Submitted:
28 November 2024
Posted:
29 November 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials & Methods
2.1. Description of Sampling Method, Bacterial Isolation and Growth Condition
2.2. Antimicrobial Susceptibility Assays
2.3. Enterococci Speciation
2.4. Detection and Identification of Resistance Genes
2.5. Insertion Sequence Detection
2.6. Statistical Analysis of Data
3. Results
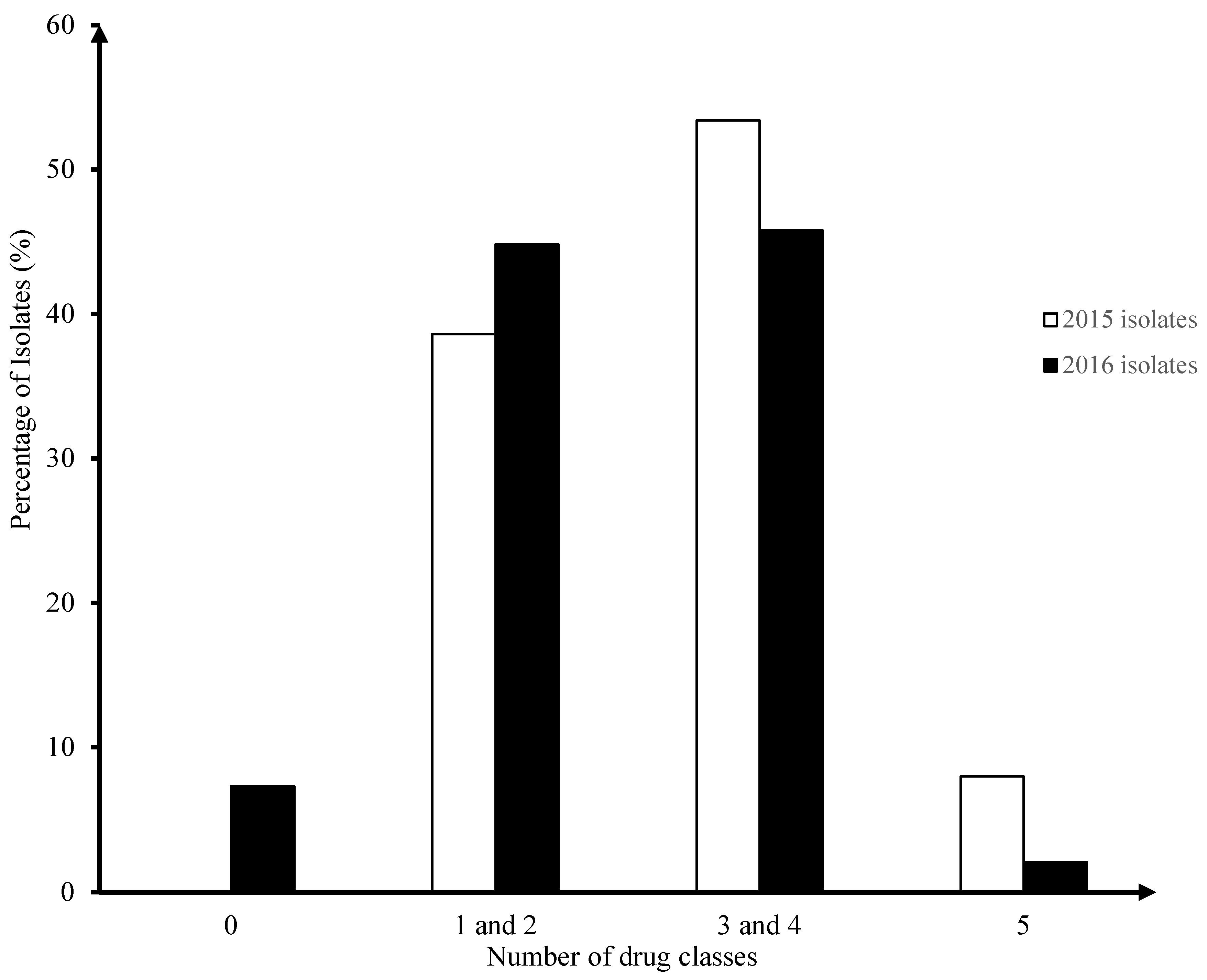
3.1. Surveillance of AMR in Enterococci Isolates from Broiler Chicken Farms in 2015-2016 (Fig. 1)
3.2. Speciation of Enterococcus Isolates from Bbroiler Chicken Farms (Table 2)
| Enterococcus species | 2015 | 2016 | ||||||
|---|---|---|---|---|---|---|---|---|
| Number of isolates | Percentage | Number of MCR isolates | Percentage | Number of isolates | Percentage | Number of MCR isolates | Percentage | |
| Enterococcus faecalis | 52 | 59% | 37 | 43% | 57 | 59% | 29 | 30% |
| Enterococcus faecium | 35 | 40% | 17 | 20% | 37 | 39% | 16 | 17% |
| Enterococcus durans | 1 | 1% | - | - | - | - | - | - |
| Enterococcus hirae | - | - | - | - | 2 | 2% | 1 | 1% |
| Total | 88 | 100% | 54 | 63% | 96 | 100% | 46 | 48% |
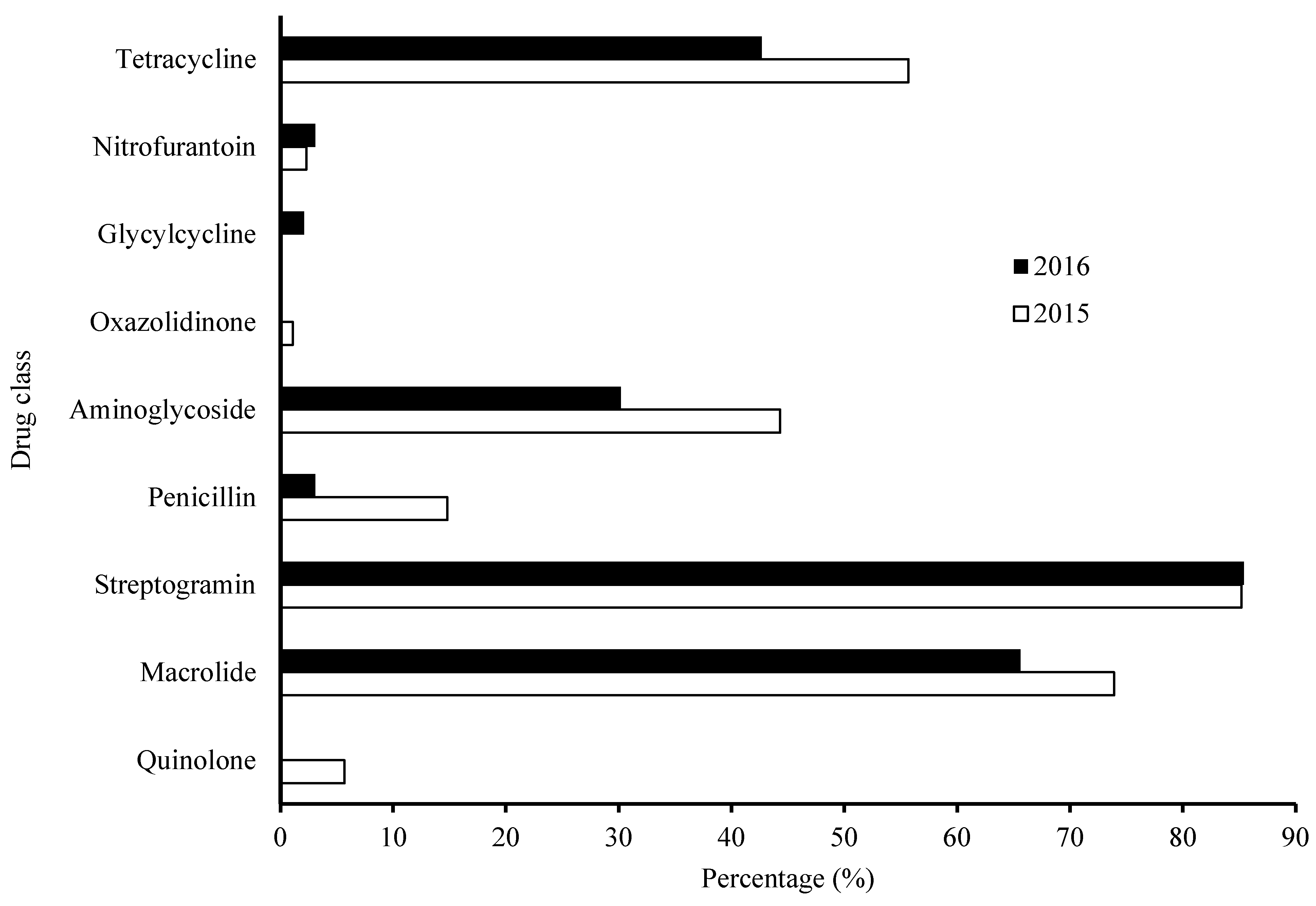
3.3. Characterization of Overall AMR Profile – Frequency Distribution per Antimicrobial Agents in Year 2015-2016 Categorized by Drug Classes (Fig. 2)
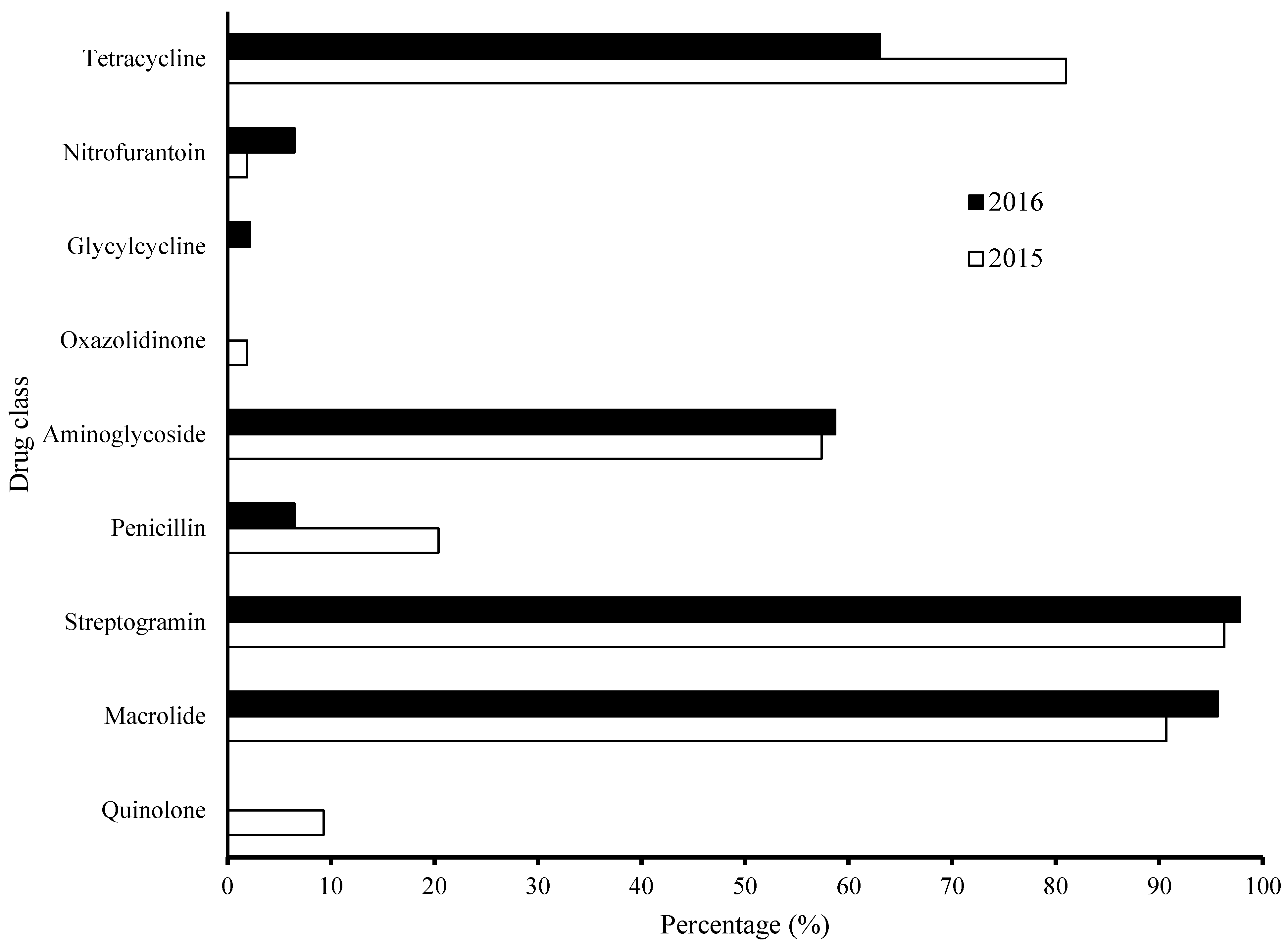
3.4. Characterization of AMR Profile – Frequency Distribution per Antimicrobial Agents in MCR Community in Year 2015-2016 Categorized by Drug Classes (Fig. 3)
3.5. Characterization of AMR Patterns of Isolates Resistant to a Combination of Three or More Antimicrobials
3.6. Characterization of AMR Patterns of Isolates with Intermediate Resistance in Combinations of Three or More Antimicrobials (Table 4)
| a. All AMR patterns of intermediate phenotypes only | |||||
| Pattern | 2015 | 2016 | Total | ||
| E. faecalis | E. faecium | E. faecalis | E. faecium | ||
| dox-ery-lzd | 1 | 0 | 0 | 0 | 1 |
| dox-ery-van | 1 | 0 | 0 | 0 | 1 |
| dox-lvx_qd | 0 | 1 | 0 | 0 | 1 |
| dox-lvx-qd-str | 0 | 1 | 0 | 0 | 1 |
| dox-lvx-str | 0 | 2 | 0 | 0 | 2 |
| ery-lvx-lzd-tgc | 1 | 0 | 0 | 0 | 1 |
| ery-str-van | 2 | 0 | 0 | 0 | 2 |
| lvx-lzd-str-tgc-van | 1 | 0 | 0 | 0 | 1 |
| lvx-lzd-van | 1 | 0 | 0 | 0 | 1 |
| lvx-nit-van | 1 | 0 | 0 | 0 | 1 |
| lvx-str-van | 1 | 0 | 0 | 0 | 1 |
| Total | 9 | 4 | 0 | 0 | 13 |
| b. Common AMR patterns of both intermediate and resistant phenotypes | |||||
| Pattern | 2015 | 2016 | Total | ||
| DOX-ERY-lvx-QD | 3 | 0 | 3 | ||
| DOX-ERY-lvx-QD-STR | 3 | 0 | 3 | ||
| DOX-ery-QD | 0 | 6 | 0 | ||
| ERY-QD-str | 0 | 5 | 5 | ||
| dox-ERY-QD | 0 | 6 | 6 | ||
| dox-ERY-lvx-QD-str | 3 | 0 | 3 | ||
3.7. The Association of Phenotype and Genotype in MCR Isolates 2015-2016
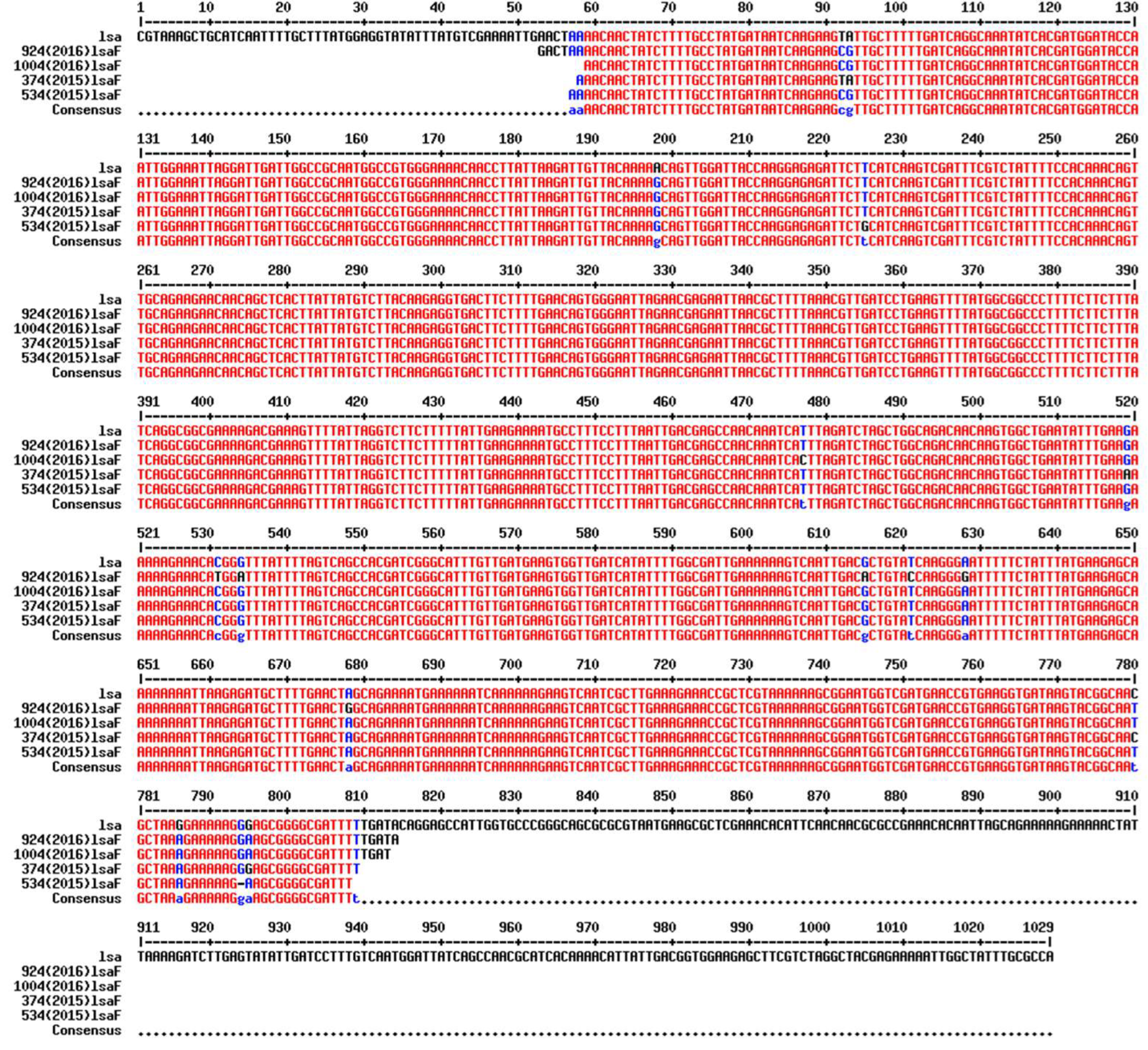
3.8. Insertion Sequence
4. Discussion
5. Conclusions
Authors’ contributions
Funding
Ethics approval and consent to participate
Consent for publication
Availability of data and materials
Acknowledgments
Competing interests
References
- Chicken Farmers of Canada. 2016.
- Grier, K. The 2015 Economic Impact of the Poultry and Egg Industries in Canada. 2016.
- Hardie, J.M.; Whiley, R.A. Classification and overview of the genera Streptococcus and Enterococcus. Society for Applied Bacteriology symposium series 1997, 26, 1S–11S. [Google Scholar] [CrossRef]
- Ruzauskas, M.; Siugzdiniene, R.; Spakauskas, V.; Povilonis, J.; Seputiene, V.; Suziedeliene, E.; Daugelavicius, R.; Pavilonis, A. Susceptibility of bacteria of the Enterococcus genus isolated on Lithuanian poultry farms. Veterinarni Medicina 2009, 54, 577–582. [Google Scholar] [CrossRef]
- Iversen, A.; Kühn, I.; Franklin, A.; Möllby, R. High Prevalence of Vancomycin-Resistant Enterococci in Swedish Sewage High Prevalence of Vancomycin-Resistant Enterococci in Swedish Sewage 2002, 68, 2838–2842. [CrossRef]
- Byappanahalli, M.N.; Nevers, M.B.; Korajkic, A.; Staley, Z.R.; Harwood, V.J. Enterococci in the Environment. Microbiology and Molecular Biology Reviews 2012, 76, 685–706. [Google Scholar] [CrossRef]
- Lu, H.; Weng, X.; Li, H.; Yin, Y.; Pang, M.; Tang, Y. Enterococcus faecium -Related Outbreak with Molecular Evidence of Transmission from Pigs to Humans 2002, 40, 913–917.
- Freitas, A.R.; Coque, T.M.; Novais, C.; Hammerum, A.M.; Lester, C.H.; Zervos, M.J.; Donabedian, S.; Jensen, L.B.; Francia, M.V.; Baquero, F.; Peixe, L. Human and swine hosts share vancomycin-resistant Enterococcus faecium CC17 and CC5 and Enterococcus faecalis CC2 clonal clusters harboring Tn1546 on indistinguishable plasmids. Journal of Clinical Microbiology 2011, 49, 925–931. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.P. The pathogenicity of enterococci. Journal of Antimicrobial Chemotherapy 1994, 33, 1083–1089. [Google Scholar] [CrossRef] [PubMed]
- Kristich, C.J.; Rice, L.B. Enterococcal Infection — Treatment and Antibiotic Resistance. 2009:1–62.
- Reik, R.; Tenover, F.C.; Klein, E.; McDonald, L.C. The burden of vancomycin-resistant enterococcal infections in US hospitals, 2003 to 2004. Diagnostic Microbiology and Infectious Disease 2008, 62, 81–85. [Google Scholar] [CrossRef]
- Low, D.E.; Keller, N.; Barth, A.; Jones, R.N. Clinical Prevalence, Antimicrobial Susceptibility, and Geographic Resistance Patterns of Enterococci: Results from the SENTRY Antimicrobial Surveillance Program, 1997-1999. Clinical Infectious Diseases 2001, 32 (Suppl. 2), 2):S133–S145. [Google Scholar] [CrossRef] [PubMed]
- Shivaprasad, H.L. An overview of anatomy, physiology and pathology of urinary system in birds. AAV Proceedings 1998:201–205.
- Jung, A.; Rautenschlein, S. Comprehensive report of an Enterococcus cecorum infection in a broiler flock in Northern Germany. BMC Veterinary Research 2014, 10, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Boerlin, P.; Nicholson, V.; Brash, M.; Slavic, D.; Boyen, F.; Sanei, B.; Butaye, P. Diversity of Enterococcus cecorum from chickens. Veterinary Microbiology 2012, 157, 405–411. [Google Scholar] [CrossRef]
- Hollenbeck, B.L.; Rice, L.B. Intrinsic and acquired resistance mechanisms in enterococcus. Virulence 2012, 3, 421–569. [Google Scholar] [CrossRef]
- Aslam, M.; Diarra, M.S.; Checkley, S.; Bohaychuk, V.; Masson, L. Characterization of antimicrobial resistance and virulence genes in Enterococcus spp. isolated from retail meats in Alberta, Canada. International Journal of Food Microbiology 2012, 156, 222–230. [Google Scholar] [CrossRef]
- Hegstad, K.; Mikalsen, T.; Coque, T.M.; Werner, G.; Sundsfjord, A. Mobile genetic elements and their contribution to the emergence of antimicrobial resistant Enterococcus faecalis and Enterococcus faecium. Clinical Microbiology and Infection 2010, 16, 541–554. [Google Scholar] [CrossRef] [PubMed]
- Dunny, G.M.; Berntsson, R.P.A. Enterococcal sex pheromones: Evolutionary Pathways to Complex, Two-Signal Systems. Journal of bacteriology 2016, 198, 1556–1562. [Google Scholar] [CrossRef] [PubMed]
- Eva, B. On Mobile Genetic Elements in Enterococci ; Adding More Facets to the Complexity. October 2010(July).
- Lett, M. Tn3-1ike elements : molecular structure, evolution. Biochimie 1988, 70, 167–176. [Google Scholar] [CrossRef]
- Rice, L.B.; Carias, L.L.; Marshall, S.H. Tn5384, a composite enterococcal mobile element conferring resistance to erythromycin and gentamicin whose ends are directly repeated copies of IS256. Antimicrobial Agents and Chemotherapy 1995, 39, 1147–1153. [Google Scholar] [CrossRef]
- Hodel-Christian, S.L.; Murray, B.E. Characterization of the gentamicin resistance transposon Tn5281 from Enterococcus faecalis and comparison to staphylococcal transposons Tn4001 and Tn4031. Antimicrobial Agents and Chemotherapy 1991, 35, 1147–1152. [Google Scholar] [CrossRef] [PubMed]
- Bonafede, M.E.; Carias, L.L.; Rice, L.B. Enterococcal transposon Tn5384: Evolution of a composite transposon through cointegration of enterococcal and staphylococcal plasmids. Antimicrobial Agents and Chemotherapy 1997, 41, 1854–1858. [Google Scholar] [CrossRef]
- Public Health Agency of Canada (PHAC): Canadian Antimicrobial Resistance Surveillance System – Report 2016. 2016.
- Clinical and Laboratory Standards Institute: Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Third Informational Supplement. M100-S23 edition. Wayne, PA; 2013.
- Vineetha, N.; Vignesh, R.; Sridhar, D. Preparation, Standardization of Antibiotic Discs and Study of Resistance Pattern for First-Line Antibiotics in Isolates from Clinical Samples. International Journal of Applied Research 2015, 1, 624–631. [Google Scholar]
- Woodford, N.; Egelton, C.M.M.D. Comparison of PCR with Phenotypic Methods for the Speciation of Enterococci. In Streptococci and the Host Advances in Experimental Medicine and Biology. Edited by Horaud T., Bouvet A., Leclercq R., de Montclos H. SM. Boston, MA; Springer; 1997:405–406.
- Zaheer, R.; Yanke, L.J.; Church, D.; Topp, E.; Read, R.R.; McAllister, T.A. High-throughput species identification of enterococci using pyrosequencing. Journal of Microbiological Methods 2012, 89, 174–178. [Google Scholar] [CrossRef] [PubMed]
- Suchiya, T.T. Functional Cloning and Expression of emeA, and Characterization of EmeA, a Multidrug Efflux Pump from Enterococcus faecalis. Biol Pharm Bull 2003, 26, 266–270. [Google Scholar]
- Jonas, B.M.; Murray, B.E.; Weinstock, G.M. Characterization of emeA,a norA Homolog and Multidrug Resistance Efflux Pump, in Enterococcus faecalis. Antimicrobial Agents and Chemotherapy 2001, 45, 3574–3579. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.; Huda, M.N.; Kuroda, T.; Mizushima, T.; Tsuchiya, T. EfrAB, an ABC Multidrug Efflux Pump in Enterococcus faecalis. Antimicrobial Agents and Chemotherapy 2003, 47, 3733–3738. [Google Scholar] [CrossRef] [PubMed]
- Lavilla Lerma, L.; Benomar, N.; Sánchez Valenzuela, A.; Casado Muñoz M del, C.; Gálvez, A.; Abriouel, H. Role of EfrAB efflux pump in biocide tolerance and antibiotic resistance of Enterococcus faecalis and Enterococcus faecium isolated from traditional fermented foods and the effect of EDTA as EfrAB inhibitor. Food Microbiology 2014, 44, 249–257. [Google Scholar] [CrossRef]
- Portillo, A.; Ruiz-Larrea, F.; Zarazaga, M.; Alonso, A.; Martinez, J.L.; Torres, C. Macrolide Resistance Genes in Enterococcus spp. Antimicrobial Agents and Chemotherapy 2000, 44, 967–971. [Google Scholar] [CrossRef] [PubMed]
- Jia, W.; Li, G.; Wang, W. Prevalence and antimicrobial resistance of Enterococcus species: A hospital-based study in China. International Journal of Environmental Research and Public Health 2014, 11, 3424–3442. [Google Scholar] [CrossRef]
- Singh, K.V.; Weinstock, G.M.; Murray, B.E. An Enterococcus faecalis ABC homologue (Lsa) is required for the resistance of this species to clindamycin and quinupristin-dalfopristin. Antimicrobial Agents and Chemotherapy 2002, 46, 1845–1850. [Google Scholar] [CrossRef]
- Jung, Y.H.; Shin, E.S.; Kim, O.; Yoo, J.S.; Lee, K.M.; Yoo, J.; Chung, G.T.; Lee, Y.S. Characterization of two newly identified genes, vgaD and vatG, conferring resistance to streptogramin A in Enterococcus faecium. Antimicrobial Agents and Chemotherapy 2010, 54, 4744–4749. [Google Scholar] [CrossRef]
- Soltani, M.; Beighton, D.; Philpott-howard, J.; Soltani, M.; Beighton, D.; Philpott-howard, J. Mechanisms of Resistance to Quinupristin-Dalfopristin among Isolates of Enterococcus faecium from Animals, Raw Meat, and Hospital Patients in Western Europe Mechanisms of Resistance to Quinupristin-Dalfopristin among Isolates of Enterococcus faecium fro. Antimicrobial Agents and Chemotherapy 2000, 44, 433–436. [Google Scholar] [CrossRef]
- Manson, J.M.; Hancock, L.E.; Gilmore, M.S. Mechanism of chromosomal transfer of Enterococcus faecalis pathogenicity island, capsule, antimicrobial resistance, and other traits. Proceedings of the National Academy of Sciences 2010, 107, 12269–12274. [Google Scholar] [CrossRef]
- Bogaard, A.E.; Willems, R.; London, N.; Top, J.; Stobberingh, E.E. Antibiotic resistance of faecal enterococci in poultry, poultry farmers and poultry slaughterers. Journal of Antimicrobial Chemotherapy 2002, 49, 497–505. [Google Scholar] [CrossRef]
- Hayes, J.R.; English, L.L.; Carr, L.E.; Wagner, D.D.; Joseph, S.W. Multiple-Antibiotic Resistance of Enterococcus spp. Isolated from Comemercial Poultry Production Environments. Apllied and Environmental Microbiology 2004, 70, 6005–6011. [Google Scholar] [CrossRef]
- Science, E.; Wong, A. Unknown Risk on the Farm : Does Agricultural Use of Ionophores Contribute to the Burden of Antimicrobial. mSphere 2019(September):1–6.
- Russell, J.B.; Houlihan, A.J. Ionophore resistance of ruminal bacteria and its potential impact on. FEMS microbiology reviews 2003, 27, 65–74. [Google Scholar] [CrossRef]
- Diarra, M.S.; Rempel, H.; Champagne, J.; Masson, L.; Pritchard, J.; Topp, E. Distribution of antimicrobial resistance and virulence genes in Enterococcus spp. and characterization of isolates from broiler chickens. Applied and Environmental Microbiology 2010, 76, 8033–8043. [Google Scholar] [CrossRef] [PubMed]
- Debnam, A.L.; Jackson, C.R.; Avellaneda, G.E.; Barrett, J.B.; Hofacre, C.L. Effect of Growth Promotant Usage on Enterococci Species on a Poultry Farm. Avian Diseases 2005, 49, 361–365. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, F.J.; Sadurski, R.; Kray, A.; Boos, M.; Geisel, R.; Koöhrer, K.; Verhoef, J.; Fluit, A.C. Prevalence of macrolide-resistance genes in Staphylococcus aureus and Enterococcus faecium isolates from 24 European university hospitals. Journal of Antimicrobial Chemotherapy 2000, 45, 891–894. [Google Scholar] [CrossRef]
- Si, H.; Zhang, W.J.; Chu, S.; Wang, X.M.; Dai, L.; Hua, X.; Dong, Z.; Schwarz, S.; Liu, S. Novel plasmid-borne multidrug resistance gene cluster including lsa(E) from a Linezolid-Resistant Enterococcus faecium Isolate of Swine Origin. Antimicrobial Agents and Chemotherapy 2015, 59, 7113–7116. [Google Scholar] [CrossRef] [PubMed]
- Wendlandt, S.; Lozano, C.; Kadlec, K.; Gómez-Sanz, E.; Zarazaga, M.; Torres, C.; Schwarz, S. The enterococcal ABC transporter gene lsa(E) confers combined resistance to lincosamides, pleuromutilins and streptogramin A antibiotics in methicillin-susceptible and methicillinresistant Staphylococcus aureus. Journal of Antimicrobial Chemotherapy 2013, 68, 473–475. [Google Scholar] [CrossRef]
- Werner, G.; Klare, I.; Heier, H.; Hinz, K.H.; Böhme, G.; Wendt, M.; Witte, W. Quinupristin/dalfopristin-resistant enterococci of the satA (vatD) and satG (vatE) genotypes from different ecological origins in Germany. Microbial drug resistance 2000, 6, 37–47. [Google Scholar] [CrossRef]
- Strain, S.T.; Kadlec, K.; Schwarz, S. Novel ABC Transporter Gene, vga(C), Located on a Multiresistance Plasmid from a Porcine Methicillin-Resistant Staphylococcus aureus. AntimicrobAgents Chemother 2009, 53, 3589–3591. [Google Scholar]
- Allignet, J.; Solh, E. Characterization of a new staphylococcal gene, vgaB, encoding a putative ABC transporter conferring resistance to streptogramin A and related compounds 1997, 202, 133–138. [CrossRef]
- Science, E.; All, P.B.V.; Cedex, P. Sequence of a staphylococcal plasmid gene, vga, encoding a putative ATP-binding protein involved in resistance to virginiamyein A-like antibiotics. Gene 1992, 117, 45–51. [Google Scholar]
- Haroche, J.; Allignet, J.; Buchrieser, C.; Pathoge, M. Characterization of a Variant of vga(A) Conferring Resistance to Streptogramin A and Related Compounds. AntimicrobAgents Chemother 2000, 44, 2271–2275. [Google Scholar] [CrossRef] [PubMed]
- Schramm, V.L.; Shi, W.; Opin, C.; Biol, S.; Allard, J.; Grochulski, P.; Sygusch, J.; Acad, P.N.; Lett, O. Role of Mobile DNA in the Evolution of Vancomycin-Resistant Enterococcus faecalis. Science 2003, 299, 2071–2075. [Google Scholar]
- Quintiliani, R.; Courvalin, P. Characterization of Tn1547, a composite transposon flanked by the IS16 and IS256-like elements, that confers vancomycin resistance in Enterococcus faecalis BM4281. Gene 1996, 172, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Rice, L.B.; Thorisdottir, A.S. The prevalence of sequences homologous to IS256 in clinical enterococcal isolates. Plasmid 1994, 32, 344–349. [Google Scholar] [CrossRef]
| Primers | PCR Product | Size (bp) | Reference |
|---|---|---|---|
| ddl1_F 5’-ATCAAGTACAGTTAGTCTT-3’ ddl1_R 5’-ACGATTCAAAGCTAACTG-3’ |
ddlE.faecalis FOd |
941 |
[28] |
| ddl2_F 5’-GCAAGGCTTCTTAGAGA-3’ ddl2_R 5’-CATCGTGTAAGCTAACTTC-3’ vanC1_F 5’- GGTATCAAGGAAACCTC-3’ vanC1_R 5’- CTTCCGCCATCATAGCT-3’ vanC2-F 5’-CTCCTACGATTCTCTTG-3’ vanC2-R 5’-CGAGCAAGACCTTTAAG-3’ |
ddlE.faecium vanC1E.gallinarum vanC2 E.casseliflavus & E.flavescens |
550 822 439 |
[28] [28] [28] |
| Ent-ES-211-233-F 5’- GHACAGAAGTRAAATAYGAAGG-3’ Ent-EL-74-95-R 5’- GGNCCTAABGTHACTTTNACTG-3’ emeA_F 5’-AGTATGATGTACTTAGCAATTTC-3’ emeA_R 5’-CATCTTATTTCGATTTAAAAATAAC-3’ efrA_F 5’-TTGGCTTTATGACGCCAGTG-3’ efrA_R 5’-CGTGCGATAGCTAAACGTTG-3’ efrB_F 5’-CCTTATTTAACTGGATTACCAAC-3’ efrB_R 5’-GAATAGTTGATAGGCGGTGG-3’ ermB_F 5’-ATTCTCAAAACTTTTTAACGAGTG -3’ ermB_R 5’- CCTCCCGTTAAATAATAGATAAC-3’ lsa_F 5’-CGTAAAGCTGCATCAATTTTGC -3’ lsa_R 5’- AATGGCTCCTGTATCAAAAATC -3’ mefA_F 5’- GGCAAGCAGTATCATTAATCAC -3’ mefA_R 5’- CATTATTGCACAGCAAACTACG -3’ vatG_F 5’-GTGGGAAAAGCATACACCT-3’ vatG_R 5’-TTGCAGGATTACCACCAAC-3’ vgaD_F 5’-CAACTGGAGCGAGCTGTTA-3’ vgaD_R 5’-GACAGCCGGATAATCTTTTG-3’ vatD_F 5’-GCTCAATAGGACCAGGTGTA-3’ vatD_R 5’-TCCAGCTAACATGTATGGCG-3’ vatE_F 5’-ACTATACCTGACGCAAATGC-3’ vatE_R 5’-GGTTCAAATCTTGGTCCG-3’ IS256c-F 5’-CATTGGTAAATTGGAATGGAAATC-3’ IS256c-R 5’-ATTCAAACATTTTTTCCTCTCC-3’ IS256d_F 5’-GATCAACTGGAGAATTAGTGTT-3’ IS256d-R 5’-CTCTAATATCCCCTAATGAAAATAATG-3’ |
groES-EL spacer region emeA efrA efrB ermB lsa mefA vatG vgaD vatD/ satA vatE/ satG Primers flanking ef0125 Primers flanking ef0529 |
variable (~200bp) 1137 1048 1513 713 825 911 200 201 272 512 ~1173 ~1173 |
[29] [30,31] [32,33] [32,33] [34,35] [36] [34,35] [37] [37] [38] [38] [39] [39] |
| IS256e_F 5’-GGCTATTTTTTAGCAAACTATGTAT-3’ IS256e_R 5’-CACAGCAACTATTGGTAACG-3’ |
Primers flanking ef2187 | ~1173 | [39] |
| IS256f_F 5’-TGTCTAGCTAAAACGAAGCC-3’ IS256f-R 5’-GACCCAACAAAAGTAACTCG-3’ |
Primers flanking ef2632 | ~1173 | [39] |
| IS256g-F 5’-CTGTTTTGTCTCGTCATTATATGA-3’ |
Primers flanking ef3100 | ~1173 | [39] |
| IS256g-R 5’-GGTTATAGTAGGAATAATTTTGCC-3’ IS256h-F 5’-CTGAACTGACACAATTCATTAAAT-3’ |
Primers flanking ef3215 |
~1173 |
[39] |
| IS256h-R 5’-AATTTAGCAACATCTTTCATTGG-3’ |
|||
| IS256t_F 5’-CTGAAAAGCGAAGAGATTCAAAGC-3’ |
IS256 transposase | 748 | This study |
| IS256t_R 5’-GAACTTGGCATCTTTGCCAACTTAC-3’ |
| Pattern | 2015 | 2016 | Total | ||
|---|---|---|---|---|---|
| E. faecalis | E. faecium | E. faecalis | E. faecium | ||
| AMP-DOX-ERY | 0 | 1 | 0 | 0 | 1 |
| AMP-DOX-ERY-GEN-QD | 1 | 0 | 0 | 0 | 1 |
| AMP-DOX-ERY-STR-QD | 1 | 1 | 0 | 1 | 3 |
| AMP-DOX-LVX-NIT-QD | 1 | 0 | 0 | 0 | 1 |
| AMP-DOX- LVX-QD | 1 | 0 | 0 | 0 | 1 |
| AMP-ERY-QD | 3 | 2 | 0 | 0 | 5 |
| AMP-ERY- STR-QD | 0 | 0 | 0 | 2 | 2 |
| DOX-ERY-GEN-QD | 3 | 0 | 1 | 0 | 4 |
| DOX-ERY-GEN-STR-QD | 2 | 0 | 1 | 0 | 3 |
| DOX-ERY-LVX-STR-QD | 2 | 0 | 0 | 0 | 2 |
| DOX-ERY-LZD-STR-QD | 1 | 0 | 0 | 0 | 1 |
| DOX-ERY-QD | 9 | 5 | 18 | 0 | 32 |
| DOX-ERY-STR | 0 | 1 | 0 | 0 | 1 |
| DOX-ERY-STR-QD | 11 | 1 | 6 | 0 | 18 |
| DOX-GEN-QD | 2 | 0 | 1 | 0 | 3 |
| DOX-STR-QD | 0 | 1 | 1 | 0 | 2 |
| ERY-LVX-QD | 0 | 1 | 0 | 0 | 1 |
| ERY-NIT-QD | 0 | 0 | 0 | 1 | 1 |
| ERY-STR-QD | 0 | 4 | 0 | 11 | 15 |
| ERY-NIT-QD-STR-TGC | 0 | 0 | 0 | 1 | 1 |
| GEN-ERY-QD | 0 | 0 | 1 | 0 | 1 |
| Total | 37 | 17 | 29 | 16 | 99 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





