Submitted:
17 December 2024
Posted:
18 December 2024
You are already at the latest version
Abstract
Glutaminase controls the first step in glutaminolysis, impacting bioenergetics, biosynthesis and oxidative stress balance. Two isoenzymes exist in humans, GLS and GLS2. GLS is considered prooncogenic and overexpressed in many tumours, while GLS2 may act as both prooncogenic or as a tumour suppressor. Glioblastoma cells usually lack GLS2 while express high GLS. We aimed to investigate how GLS2 expression modifies the metabolism of glioblastoma cells, looking for changes that may explain GLS2’s potential tumour suppressive role. We developed LN-229 human glioblastoma cells stably expressing GLS2 and performed isotope tracing using U-13C-glutamine and metabolomic quantification to analyse metabolic changes. Treatment with the GLS inhibitor CB-839 was also included to concomitantly inhibit endogenous GLS activity. GLS2 overexpression resulted in extensive metabolic changes, altering the TCA cycle in an isoenzyme-specific and previously unsuspected way, by upregulating part of the cycle but blocking the synthesis of the 6-carbon intermediates from acetyl-CoA. Expression of GLS2 caused downregulation of PDH activity through phosphorylation of S293 of PDHA1. GLS2 also altered nucleotide levels as well as induced the accumulation of methylated metabolites and methyl donor S-adenosyl methionine. These changes suggest that GLS2 may be a key regulator linking glutamine and glucose metabolism, also impacting nucleotides and epigenetics.
Keywords:
1. Introduction
2. Results
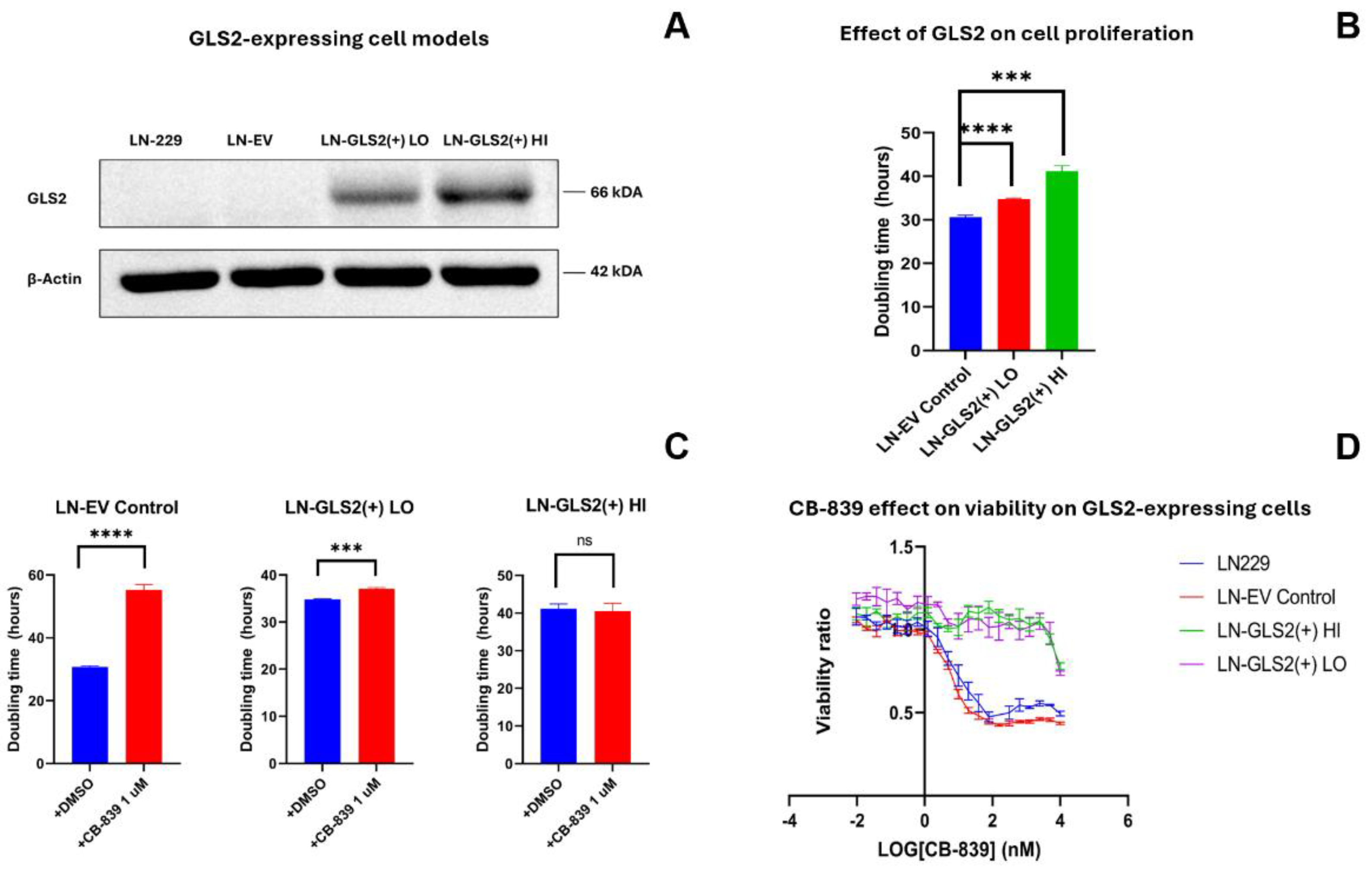
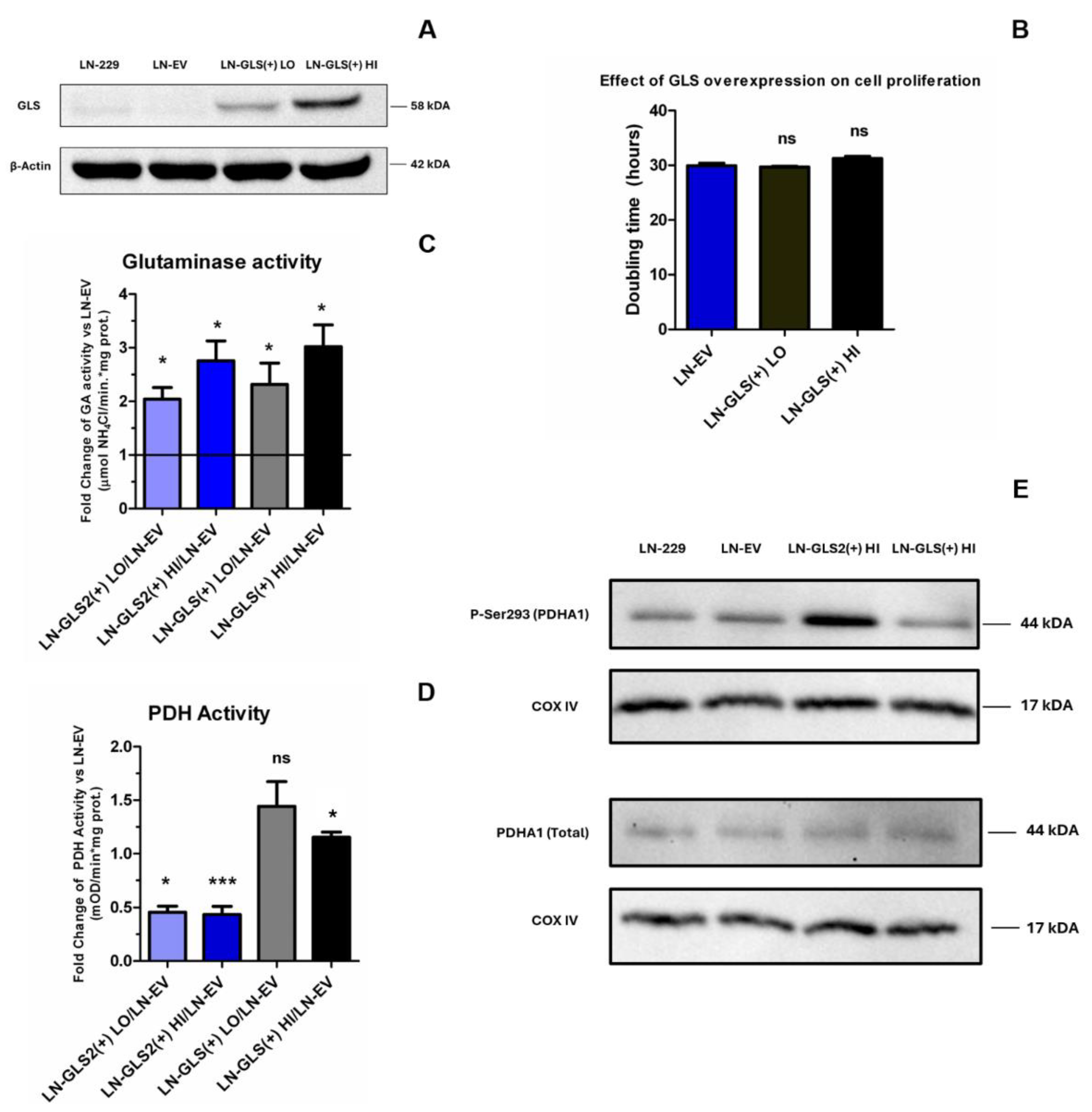
2.1. GLS2 Overexpression Slows Cell Proliferation in Human LN-229 Cell Line and Confers Resistance to CB-839 Treatment
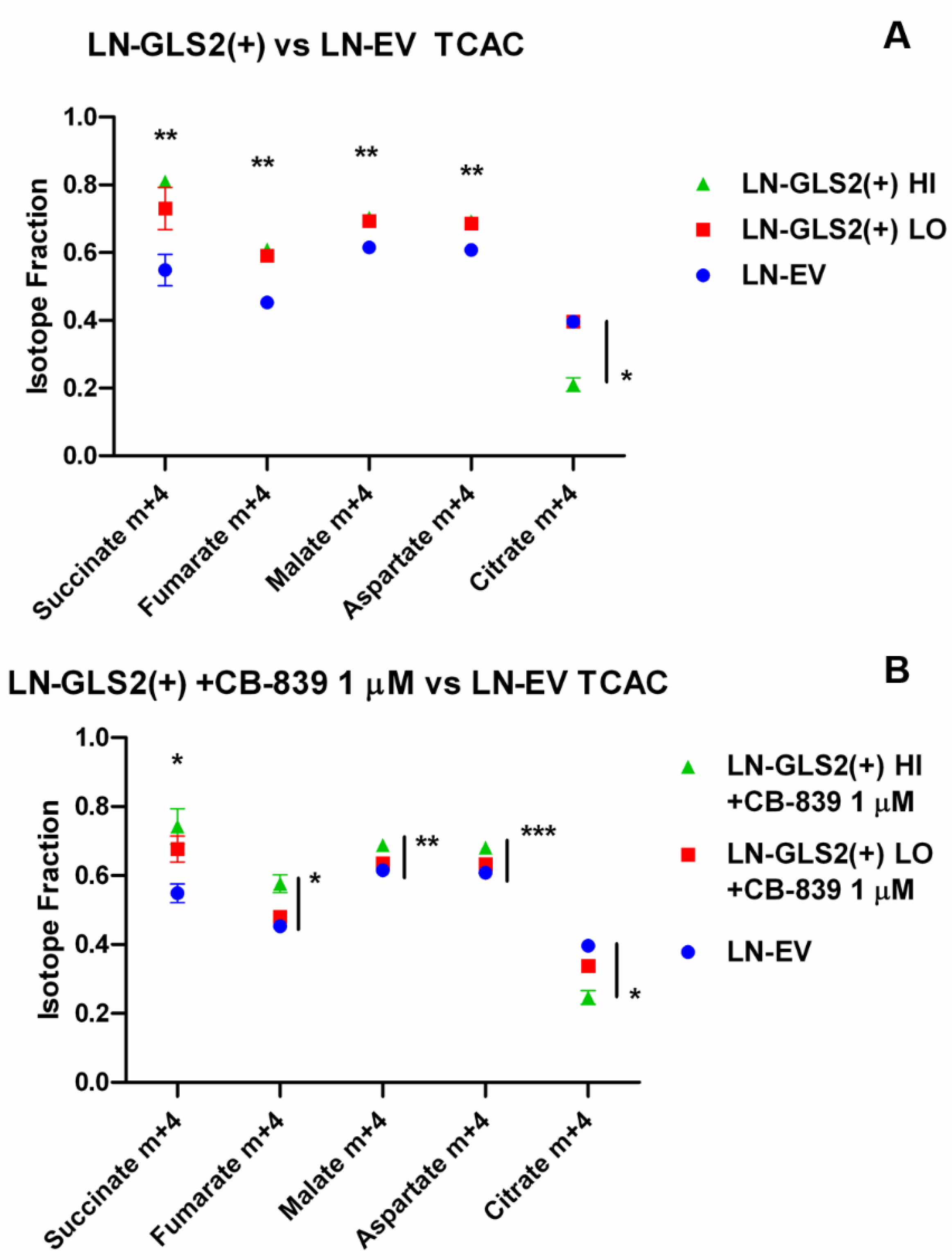
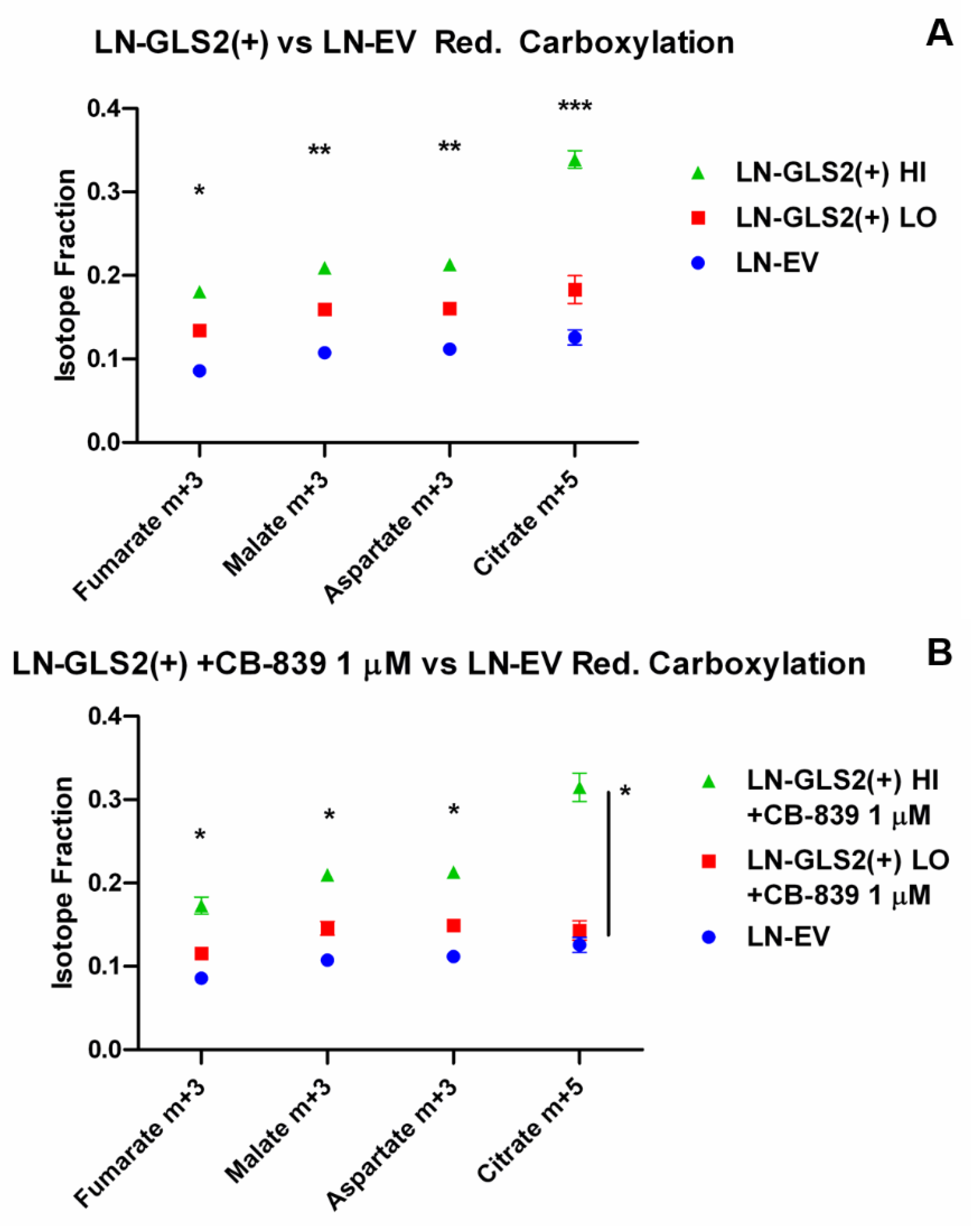
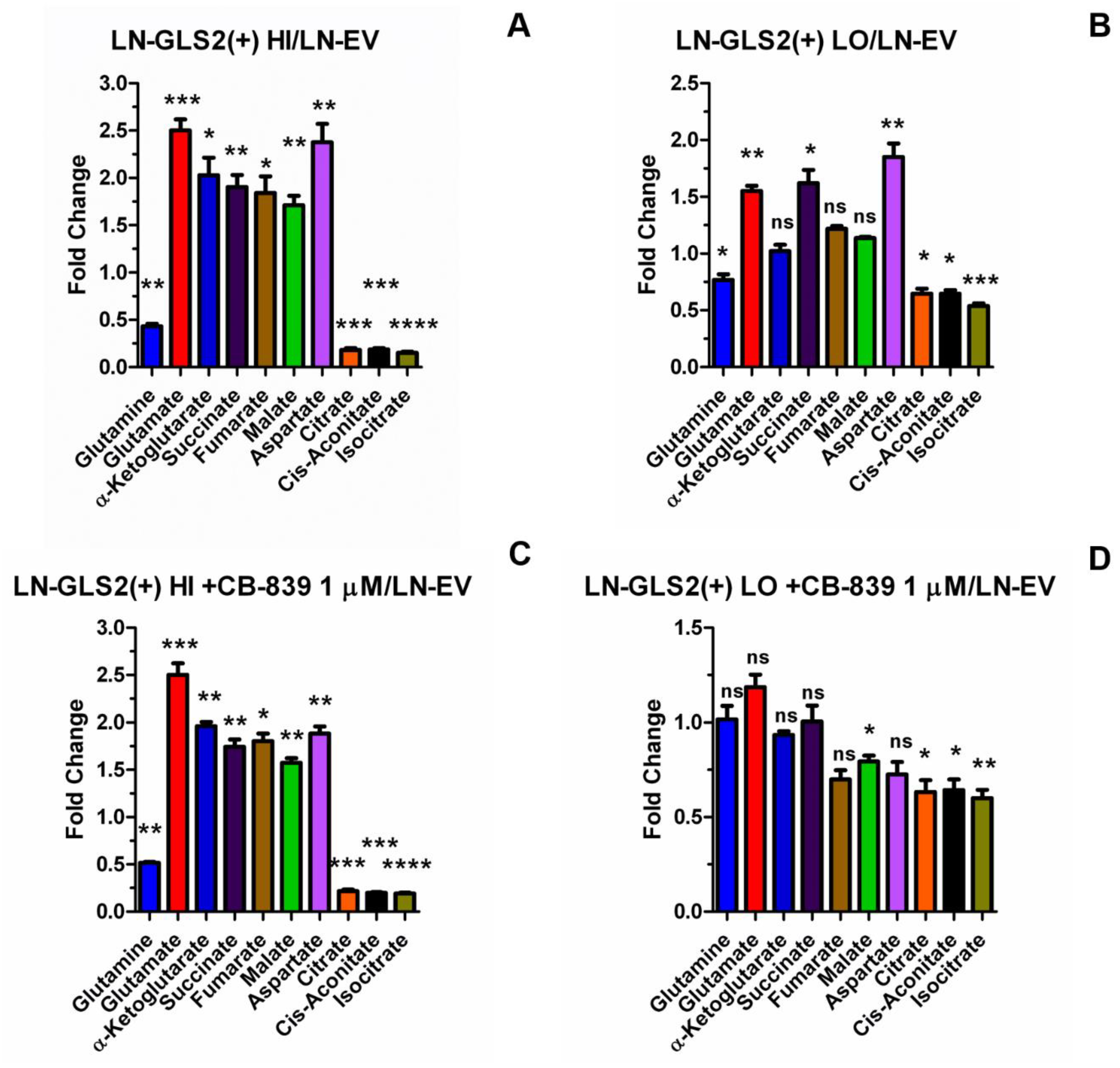
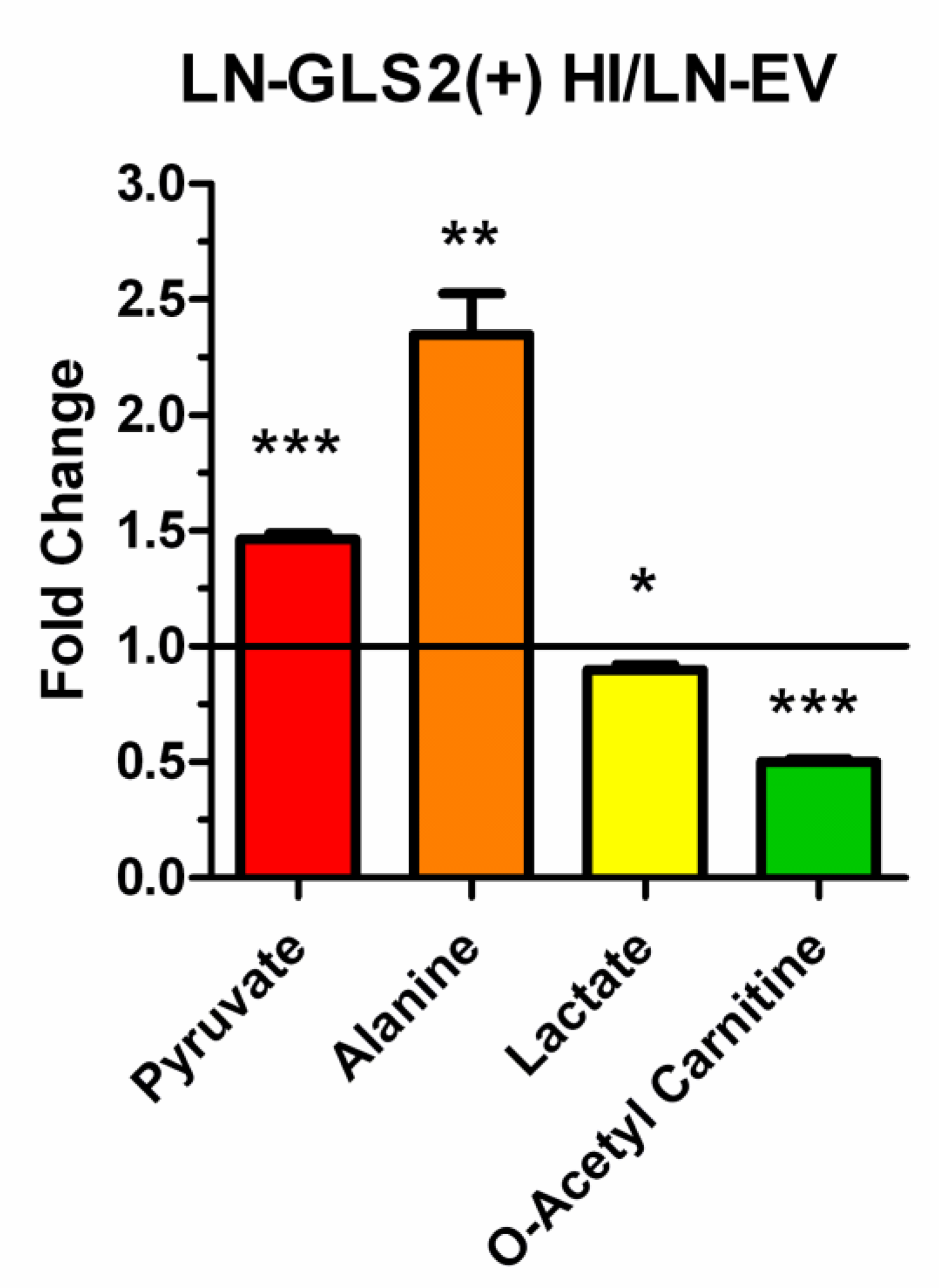
2.2. GLS2 Alters Both Oxidative Decarboxylation and Reductive Carboxylation of Gln-Derived AKG in the TCA Cycle
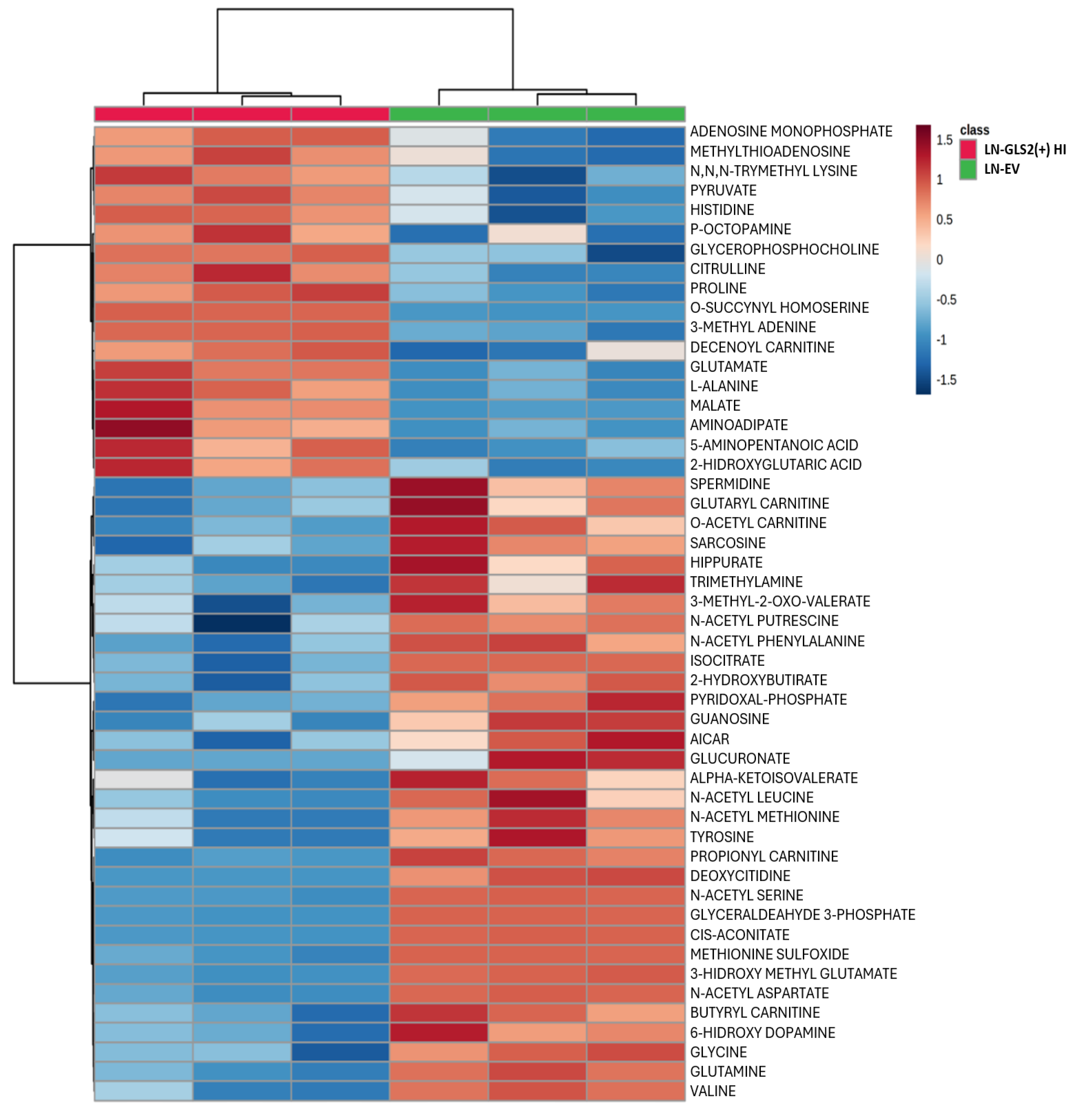
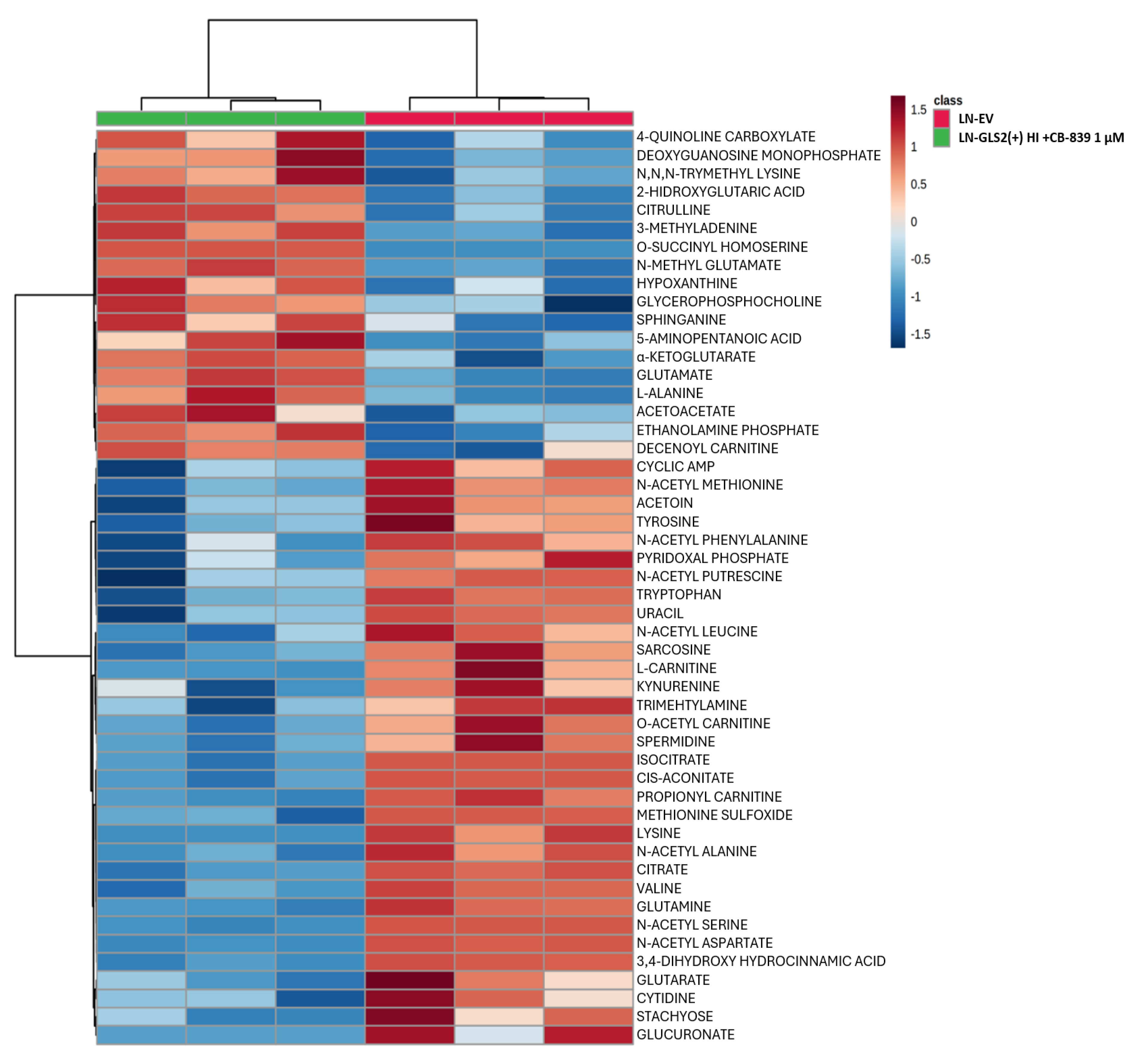
2.3. Effect of GLS2 Overexpression on the Metabolome
2.4. GLS2 Reshapes Key Metabolic Pathways in Human GBM Cells
2.5. GLS2 Modulates PDH Activity and Phosphorylation Pattern
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Cell Lines, Culture Conditions and Stable Transfections
4.3. Cell Proliferation and Viability Assays
4.4. Metabolomics
4.5. [U-13C]Glutamine Tracing Experiments
4.6. Protein Expression and Mitochondria Isolation
4.7. Glutaminase Enzymatic Activity
4.8. PDH Enzymatic Activity
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–74. [Google Scholar] [CrossRef] [PubMed]
- Cairns, R.A.; Mak, T.W. Lung cancer resets the liver’s metabolic clock. Cell Metab 2016, 23, 767–9. [Google Scholar] [CrossRef] [PubMed]
- DeBerardinis, R.J.; Chandel, N.S. Fundamentals of cancer metabolism. Sci Adv 2016, 2, e1600200. [Google Scholar] [CrossRef] [PubMed]
- Faubert, B.; Solmonson, A.; DeBerardinis, R.J. Metabolic reprogramming and cancer progression. Science 2020, 368, eaaw5473. [Google Scholar] [CrossRef] [PubMed]
- Bott, A.J.; Maimouni, S.; Zong, W.X. The pleiotropic effects of glutamine metabolism in cancer. Cancers (Basel) 2019, 11, 770. [Google Scholar] [CrossRef]
- Hensley, C.T.; Wasti, A.T.; DeBerardinis, R.J. Glutamine and cancer: cell biology, physiology, and clinical opportunities. J Clin Invest 2013, 123, 3678–84. [Google Scholar] [CrossRef] [PubMed]
- DeBerardinis, R.J.; Cheng, T. Q's next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene 2010, 29, 313–24. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Copeland, C.; Le, A. Glutamine Metabolism in Cancer. Adv Exp Med Biol 2021, 1311, 17–38. [Google Scholar] [PubMed]
- Tong, X.; Zhao, F.; Thompson, C.B. The molecular determinants of de novo nucleotide biosynthesis in cancer cells. Curr Opin Genet Dev 2009, 19, 32–7. [Google Scholar] [CrossRef]
- Mullen, A.R.; Wheaton, W.W.; Jin, E.S.; Chen, P.H.; Sullivan, L.B.; Cheng, T.; et al. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature 2011, 481, 385–8. [Google Scholar] [CrossRef] [PubMed]
- De Los Santos-Jiménez, J.; Campos-Sandoval, J.A.; Alonso, F.J.; Márquez, J.; Matés, J.M. GLS and GLS2 Glutaminase Isoenzymes in the Antioxidant System of Cancer Cells. Antioxidants (Basel) 2024, 13, 745. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, N.N.; Zhu, J.; Thompson, C.B. The hallmarks of cancer: still emerging. Cell Metab 2022, 34, 355–77. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: a summary. Neuro Oncol 2021, 23, 1231–51. [Google Scholar] [CrossRef]
- Samad, A.; Samant, R.; Venkateshwara Rao, K.; Bhargava, V.; Sadique, S.I.; Yadav, R. Oxaloacetate as a Holy Grail Adjunctive Treatment in Gliomas: A Revisit to Metabolic Pathway. Cureus 2023, 15, e48821. [Google Scholar] [CrossRef] [PubMed]
- Esemen, Y.; Awan, M.; Parwez, R.; Baig, A.; Rahman, S.; Masala, I.; et al. Molecular Pathogenesis of Glioblastoma in Adults and Future Perspectives: A Systematic Review. Int J Mol Sci 2022, 23, 2607. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol 2016, 131, 803–20. [Google Scholar] [CrossRef] [PubMed]
- Campos-Sandoval, J.A.; Gómez-García, M.C.; Santos-Jiménez, J.L.; Matés, J.M.; Alonso, F.J.; Márquez, J. Antioxidant responses related to temozolomide resistance in glioblastoma. Neurochem Int 2021, 149, 105136. [Google Scholar] [CrossRef]
- Dang, L.; White, D.W.; Gross, S.; Bennett, B.D.; Bittinger, M.A.; Driggers, E.M.; et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 2009, 462, 739–44. [Google Scholar] [CrossRef] [PubMed]
- An, Z.; Aksoy, O.; Zheng, T.; Fan, Q.W.; Weiss, W.A. Epidermal growth factor receptor and EGFRvIII in glioblastoma: signaling pathways and targeted therapies. Oncogene 2018, 37, 1561–1575. [Google Scholar] [CrossRef]
- Obara-Michlewska, M.; Szeliga, M. Targeting Glutamine Addiction in Gliomas. Cancers (Basel) 2020, 12, 310. [Google Scholar] [CrossRef] [PubMed]
- Szeliga, M.; Obara-Michlewska, M. Glutamine in neoplastic cells: focus on the expression and roles of glutaminases. Neurochem Int 2009, 55, 71–5. [Google Scholar] [CrossRef]
- Márquez, J.; Matés, J.M.; Campos-Sandoval, J.A. Glutaminases. Adv Neurobiol 2016, 13, 133–71. [Google Scholar] [PubMed]
- Maley, J.A.; Davidson, J.N. Identification of the junction between the glutamine amidotransferase and carbamyl phosphate synthetase domains of the mammalian CAD protein. Biochem Biophys Res Commun 1988, 154, 1047–53. [Google Scholar] [CrossRef]
- Muir, A.; Danai, L.V.; Gui, D.Y.; Waingarten, C.Y.; Lewis, C.A.; Vander Heiden, M.G. Environmental cystine drives glutamine anaplerosis and sensitizes cancer cells to glutaminase inhibition. Elife 2017, 6, e27713. [Google Scholar] [CrossRef] [PubMed]
- Márquez, J.; Alonso, F.J.; Matés, J.M.; Segura, J.A.; Martín-Rufián, M.; Campos-Sandoval, J.A. Glutamine addiction in gliomas. Neurochem Res 2017, 42, 1735–46. [Google Scholar] [CrossRef] [PubMed]
- Matés, J.M. , Campos-Sandoval, J.A., Santos-Jiménez, J.L., Márquez, J. Dysregulation of glutaminase and glutamine synthetase in cancer. Cancer Lett 2019, 467, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Matés, J.M.; Campos-Sandoval, J.A.; Márquez, J. Glutaminase isoenzymes in the metabolic therapy of cancer. Biochim Biophys Acta Rev Cancer 2018, 1870, 158–64. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Tchernyshyov, I.; Chang, T.C.; Lee, Y.S.; Kita, K.; Ochi, T.; et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 2009, 458, 762–5. [Google Scholar] [CrossRef]
- Suzuki, S.; Tanaka, T.; Poyurovsky, M.V.; Nagano, H.; Mayama, T.; Ohkubo, S.; et al. Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc Natl Acad Sci U S A 2010, 107, 7461–6. [Google Scholar] [CrossRef] [PubMed]
- Matés, J.M.; Di Paola, F.J.; Campos-Sandoval, J.A. Mazurek, S.; Márquez, J. Therapeutic targeting of glutaminolysis as an essential strategy to combat cancer. Semin Cell Dev Biol 2020, 98, 34–43. [Google Scholar] [CrossRef]
- Katt, W.P.; Lukey, M.J.; Cerione, R.A. A tale of two glutaminases: homologous enzymes with distinct roles in tumorigenesis. Future Med Chem 2017, 9, 223–43. [Google Scholar] [CrossRef]
- Szeliga, M.; Bogacińska-Karaś, M.; Kuźmicz, K.; Rola, R.; Albrecht, J. Downregulation of GLS2 in glioblastoma cells is related to DNA hypermethylation but not to the p53 status. Mol Carcinog 2016, 55, 1309–16. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, C.; Chen, M.; Cao, J.; Zhong, Y.; Chen, L.; et al. Epigenetic silencing of glutaminase 2 in human liver and colon cancers. BMC Cancer 2013, 13, 601. [Google Scholar] [CrossRef] [PubMed]
- Koch, K.; Hartmann, R.; Tsiampali, J.; Uhlmann, C.; Nickel, A.C.; He, X.; et al. A comparative pharmaco-metabolomic study of glutaminase inhibitors in glioma stem-like cells confirms biological effectiveness but reveals differences in target-specificity. Cell Death Discov 2020, 6, 20. [Google Scholar] [CrossRef] [PubMed]
- De Los Santos-Jiménez, J.; Rosales, T.; Ko, B.; Campos-Sandoval, JA.; Alonso, F.J.; Márquez, J.; et al. Metabolic Adjustments following Glutaminase Inhibition by CB-839 in Glioblastoma Cell Lines. Cancers (Basel) 2023, 15, 531. [Google Scholar] [CrossRef]
- Majewska, E.; Márquez, J.; Albrecht, J.; Szeliga, M. Transfection with GLS2 Glutaminase (GAB) Sensitizes Human Glioblastoma Cell Lines to Oxidative Stress by a Common Mechanism Involving Suppression of the PI3K/AKT Pathway. Cancers (Basel) 2019, 11, 115. [Google Scholar] [CrossRef] [PubMed]
- López de la Oliva, A.R.; Campos-Sandoval, J.A.; Gómez-García, M.C.; Cardona, C.; Martín-Rufián, M.; Sialana, F.J.; et al. Nuclear Translocation of Glutaminase GLS2 in Human Cancer Cells Associates with Proliferation Arrest and Differentiation. Sci Rep 2020, 10, 2259. [Google Scholar] [CrossRef]
- Kuo, T.C.; Chen, C.K.; Hua, K.T.; Yu, P.; Lee, W.J.; Chen, M.W.; et al. Glutaminase 2 stabilizes Dicer to repress Snail and metastasis in hepatocellular carcinoma cells. Cancer Lett 2016, 383, 282–94. [Google Scholar] [CrossRef]
- Phang, J.M.; Liu, W.; Hancock, C. Bridging epigenetics and metabolism: role of non-essential amino acids. Epigenetics 2013, 8, 231–6. [Google Scholar] [CrossRef]
- Melone, M.A.B.; Valentino, A.; Margarucci, S.; Galderisi, U.; Giordano, A.; Peluso, G. The carnitine system and cancer metabolic plasticity. Cell Death Dis 2018, 9, 228. [Google Scholar] [CrossRef] [PubMed]
- Pietrocola, F.; Galluzzi, L.; Bravo-San Pedro, J.M.; Madeo, F.; Kroemer, G. Acetyl coenzyme A: a central metabolite and second messenger. Cell Metab 2015, 21, 805–21. [Google Scholar] [CrossRef] [PubMed]
- Ree, R.; Varland, S.; Arnesen, T. Spotlight on protein N-terminal acetylation. Exp Mol Med 2018, 50, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Patel, M.S.; Korotchkina, L.G. Regulation of the pyruvate dehydrogenase complex. Biochem Soc Trans 2006, 34, 217–22. [Google Scholar] [CrossRef]
- Saunier, E.; Benelli, C.; Bortoli, S. The pyruvate dehydrogenase complex in cancer: An old metabolic gatekeeper regulated by new pathways and pharmacological agents. Int J Cancer 2016, 138, 809–17. [Google Scholar] [CrossRef]
- Porporato, P.E.; Filigheddu, N.; Pedro, J.M.B.; Kroemer, G.; Galluzzi, L. Mitochondrial metabolism and cancer. Cell Res 2018, 28, 265–280. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Shen, X.; Yan, Y.; Li, H. Pyruvate dehydrogenase kinases (PDKs): an overview toward clinical applications. Biosci Rep 2021, 41, BSR20204402. [Google Scholar] [CrossRef] [PubMed]
- Linn, T.C.; Pettit, F.H.; Reed, L.J. Alpha-keto acid dehydrogenase complexes. X. Regulation of the activity of the pyruvate dehydrogenase complex from beef kidney mitochondria by phosphorylation and dephosphorylation. Proc Natl Acad Sci U S A 1969, 62, 234–41. [Google Scholar] [CrossRef]
- Popov, K.M.; Kedishvili, N.Y.; Zhao, Y.; Shimomura, Y.; Crabb, D.W.; Harris, R.A. Primary structure of pyruvate dehydrogenase kinase establishes a new family of eukaryotic protein kinases. J Biol Chem 1993, 268, 26602–6. [Google Scholar] [CrossRef]
- Korotchkina, L.G.; Patel, M.S. Site specificity of four pyruvate dehydrogenase kinase isoenzymes toward the three phosphorylation sites of human pyruvate dehydrogenase. J Biol Chem 2001, 276, 37223–9. [Google Scholar] [CrossRef] [PubMed]
- Korotchkina, L.G.; Patel, M.S. Mutagenesis studies of the phosphorylation sites of recombinant human pyruvate dehydrogenase. Site-specific regulation. J Biol Chem 1995, 270, 14297–304. [Google Scholar] [CrossRef]
- Echeverri Ruiz, N.P.; Mohan, V.; Wu, J.; Scott, S.; Kreamer, M.; Benej, M.; et al. Dynamic regulation of mitochondrial pyruvate metabolism is necessary for orthotopic pancreatic tumor growth. Cancer Metab 2021, 9, 39. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.M.; Watford, M. Molecular cloning of a cDNA for rat hepatic glutaminase. Sequence similarity to kidney-type glutaminase. J Biol Chem 1990, 265, 10631–6. [Google Scholar] [CrossRef]
- Campos-Sandoval, J.A.; Martín-Rufián, M.; Cardona, C.; Lobo, C.; Peñalver, A.; Márquez, J. Glutaminases in brain: Multiple isoforms for many purposes. Neurochem Int 2015, 88, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.T.; Chacko, S.K.; Sunehag, A.L.; Haymond, M.W. Measurements of Gluconeogenesis and Glycogenolysis: A Methodological Review. Diabetes 2015, 64, 3996–4010. [Google Scholar] [CrossRef]
- Wyatt, M.D.; Allan, J.M.; Lau, A.Y.; Ellenberger, T.E.; Samson, L.D. 3-methyladenine DNA glycosylases: structure, function, and biological importance. Bioessays 1999, 21, 668–76. [Google Scholar] [CrossRef]
- Binda, O. On your histone mark, SET, methylate! Epigenetics 2013, 8, 457–63. [Google Scholar] [CrossRef]
- Maas, M.N.; Hintzen, J.C.J.; Porzberg, M.R.B.; Mecinović, J. Trimethyllysine: From carnitine biosynthesis to epigenetics. Int J Mol Sci 2020, 21, 9451. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lv, J.Y.; Shi, J.F.; Yang, M.; Liu, S.H.; Li, Z.W.; et al. Targeting the raft-associated Akt signaling in hepatocellular carcinoma. Biomed Res Int 2014, 2014, 836025. [Google Scholar] [CrossRef]
- De los Santos-Jiménez, J.; Campos-Sandoval, J.A.; Márquez-Torres, C.; Urbano-Polo, N.; Brøndegaard, D.; Martín-Rufián, M.; et al. Glutaminase isoforms expression switches microRNA levels and oxidative status in glioblastoma cells. J Biomed Sci 2021, 28, 14. [Google Scholar] [CrossRef] [PubMed]
- Martín-Rufián, M.; Nascimento-Gomes, R.; Higuero, A.; Crisma, A.R. .; Campos-Sandoval, J.A.; Gómez-García, M.C.; et al. Both GLS silencing and GLS2 overexpression synergize with oxidative stress against proliferation of glioma cells. J Mol Med (Berl) 2014, 92, 277–90. [Google Scholar] [CrossRef] [PubMed]
- Cerniglia, G.J.; Dey, S.; Gallagher-Colombo, S.M.; Daurio, N.A.; Tuttle, S.; Busch, TM; et al. The PI3K/Akt Pathway Regulates Oxygen Metabolism via Pyruvate Dehydrogenase (PDH)-E1α Phosphorylation. Mol Cancer Ther 2015, 14, 1928–38. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Hu, Y.Z. PI3K/Akt/mTOR pathway inhibitors in cancer: a perspective on clinical progress. Curr Med Chem 2010, 17, 4326–41. [Google Scholar] [CrossRef] [PubMed]
- Chiang, P.K.; Gordon, R.K.; Tal, J.; Zeng, G.C.; Doctor, B.P.; Pardhasaradhi, K.; et al. S-adenosylmethionine and methylation. FASEB J 1996, 10, 471–80. [Google Scholar] [CrossRef] [PubMed]
- McGee, M.M.; Knox, W.E. Phosphate-activated isoenzymes of glutaminase in rat liver and kidney. Enzyme 1971, 12, 618–21. [Google Scholar] [CrossRef] [PubMed]
- Heini, H.G.; Gebhardt, R.; Brecht, A.; Mecke, D. Purification and characterization of rat liver glutaminase. Eur J Biochem 1987, 162, 541–6. [Google Scholar] [CrossRef]
- Shukla, S.K.; Purohit, V.; Mehla, K.; Gunda, V.; Chaika, N.V.; Vernucci, E; et al. MUC1 and HIF-1alpha Signaling Crosstalk Induces Anabolic Glucose Metabolism to Impart Gemcitabine Resistance to Pancreatic Cancer. Cancer Cell 2017, 32, 71–87.e7. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




