Submitted:
02 December 2024
Posted:
03 December 2024
You are already at the latest version
Abstract
Keywords:
Introduction
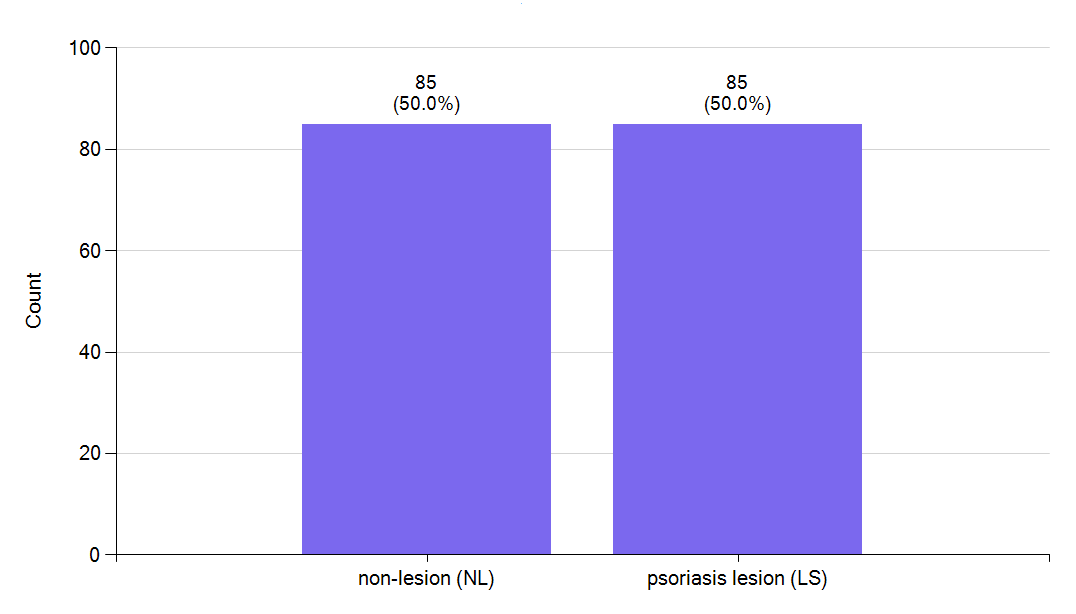
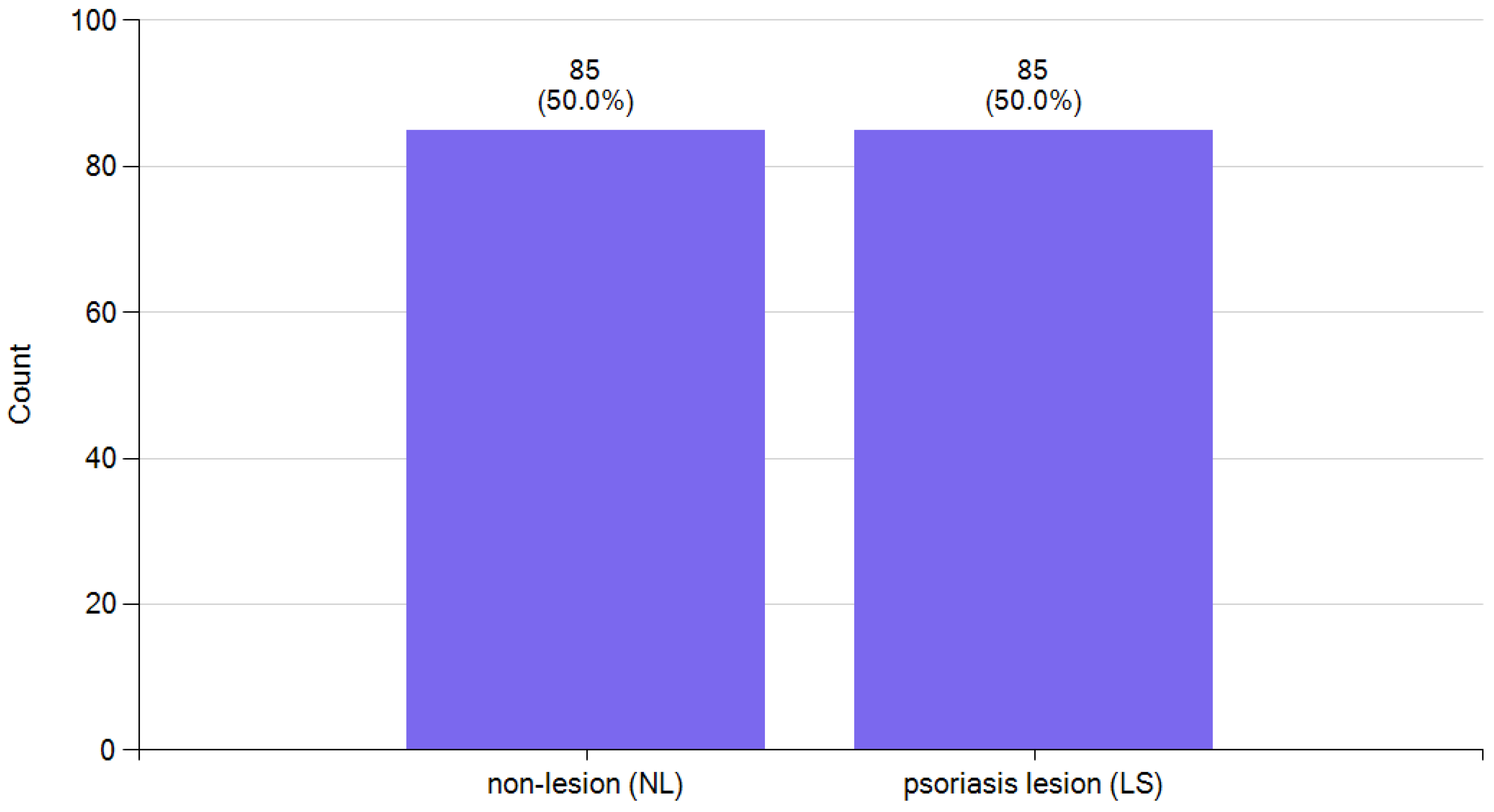
Data
Overexpressed Genes in Psoriasis and LLM-Guided Natural Treatments
Discussion
Summary
References
- Wilsmann-Theis, D.; Wagenpfeil, J.; Holzinger, D.; Roth, J.; Koch, S.; Schnautz, S.; Bieber, T.; Wenzel, J. Among the S100 proteins, S100A12 is the most significant marker for psoriasis disease activity. J Eur Acad Dermatol Venereol. 2016, 30, 1165–1170. [Google Scholar] [CrossRef] [PubMed]
- Kantaputra, P.; Daroontum, T.; Chuamanochan, M.; Chaowattanapanit, S.; Kiratikanon, S.; Choonhakarn, C.; Intachai, W.; Olsen, B.; Tongsima, S.; Ngamphiw, C.; Pontisso, P.; Cox, T.C.; Ounjai, P. SERPINB3, Adult-Onset Immunodeficiency, and Generalized Pustular Psoriasis. Genes (Basel). 2023, 14, 266. [Google Scholar] [CrossRef]
- Batycka-Baran, A.; Hattinger, E.; Zwicker, S.; Summer, B.; Zack Howard, O.M.; Thomas, P.; Szepietowski, J.C.; Ruzicka, T.; Prinz, J.C.; Wolf, R. Leukocyte-derived koebnerisin (S100A15) and psoriasin (S100A7) are systemic mediators of inflammation in psoriasis. J Dermatol Sci. 2015, 79, 214–221. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Wang, Y.; Zhang, M.; Duan, Q.; Chen, C.; Sun, Q.; Liu, M.; Zheng, Y.; Shao, Y. Kynureninase contributes to the pathogenesis of psoriasis through pro-inflammatory effect. J Cell Physiol. 2022, 237, 1044–1056. [Google Scholar] [CrossRef]
- Jansen, P.A.; Kamsteeg, M.; Rodijk-Olthuis, D.; van Vlijmen-Willems, I.M.; de Jongh, G.J.; Bergers, M.; Tjabringa, G.S.; Zeeuwen, P.L.; Schalkwijk, J. Expression of the vanin gene family in normal and inflamed human skin: induction by proinflammatory cytokines. J Invest Dermatol. 2009, 129, 2167–2174. [Google Scholar] [CrossRef] [PubMed]
- Dubertret, L.; Bertaux, B.; Fosse, M.; Touraine, R. Psoriasis: a defect in the regulation of epidermal proteases, as shown by serial biopsies after cantharidin application. Br J Dermatol. 1984, 110, 405–410. [Google Scholar] [CrossRef]
- Yin, X.; Yang, Z.; Zhu, M.; Chen, C.; Sun, Q. Role of the long non-coding RNA, SPRR2C, based on an in vitro psoriatic keratinocyte cell model. Eur J Dermatol. 2022, 32, 171–180. [Google Scholar] [PubMed]
- Huang, Y.Z.; Zheng, Y.X.; Zhou, Y.; Xu, F.; Cui, Y.Z.; Chen, X.Y.; Wang, Z.Y.; Yan, B.X.; Zheng, M.; Man, X.Y. OAS1, OAS2, and OAS3 Contribute to Epidermal Keratinocyte Proliferation by Regulating Cell Cycle and Augmenting IFN-1‒Induced Jak1‒Signal Transducer and Activator of Transcription 1 Phosphorylation in Psoriasis. J Invest Dermatol. 2022, 142, 2635–2645.e9. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Lu, C.; Liu, H.; Wang, M.; Zhao, H.; Yan, Y.; Han, L. Quercetin ameliorates imiquimod-induced psoriasis-like skin inflammation in mice via the NF-κB pathway. Int Immunopharmacol. 2017, 48, 110–117. [Google Scholar] [CrossRef]
- Marko, M.; Pawliczak, R. Resveratrol and Its Derivatives in Inflammatory Skin Disorders-Atopic Dermatitis and Psoriasis: A Review. Antioxidants (Basel). 2023, 12, 1954. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, J.; Liu, L.; Sun, X.; Zhou, Y.; Chen, S.; Lu, Y.; Cai, X.; Hu, M.; Yan, G.; Miao, X.; Li, X. Efficacy and safety of curcumin in psoriasis: preclinical and clinical evidence and possible mechanisms. Front Pharmacol. 2022, 13, 903160. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Meng, X.; Song, Z. Homocysteine and psoriasis. Biosci Rep. 2019, 39, BSR20190867. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Huang, Z.; Xuan, J.; Yang, L.; Zhao, T.; Peng, W. Anti-Psoriatic Activity of Black, Green and White Tea Extracts from Southeastern China. Molecules. 2024, 29, 1279. [Google Scholar] [CrossRef] [PubMed]
- Balbás, G.M.; Regaña, M.S.; Millet, P.U. Study on the use of omega-3 fatty acids as a therapeutic supplement in treatment of psoriasis. Clin Cosmet Investig Dermatol. 2011, 4, 73–77. [Google Scholar] [CrossRef]
- Togni, S.; Maramaldi, G.; Di Pierro, F.; Biondi, M. A cosmeceutical formulation based on boswellic acids for the treatment of erythematous eczema and psoriasis. Clin Cosmet Investig Dermatol. 2014, 7, 321–327. [Google Scholar]
- Kamel, N.M.; El-Sayed, S.S.; El-Said, Y.A.M.; El-Kersh, D.M.; Hashem, M.M.; Mohamed, S.S. Unlocking milk thistle's anti-psoriatic potential in mice: Targeting PI3K/AKT/mTOR and KEAP1/NRF2/NF-κB pathways to modulate inflammation and oxidative stress. Int Immunopharmacol. 2024, 139, 112781. [Google Scholar] [CrossRef]
- Balkrishna, A.; Nain, P.; Chauhan, A.; Sharma, N.; Gupta, A.; Ranjan, R.; Varshney, A. Super Critical Fluid Extracted Fatty Acids from Withania somnifera Seeds Repair Psoriasis-Like Skin Lesions and Attenuate Pro-Inflammatory Cytokines (TNF-α and IL-6) Release. Biomolecules. 2020, 10, 185. [Google Scholar] [CrossRef]
- Mikkelsen, K.; Dargahi, N.; Fraser, S.; Apostolopoulos, V. High-Dose Vitamin B6 (Pyridoxine) Displays Strong Anti-Inflammatory Properties in Lipopolysaccharide-Stimulated Monocytes. Biomedicines. 2023, 11, 2578. [Google Scholar] [CrossRef]
- Ballester, P.; Cerdá, B.; Arcusa, R.; Marhuenda, J.; Yamedjeu, K.; Zafrilla, P. Effect of Ginger on Inflammatory Diseases. Molecules. 2022, 27, 7223. [Google Scholar] [CrossRef]
- Kumar, V.; Mangla, B.; Javed, S.; Ahsan, W.; Kumar, P.; Garg, V.; Dureja, H. Bromelain: a review of its mechanisms, pharmacological effects and potential applications. Food Funct. 2023, 14, 8101–8128. [Google Scholar] [CrossRef]
- Syed, T.A.; Ahmad, S.A.; Holt, A.H.; Ahmad, S.A.; Ahmad, S.H.; Afzal, M. Management of psoriasis with Aloe vera extract in a hydrophilic cream: a placebo-controlled, double-blind study. Trop Med Int Health. 1996, 1, 505–509. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.H.; Oh, I.; Kim, J.H.; Jeon, J.E.; Jeon, B.; Shin, J.; Kim, T.Y. Anti-inflammatory activity of compounds isolated from Astragalus sinicus L. in cytokine-induced keratinocytes and skin. Exp Mol Med. 2014, 46, e87. [Google Scholar] [CrossRef] [PubMed]
| Overexpressed Genes in Psoriasis and Natural Treatments by ChatGPT | ||
| Gene | Natural substance | Mechanism of Action |
| S100A12 | Quercetin | A flavonoid with strong anti-inflammatory effects, known to inhibit the S100 family of proteins |
| SERPINB4 | Resveratrol | Known for its anti-inflammatory and antioxidant properties, which may downregulate serpin family proteins |
| S100A7A | Curcumin | The active component of turmeric, which has anti-inflammatory effects and modulates S100 protein activity |
| TCN1 | Folic Acid and Vitamin B12 | Both vitamins, important for modulating the immune system, have been linked to psoriasis management |
| KYNU | Green Tea Extract | Epigallocatechin gallate from green tea inhibits kynurenine pathways and reduces inflammation |
| VNN3P | Omega-3 Fatty Acids | Fish oil has anti-inflammatory properties and may influence genes involved in immune responses |
| TMPRSS11D | N-Acetylcysteine | Known to modulate inflammation and may influence serine protease activity |
| SERPINB3 | Boswellia | Shown to reduce inflammation by modulating immune pathways, potentially affecting serpin expression |
| SPRR2C | Milk Thistle | Known for its anti-inflammatory and antioxidant properties, which may influence skin barrier-related genes |
| OASL | Ashwagandha | An adaptogen that modulates the immune system and reduces stress, potentially influencing OASL expression |
| Overexpressed Genes in Psoriasis and Natural Treatments by Gemini | ||
| Gene | Natural substance | Mechanism of Action |
| S100A12 | Curcumin | Found in turmeric, with anti-inflammatory and antioxidant properties that may downregulate S100A12 expression |
| SERPINB4 | Green tea extract | Rich in epigallocatechin gallate, this extract has been shown to inhibit SERPINB4 expression in vitro |
| S100A7A | Boswellia serrata | An Ayurvedic herb, boswellia serrata contains boswellic acids that have anti-inflammatory effects and may reduce S100A7A expression |
| TCN1 | Quercetin | A flavonoid found in onions, apples, and berries, quercetin has antioxidant and anti-inflammatory properties that may downregulate TCN1 expression |
| KYNU | Vitamin B6 | A cofactor for enzymes involved in tryptophan metabolism, which can regulate KYNU expression |
| VNN3P | Ginger | Ginger contains gingerols, which have anti-inflammatory and antioxidant properties that may reduce VNN3P expression |
| TMPRSS11D | Resveratrol | A polyphenol found in grapes and red wine, resveratrol has been shown to inhibit TMPRSS11D expression in vitro |
| SERPINB3 | Bromelain | An enzyme found in pineapple, bromelain has anti-inflammatory and fibrinolytic properties that may downregulate SERPINB3 expression |
| SPRR2C | Aloe vera | Aloe vera contains aloin, which has anti-inflammatory and wound-healing properties that may reduce SPRR2C expression |
| OASL | Astragalus | An adaptogenic herb, astragalus contains saponins that have antioxidant and immune-modulating properties that may downregulate OASL expression |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





