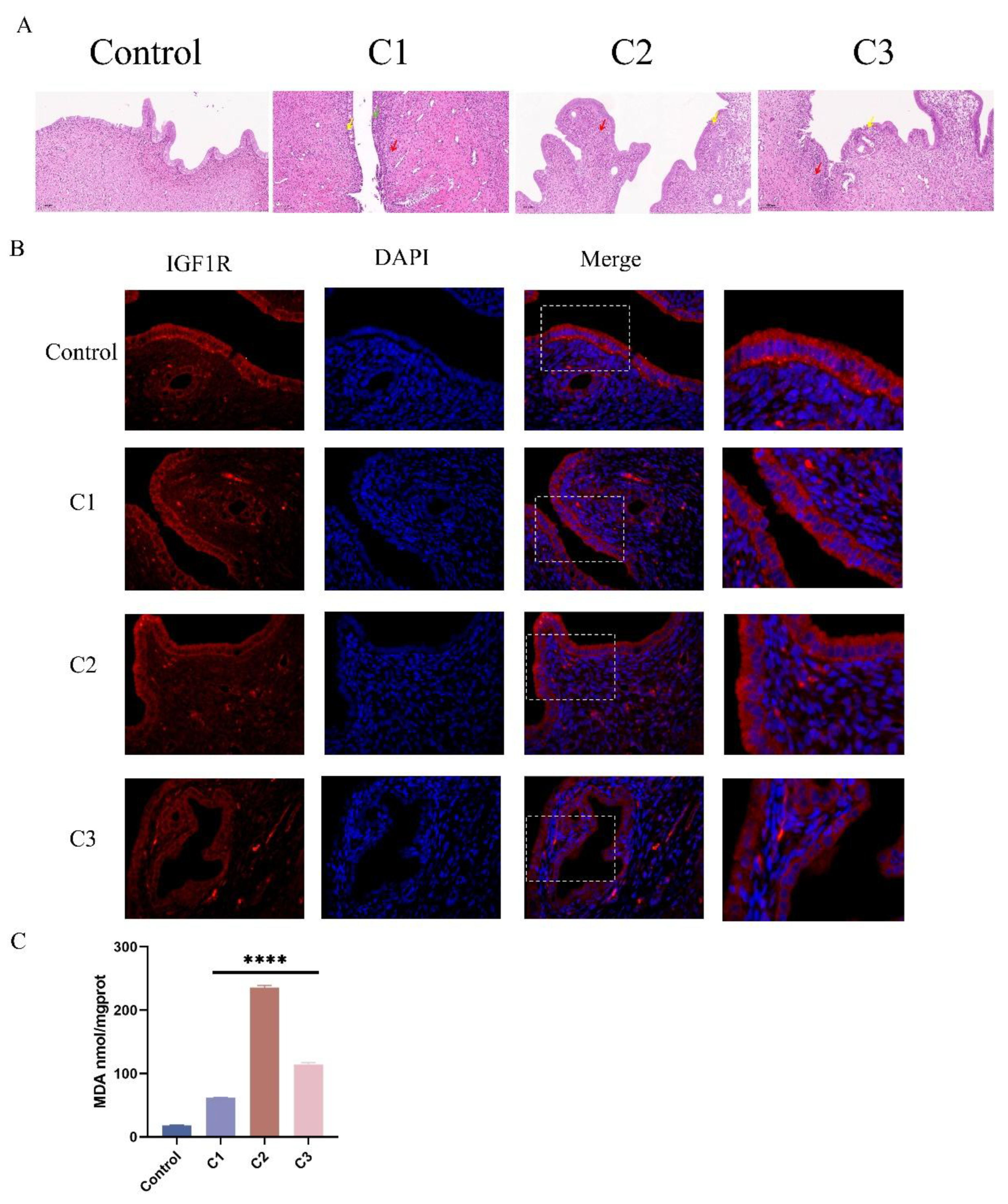
Figure 8.
Potassium Permanganate-Induced Uterine Injury is Associated with IGF1R Expression. (A) Histopathological analysis of rat uterine tissues stained with H&E after potassium permanganate lavage for 12 hours (HE, ×100), scale bar = 100 μm. (B) Quantifying IGF1R protein levels and immunofluorescence staining of IGF1R expression in the tissues, scale bar = 100 μm. Blue indicates nuclei, and red indicates IGF1R-positive staining. (C) Lipid peroxidation (MDA) levels in the tissues above were measured using a commercial kit. Data are presented as the mean ± SEM of three independent experiments. *P < 0.05; **P < 0.01,***P < 0.001,****P < 0.0001.
Figure 8.
Potassium Permanganate-Induced Uterine Injury is Associated with IGF1R Expression. (A) Histopathological analysis of rat uterine tissues stained with H&E after potassium permanganate lavage for 12 hours (HE, ×100), scale bar = 100 μm. (B) Quantifying IGF1R protein levels and immunofluorescence staining of IGF1R expression in the tissues, scale bar = 100 μm. Blue indicates nuclei, and red indicates IGF1R-positive staining. (C) Lipid peroxidation (MDA) levels in the tissues above were measured using a commercial kit. Data are presented as the mean ± SEM of three independent experiments. *P < 0.05; **P < 0.01,***P < 0.001,****P < 0.0001.
Table 1.
Primers used for qPCR.
Table 1.
Primers used for qPCR.
| Species |
Gene name |
Primer sequence (5'–3') |
GenBank accession number |
Product size (bp) |
| Bos taurus |
IGF1R |
Forward: CCAAAACCGAAGCTGAGAAG |
XM_054377836.1 |
199 |
| |
|
Reverse: TCCGGGTCTGTGATGTTGTA |
|
|
| |
GPx1 |
Forward: CTTGCTGCTTGGCGGTCA |
NM_174076.3 |
139 |
| |
|
Reverse: AGGGGAGGCTGGGATGGAT |
|
|
| |
SOD3 |
Forward: CTTCTTCCACCTTGAGGGCTTC |
NM_001428313.1 |
125 |
| |
|
Reverse: CGGACATCGGGTTGTAGTGC |
|
|
| |
CAT |
Forward: CTGGGACCCAACTATCTCCA |
NM_001035386.2 |
148 |
| |
|
Reverse: GATGCTCGGGAGCACTAAAG |
|
|
| |
Nrf2 |
Forward: GGTTGCCCACATTCCCAAATC |
XM_005202314.5 |
119 |
| |
|
Reverse: CAAGTGACTGAAACGTAGCCG |
|
|
| |
NOS2 |
Forward: ACCTACCAGCTGACGGGAGAT |
XM_024979646.2 |
316 |
| |
|
Reverse: TGGCAGGGTCCCCTCTGATG |
|
|
| |
Ptgs2 |
Forward: TCCTGAAACCCACTCCCAACA |
NM_174445.2 |
242 |
| |
|
Reverse: TGGGCAGTCAGGCACAG |
|
|
| |
Bax |
Forward: CAGATCATGAAGACAGGGGC |
NM_173894.1 |
389 |
| |
|
Reverse: CGCTCTCGAAGGAAGTCCAA |
|
|
| |
Bcl-2 |
Forward: ATGTGTGTGGAGAGCGTCAA |
NM_001166486.1 |
143 |
| |
|
Reverse: GGGCCATACAGCTCCACAAA |
|
|
| |
Caspase-3 |
Forward: AAGCCATGGTGAAGAAGGAA |
XM_010820245.4 |
134 |
| |
|
Reverse: GGCAGGCCTGAATAATGAAA |
|
|
| |
Caspase-9 |
Forward: CGCCACCATCTTCTCCCTG |
XM_024999238.2 |
83 |
| |
|
Reverse: CCAACGTCTCCTTCTCCTCC |
|
|
| |
β-actin |
Forward: CATCACCATCGGCAATGAGC |
NM_173979.3 |
156 |
| |
|
Reverse: AGCACCGTGTTGGCGTAGAG |
|
|
Table 2.
Histological score for Figure A.
Table 2.
Histological score for Figure A.
| Name |
Edema |
Necrosis |
Hemorrhages |
Endothelium Damage |
Leukocyte Infiltration |
Total |
| C0 |
0 |
0 |
0 |
0 |
0 |
0 |
| C1 |
0 |
0 |
0 |
2 |
1 |
3 |
| C2 |
1 |
0 |
0 |
2 |
1 |
4 |
| C3 |
0 |
0 |
0 |
2 |
1 |
3 |