Submitted:
22 December 2024
Posted:
23 December 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
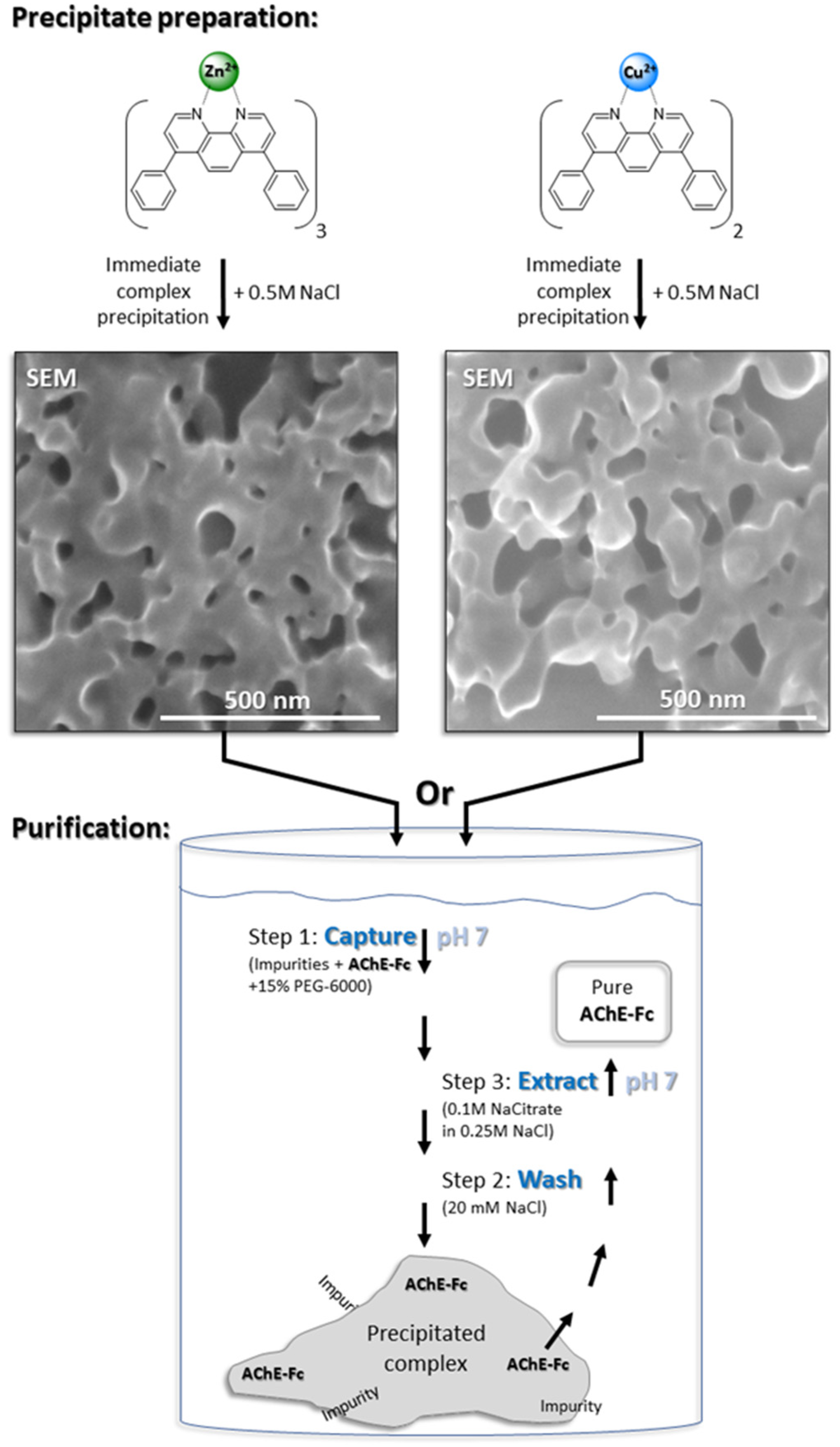
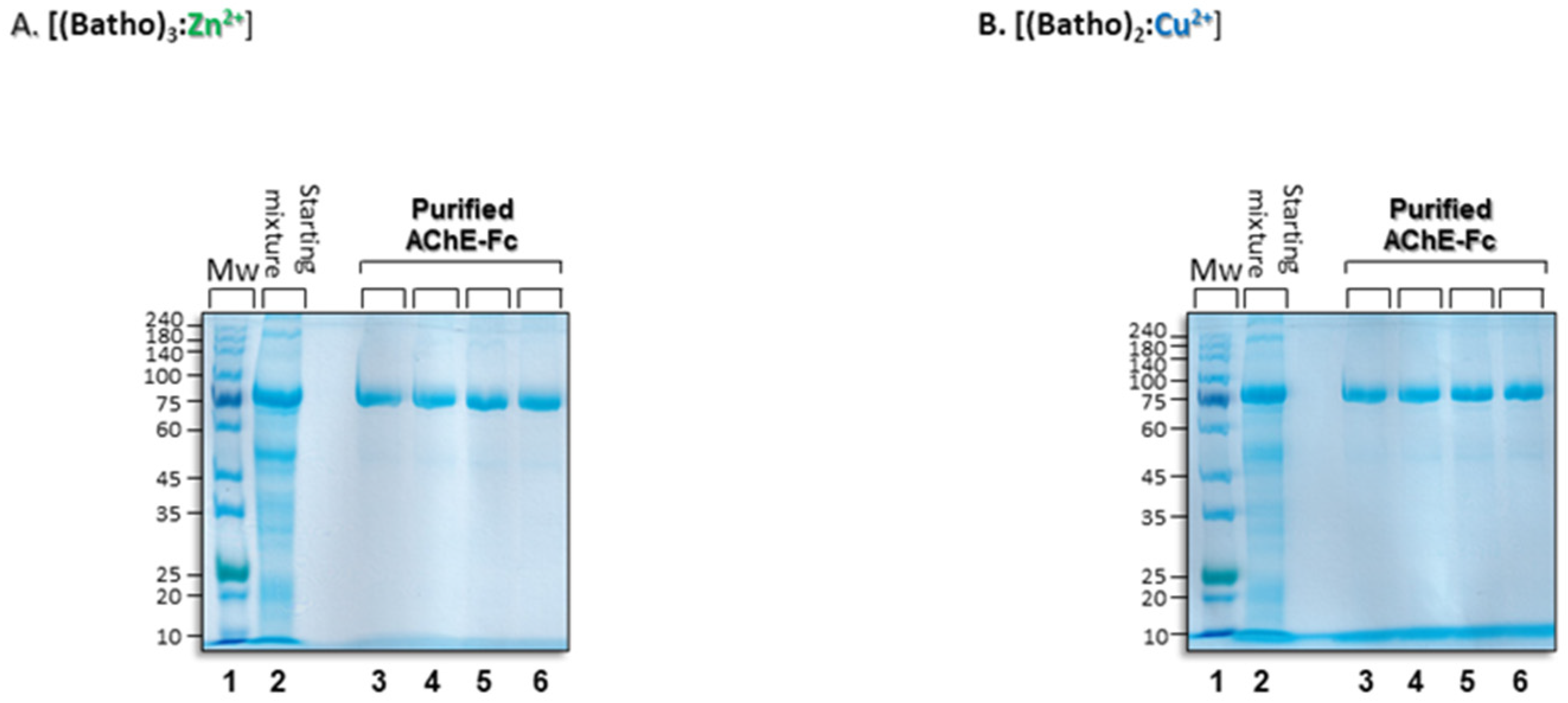
2.2.2. Purification of AChE-Fc with [(Batho)3:Zn2+] or [(Batho)2:Cu2+] Complexes.
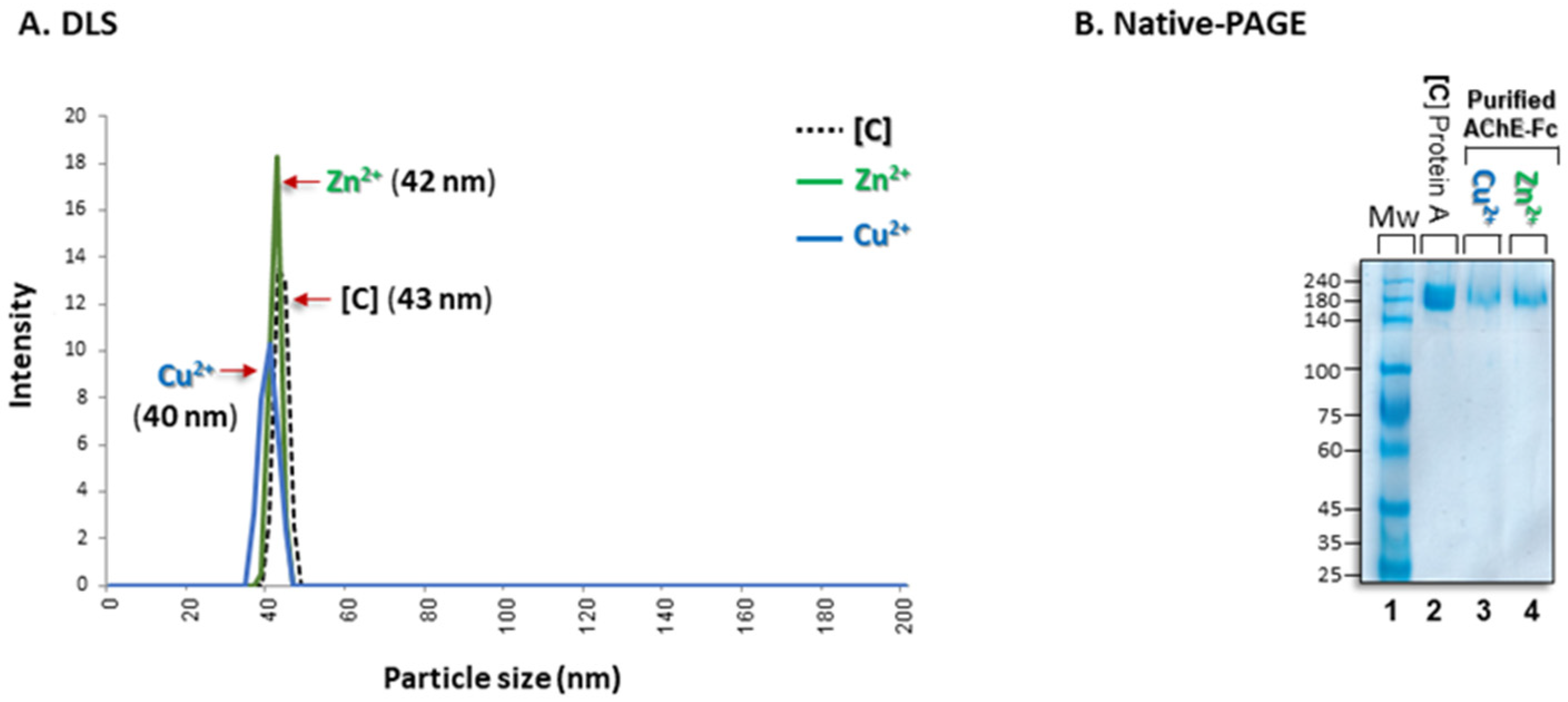
2.2.3. Dynamic Light Scattering (DLS)
2.2.4. Circular Dichroism (CD) Spectroscopy
2.2.5. Native PAGE
2.2.6. Batho Recrystallization and Quantitation
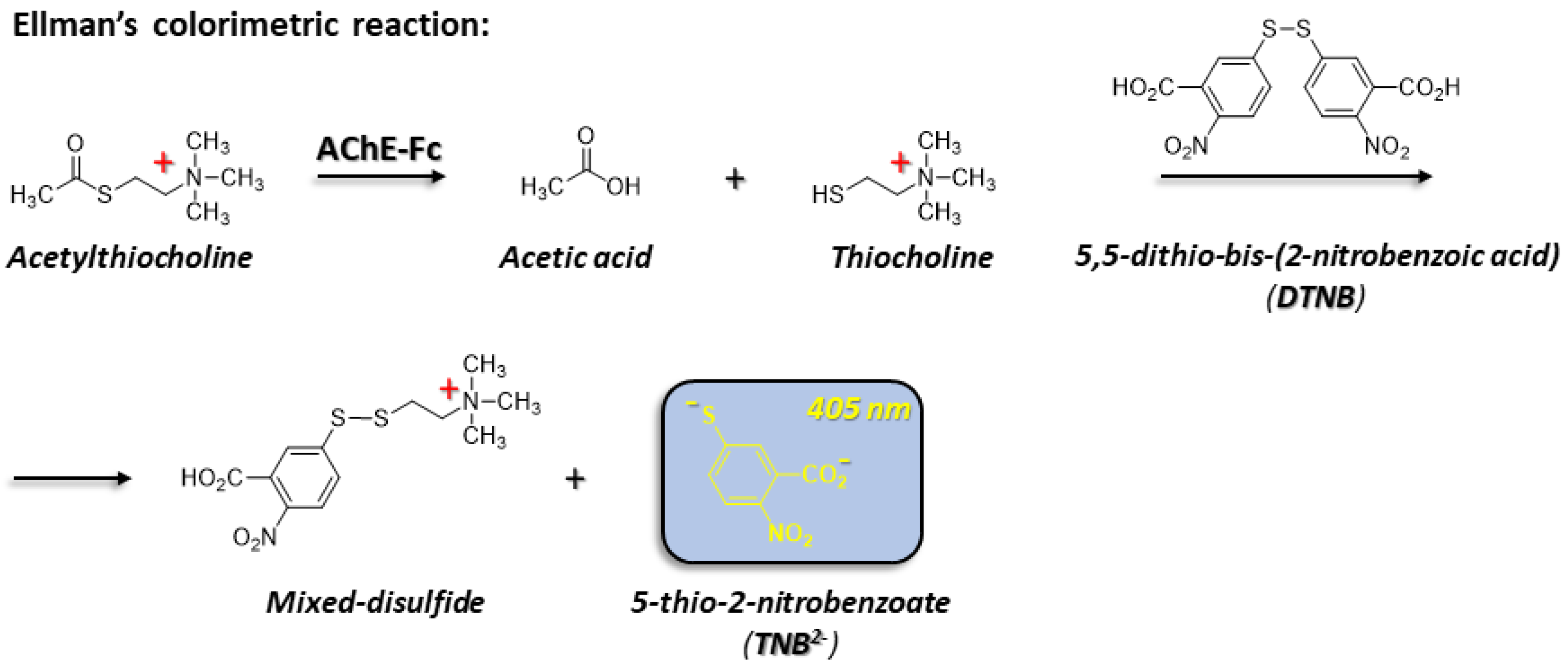
2.2.7. AChE-Fc Activity
2.2.8. Leaching Assessment
2.2.9. Scanning Electron Microscopy (SEM)
3. Results and Discussion
3.1. Precipitation of the Metal-Chelator Complexes
3.2. The Role of PEG 6000
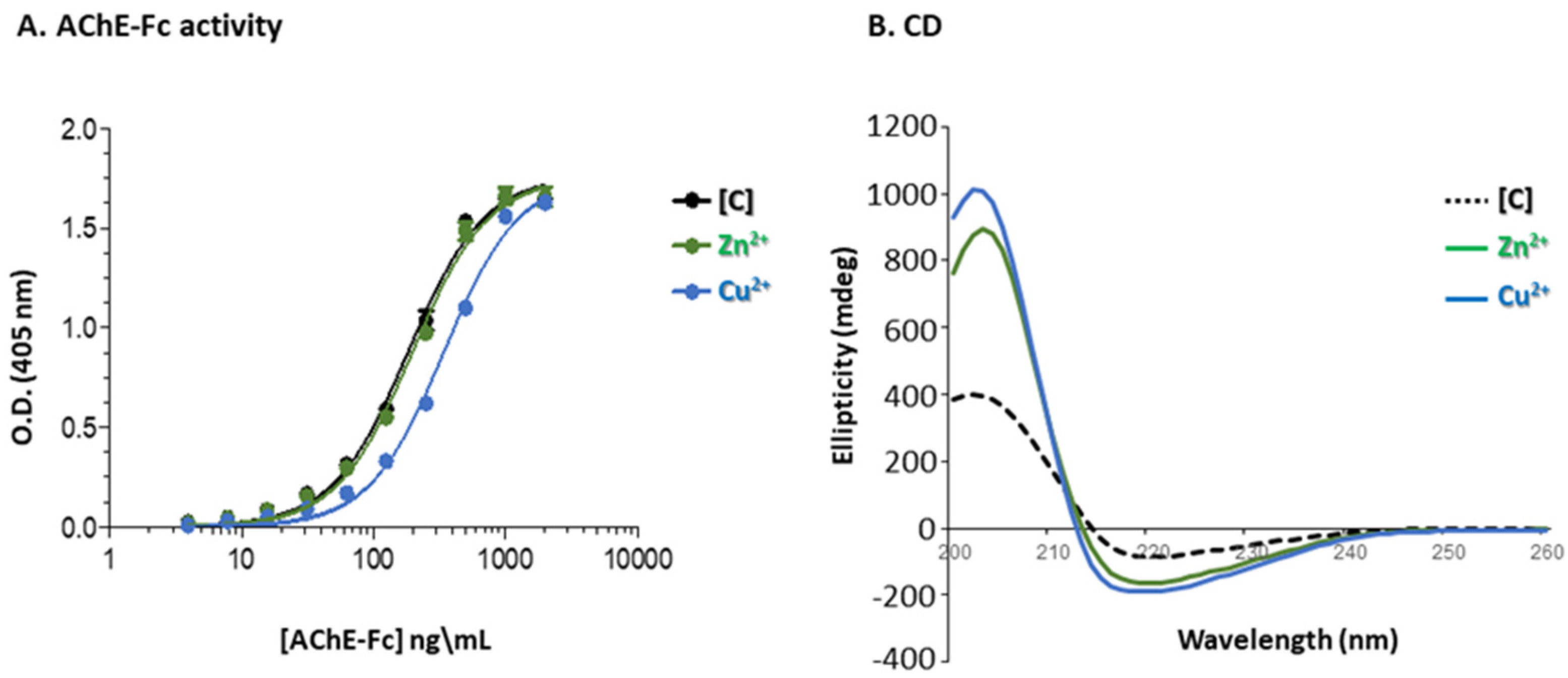
3.3. Aggregational State of the Recovered Fc-AChE
3.4. Enzymatic Activity
3.5. Binding Interactions and Process Yield
3.6. Binding Capacity
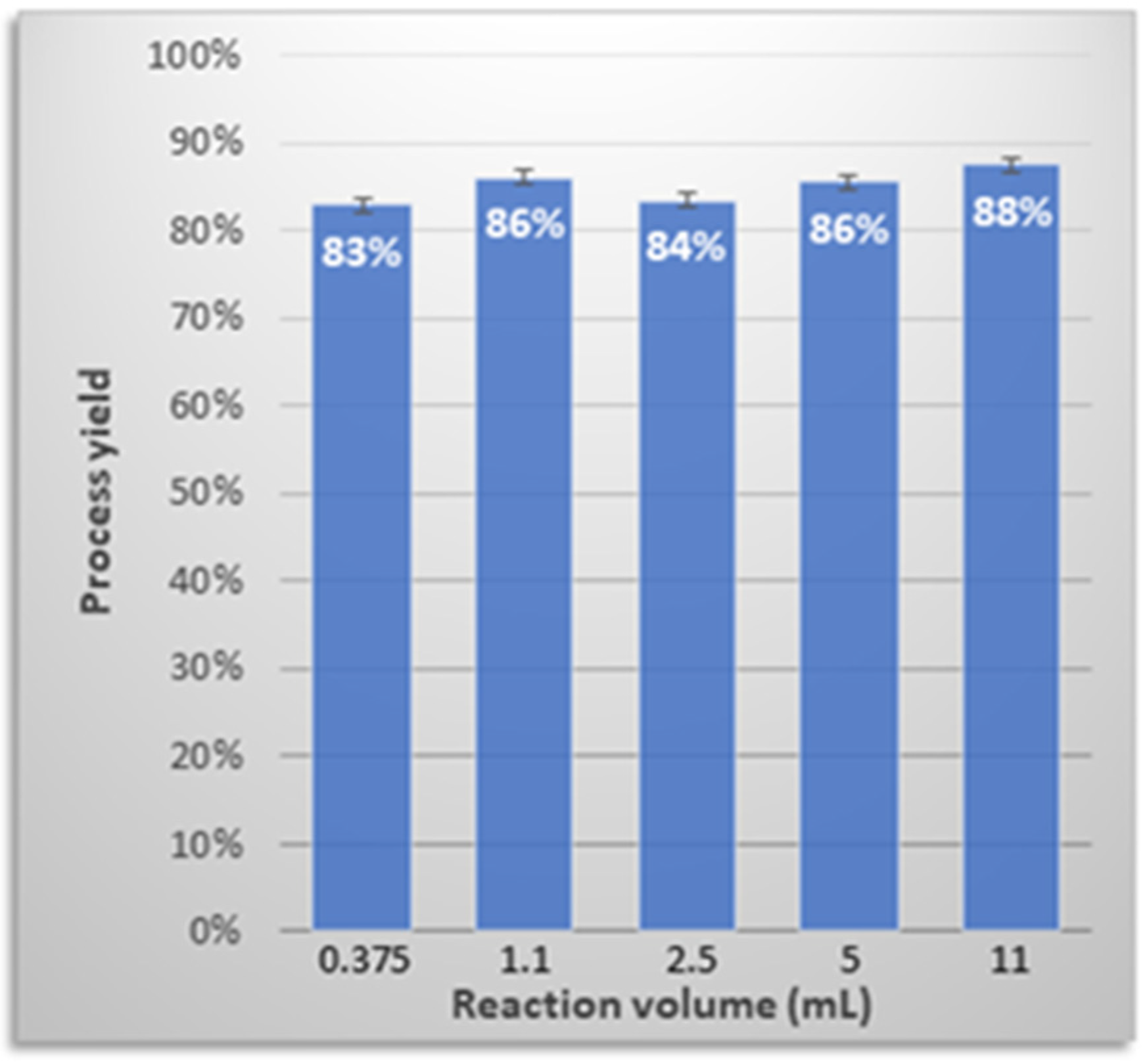
3.7. Process Upscaling
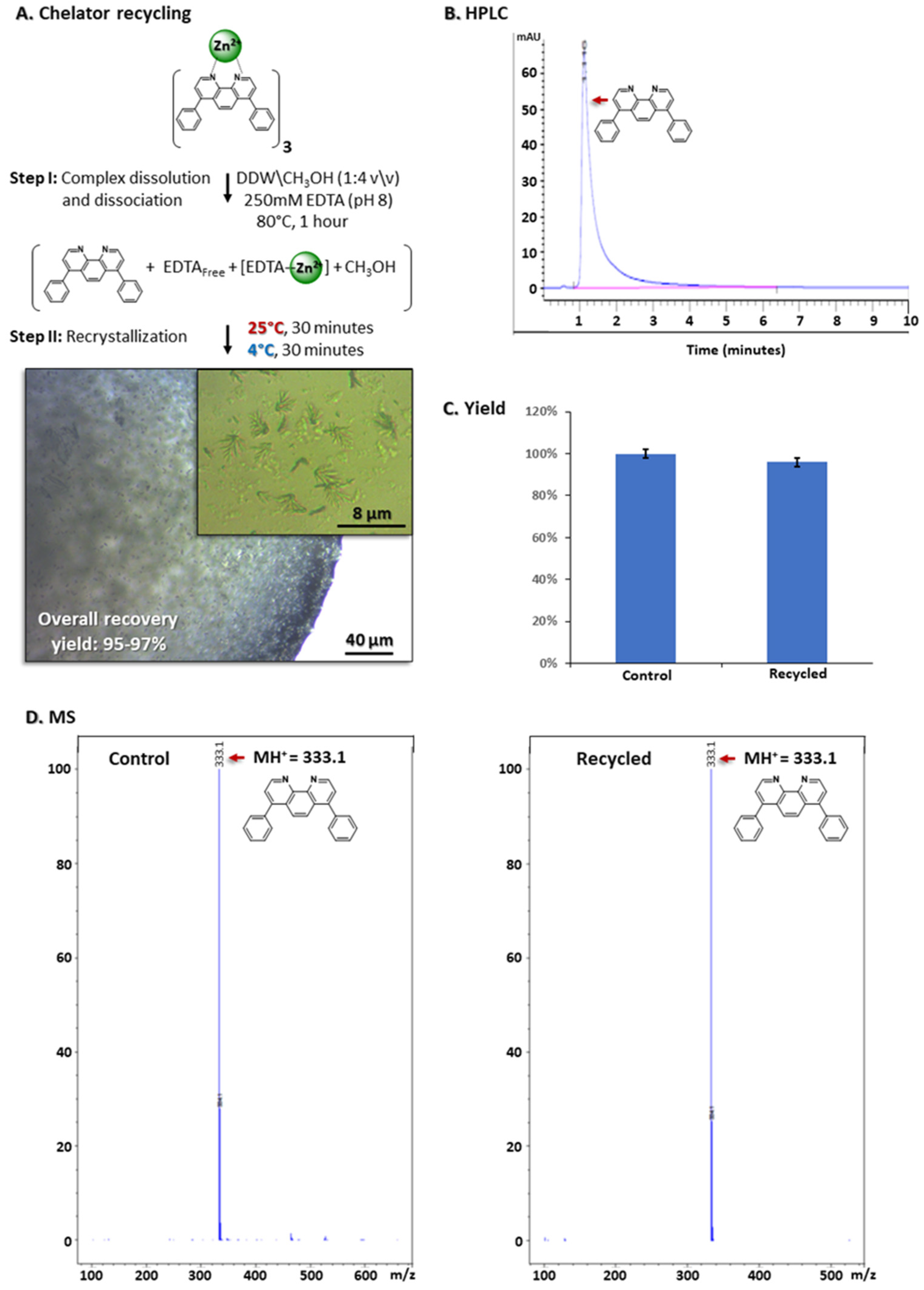
3.8. Chelator Recycling
3.9. Chelator Leaching
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Capon, D.J., et al., Designing CD4 immunoadhesins for AIDS therapy. Nature, 1989. 337(6207): p. 525-31. [CrossRef]
- Sokolowska-Wedzina, A., et al., Efficient production and purification of extracellular domain of human FGFR-Fc fusion proteins from Chinese hamster ovary cells. Protein Expression and Purification, 2014. 99: p. 50-57. [CrossRef]
- Ning, L., et al., Molecular Design of Peptide-Fc Fusion Drugs. Curr Drug Metab, 2019. 20(3): p. 203-208. [CrossRef]
- Dwyer, M.A., et al., Expression and Characterization of a DNase I-Fc Fusion Enzyme*. Journal of Biological Chemistry, 1999. 274(14): p. 9738-9743. [CrossRef]
- Noy-Porat, T., et al., Acetylcholinesterase-Fc Fusion Protein (AChE-Fc): A Novel Potential Organophosphate Bioscavenger with Extended Plasma Half-Life. Bioconjug Chem, 2015. 26(8): p. 1753-8. [CrossRef]
- Jazayeri, J.A. and G.J. Carroll, Fc-Based Cytokines. BioDrugs, 2008. 22(1): p. 11-26.
- Duivelshof, B.L., et al., Therapeutic Fc-fusion proteins: Current analytical strategies. J Sep Sci, 2021. 44(1): p. 35-62. [CrossRef]
- Zhang, J., et al., Fusion partners as a tool for the expression of difficult proteins in mammalian cells. Curr Pharm Biotechnol, 2010. 11(3): p. 241-5. [CrossRef]
- Sato, A.K., et al., Therapeutic peptides: technological advances driving peptides into development. Curr Opin Biotechnol, 2006. 17(6): p. 638-42. [CrossRef]
- Meibohm, B. and H. Zhou, Characterizing the impact of renal impairment on the clinical pharmacology of biologics. J Clin Pharmacol, 2012. 52(1 Suppl): p. 54s-62s. [CrossRef]
- Nardella, F.A. and D.C. Teller, Fc intermediate (Fci), a papain-generated fragment of human IgG, intermediate in charge, molecular weight and cleavage between the Fc and Fc' fragments of IgG. Mol Immunol, 1985. 22(6): p. 705-13. [CrossRef]
- Jevsevar, S., M. Kunstelj, and V.G. Porekar, PEGylation of therapeutic proteins. Biotechnol J, 2010. 5(1): p. 113-28. [CrossRef]
- Meibohm, B. and H. Zhou, Characterizing the Impact of Renal Impairment on the Clinical Pharmacology of Biologics. The Journal of Clinical Pharmacology, 2012. 52(S1): p. 54S-62S. [CrossRef]
- Ko, S., et al., An Fc variant with two mutations confers prolonged serum half-life and enhanced effector functions on IgG antibodies. Experimental & Molecular Medicine, 2022. 54(11): p. 1850-1861. [CrossRef]
- Kontermann, R.E., Strategies for extended serum half-life of protein therapeutics. Curr Opin Biotechnol, 2011. 22(6): p. 868-76. [CrossRef]
- Rath, T., et al., Fc-fusion proteins and FcRn: structural insights for longer-lasting and more effective therapeutics. Crit Rev Biotechnol, 2015. 35(2): p. 235-54. [CrossRef]
- Roopenian, D.C. and S. Akilesh, FcRn: the neonatal Fc receptor comes of age. Nat Rev Immunol, 2007. 7(9): p. 715-25. [CrossRef]
- Weflen, A.W., et al., Multivalent immune complexes divert FcRn to lysosomes by exclusion from recycling sorting tubules. Mol Biol Cell, 2013. 24(15): p. 2398-405. [CrossRef]
- Strohl, W.R., Fusion Proteins for Half-Life Extension of Biologics as a Strategy to Make Biobetters. BioDrugs, 2015. 29(4): p. 215-39. [CrossRef]
- Czajkowsky, D.M., et al., Fc-fusion proteins: new developments and future perspectives. EMBO Mol Med, 2012. 4(10): p. 1015-28. [CrossRef]
- Nimmerjahn, F. and J.V. Ravetch, Fcgamma receptors as regulators of immune responses. Nat Rev Immunol, 2008. 8(1): p. 34-47.
- Mekhaiel, D.N., et al., Polymeric human Fc-fusion proteins with modified effector functions. Sci Rep, 2011. 1: p. 124. [CrossRef]
- Carter, P.J., Introduction to current and future protein therapeutics: a protein engineering perspective. Exp Cell Res, 2011. 317(9): p. 1261-9. [CrossRef]
- Chen, X., J.L. Zaro, and W.C. Shen, Fusion protein linkers: property, design and functionality. Adv Drug Deliv Rev, 2013. 65(10): p. 1357-69. [CrossRef]
- Strohl, W.R., Current progress in innovative engineered antibodies. Protein Cell, 2018. 9(1): p. 86-120. [CrossRef]
- Kanje, S., et al., Chapter 2 - Engineering of Protein A for improved purification of antibodies and Fc-fused proteins, in Approaches to the Purification, Analysis and Characterization of Antibody-Based Therapeutics, A. Matte, Editor. 2020, Elsevier. p. 35-54.
- Roque, A.C., C.S. Silva, and M.A. Taipa, Affinity-based methodologies and ligands for antibody purification: advances and perspectives. J Chromatogr A, 2007. 1160(1-2): p. 44-55. [CrossRef]
- Tscheliessnig, A., et al., Ethanol precipitation for purification of recombinant antibodies. J Biotechnol, 2014. 188: p. 17-28. [CrossRef]
- Mao, L.N., et al., Downstream antibody purification using aqueous two-phase extraction. Biotechnol Prog, 2010. 26(6): p. 1662-70. [CrossRef]
- Smejkal, B., et al., Fast and scalable purification of a therapeutic full-length antibody based on process crystallization. Biotechnol Bioeng, 2013. 110(9): p. 2452-61. [CrossRef]
- Bolton, G.R. and K.K. Mehta, The role of more than 40 years of improvement in protein A chromatography in the growth of the therapeutic antibody industry. Biotechnol Prog, 2016. 32(5): p. 1193-1202. [CrossRef]
- Hober, S., K. Nord, and M. Linhult, Protein A chromatography for antibody purification. J Chromatogr B Analyt Technol Biomed Life Sci, 2007. 848(1): p. 40-7. [CrossRef]
- Deisenhofer, J., Crystallographic refinement and atomic models of a human Fc fragment and its complex with fragment B of protein A from Staphylococcus aureus at 2.9- and 2.8-A resolution. Biochemistry, 1981. 20(9): p. 2361-70.
- Tashiro, M. and G.T. Montelione, Structures of bacterial immunoglobulin-binding domains and their complexes with immunoglobulins. Curr Opin Struct Biol, 1995. 5(4): p. 471-81. [CrossRef]
- Tashiro, M., et al., High-resolution solution NMR structure of the Z domain of staphylococcal protein A. J Mol Biol, 1997. 272(4): p. 573-90. [CrossRef]
- Gagnon, P., Technology trends in antibody purification. J Chromatogr A, 2012. 1221: p. 57-70. [CrossRef]
- Hale, G., et al., Repeated cleaning of protein A affinity column with sodium hydroxide. J Immunol Methods, 1994. 171(1): p. 15-21. [CrossRef]
- Shukla, A.A. and P. Hinckley, Host cell protein clearance during protein A chromatography: development of an improved column wash step. Biotechnol Prog, 2008. 24(5): p. 1115-21. [CrossRef]
- Pabst, T.M., J. Thai, and A.K. Hunter, Evaluation of recent Protein A stationary phase innovations for capture of biotherapeutics. J Chromatogr A, 2018. 1554: p. 45-60. [CrossRef]
- Liu, Z., S.S. Mostafa, and A.A. Shukla, A comparison of protein A chromatographic stationary phases: performance characteristics for monoclonal antibody purification. Biotechnol Appl Biochem, 2015. 62(1): p. 37-47. [CrossRef]
- Hahn, R., R. Schlegel, and A. Jungbauer, Comparison of protein A affinity sorbents. J Chromatogr B Analyt Technol Biomed Life Sci, 2003. 790(1-2): p. 35-51.
- Pfaunmiller, E.L., et al., Affinity monolith chromatography: a review of principles and recent analytical applications. Anal Bioanal Chem, 2013. 405(7): p. 2133-45. [CrossRef]
- Boi, C., Membrane adsorbers as purification tools for monoclonal antibody purification. J Chromatogr B Analyt Technol Biomed Life Sci, 2007. 848(1): p. 19-27. [CrossRef]
- Farid, S.S., Process economics of industrial monoclonal antibody manufacture. J Chromatogr B Analyt Technol Biomed Life Sci, 2007. 848(1): p. 8-18. [CrossRef]
- Love, J.C., K.R. Love, and P.W. Barone, Enabling global access to high-quality biopharmaceuticals. Current Opinion in Chemical Engineering, 2013. 2(4): p. 383-390. [CrossRef]
- Withanage, T.J., et al., The [(bathophenanthroline)(3):Fe(2+)] complex as an aromatic non-polymeric medium for purification of human lactoferrin. J Chromatogr A, 2024. 1732: p. 465218. [CrossRef]
- Trudel, J. and A. Asselin, Protein purification for microsequencing by sequential native and denaturing polyacrylamide gel electrophoresis: application to one chitinase. Anal Biochem, 1994. 221(1): p. 214-6. [CrossRef]
- Smith, G.L., et al., Complexation of ferrous ions by ferrozine, 2,2'-bipyridine and 1,10-phenanthroline: Implication for the quantification of iron in biological systems. J Inorg Biochem, 2021. 220: p. 111460. [CrossRef]
- Ellman, G.L., et al., A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem Pharmacol, 1961. 7: p. 88-95. [CrossRef]
- O'Laughlin, J.W., Separation of cationic metal chelates of 1,10-phenanthroline by liquid chromatography. Analytical Chemistry, 1982. 54(2): p. 178-181. [CrossRef]
- Ng, N.S., et al., The antimicrobial efficacy and DNA binding activity of the copper(II) complexes of 3,4,7,8-tetramethyl-1,10-phenanthroline, 4,7-diphenyl-1,10-phenanthroline and 1,2-diaminocyclohexane. J Inorg Biochem, 2016. 162: p. 62-72.
- Hellawell, J.M., Biological indicators of freshwater pollution and environmental management. 2012: Springer Science & Business Media. [CrossRef]
- Dhandapani, G., et al., Nonionic detergent micelle aggregates: An economical alternative to protein A chromatography. N Biotechnol, 2021. 61: p. 90-98. [CrossRef]
- Lavery, P.E. and S.C. Kowalczykowski, Enhancement of recA protein-promoted DNA strand exchange activity by volume-occupying agents. J Biol Chem, 1992. 267(13): p. 9307-14. [CrossRef]
- Wang, X., et al., Polyethylene Glycol Crowder’s Effect on Enzyme Aggregation, Thermal Stability, and Residual Catalytic Activity. Langmuir, 2021. 37(28): p. 8474-8485.
- Jiang, M. and Z. Guo, Effects of Macromolecular Crowding on the Intrinsic Catalytic Efficiency and Structure of Enterobactin-Specific Isochorismate Synthase. Journal of the American Chemical Society, 2007. 129(4): p. 730-731. [CrossRef]
- Aoki, K., et al., A quantitative model of ERK MAP kinase phosphorylation in crowded media. Sci Rep, 2013. 3: p. 1541. [CrossRef]
- Khodabandehloo, A. and D.D.Y. Chen, Particle Sizing Methods for The Detection of Protein Aggregates in Biopharmaceuticals. Bioanalysis, 2017. 9(3): p. 313-326. [CrossRef]
- Nobbmann, U., et al., Dynamic light scattering as a relative tool for assessing the molecular integrity and stability of monoclonal antibodies. Biotechnol Genet Eng Rev, 2007. 24: p. 117-28. [CrossRef]
- Conner, C.G., et al., An accelerated antibody aggregation test based on time sequenced dynamic light scattering. Colloids and Surfaces A: Physicochemical and Engineering Aspects, 2022. 653: p. 129833.
- Paul, A.J., K. Schwab, and F. Hesse, Direct analysis of mAb aggregates in mammalian cell culture supernatant. BMC Biotechnol, 2014. 14: p. 99. [CrossRef]
- Eyer, P., et al., Molar absorption coefficients for the reduced Ellman reagent: reassessment. Analytical Biochemistry, 2003. 312(2): p. 224-227. [CrossRef]
- Smith, R.M. and A.E. Martell, Critical stability constants: volume 2: amines. 1975: Springer.
- Greenfield, N.J., Using circular dichroism spectra to estimate protein secondary structure. Nat Protoc, 2006. 1(6): p. 2876-90. [CrossRef]
- Tetin, S.Y., F.G. Prendergast, and S.Y. Venyaminov, Accuracy of protein secondary structure determination from circular dichroism spectra based on immunoglobulin examples. Analytical Biochemistry, 2003. 321(2): p. 183-187. [CrossRef]
- Sussman, J.L., et al., Atomic structure of acetylcholinesterase from Torpedo californica: a prototypic acetylcholine-binding protein. Science, 1991. 253(5022): p. 872-9.
- Ellman, G.L., Tissue sulfhydryl groups. Archives of Biochemistry and Biophysics, 1959. 82(1): p. 70-77.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




