Submitted:
27 November 2024
Posted:
29 November 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
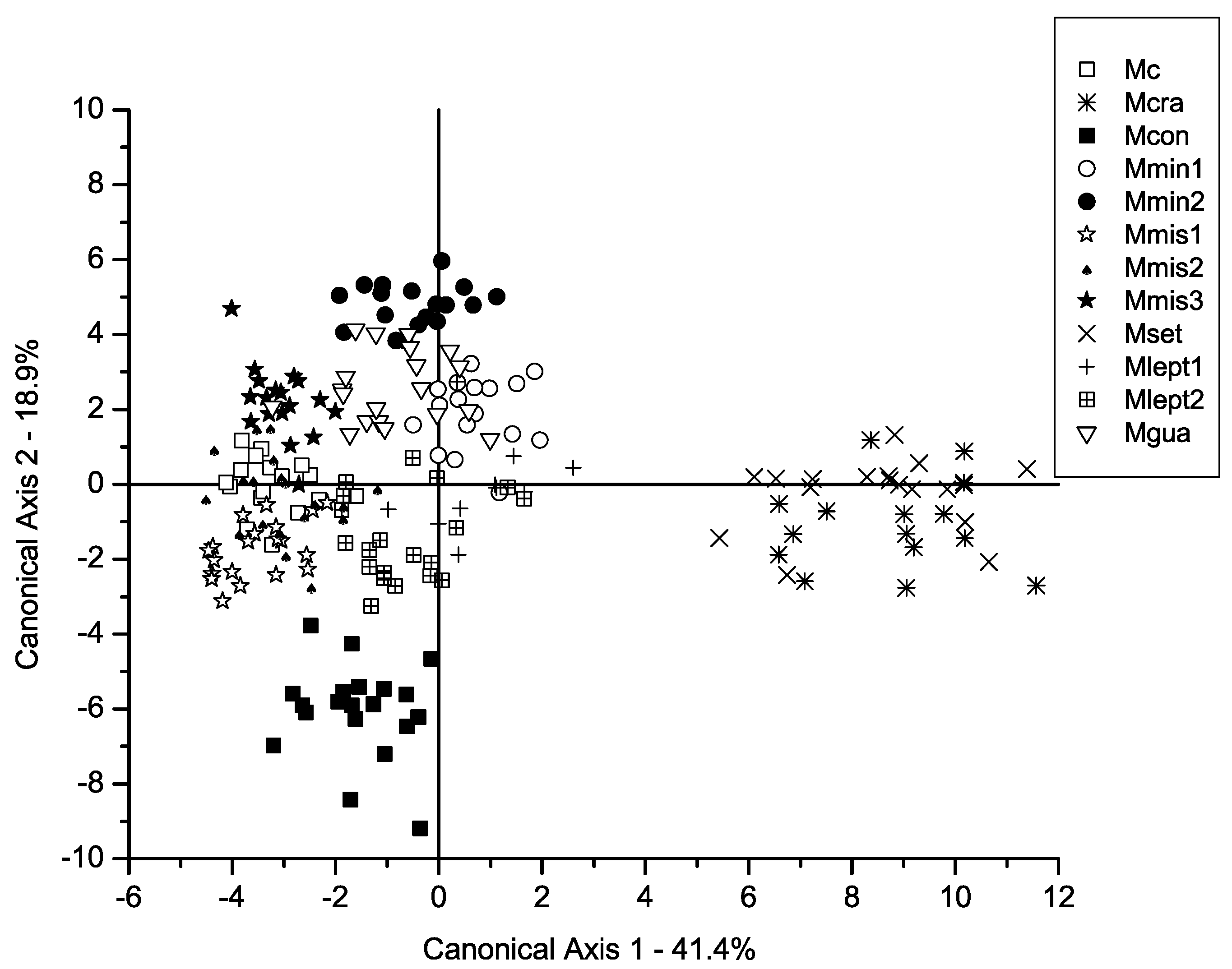
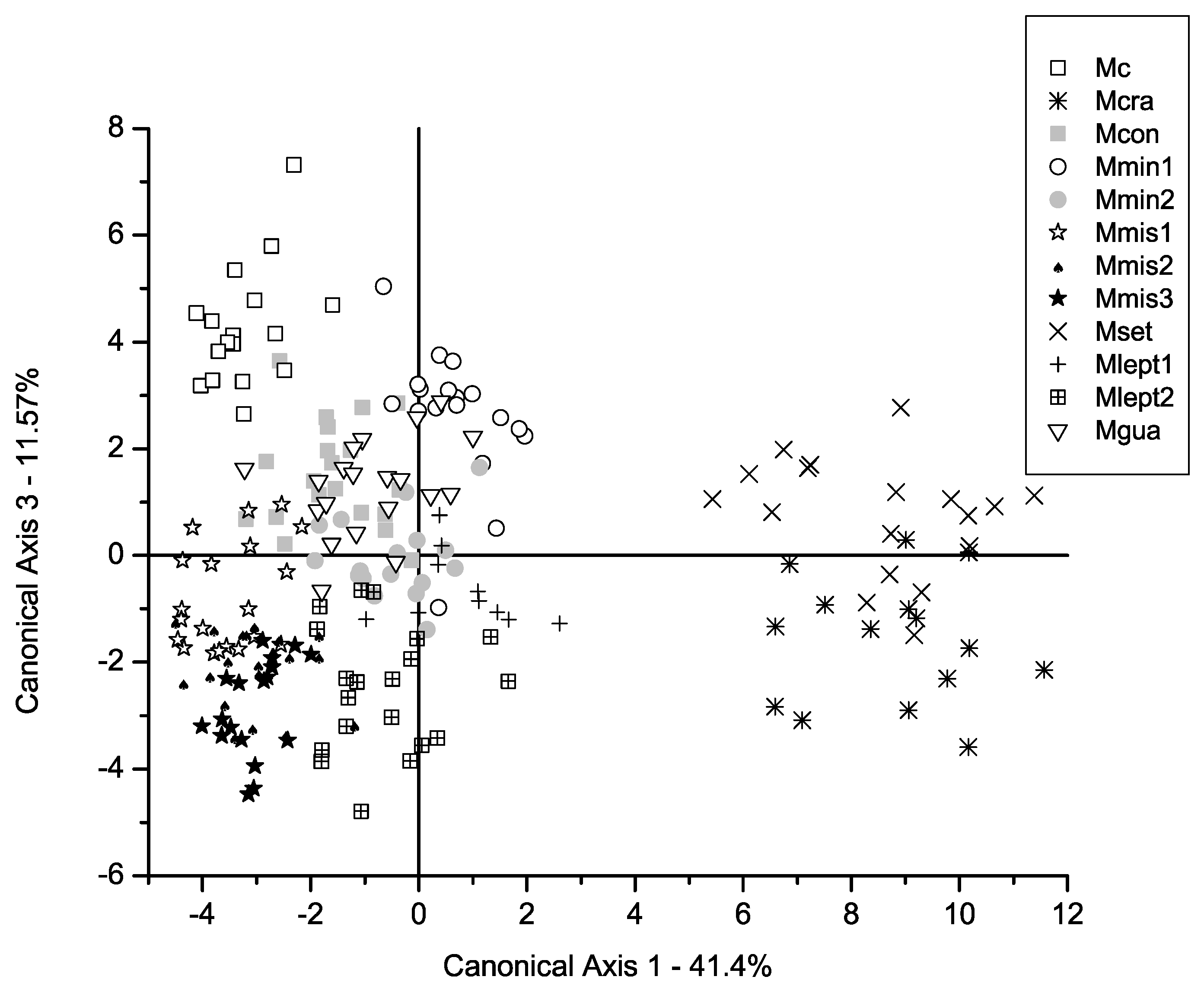
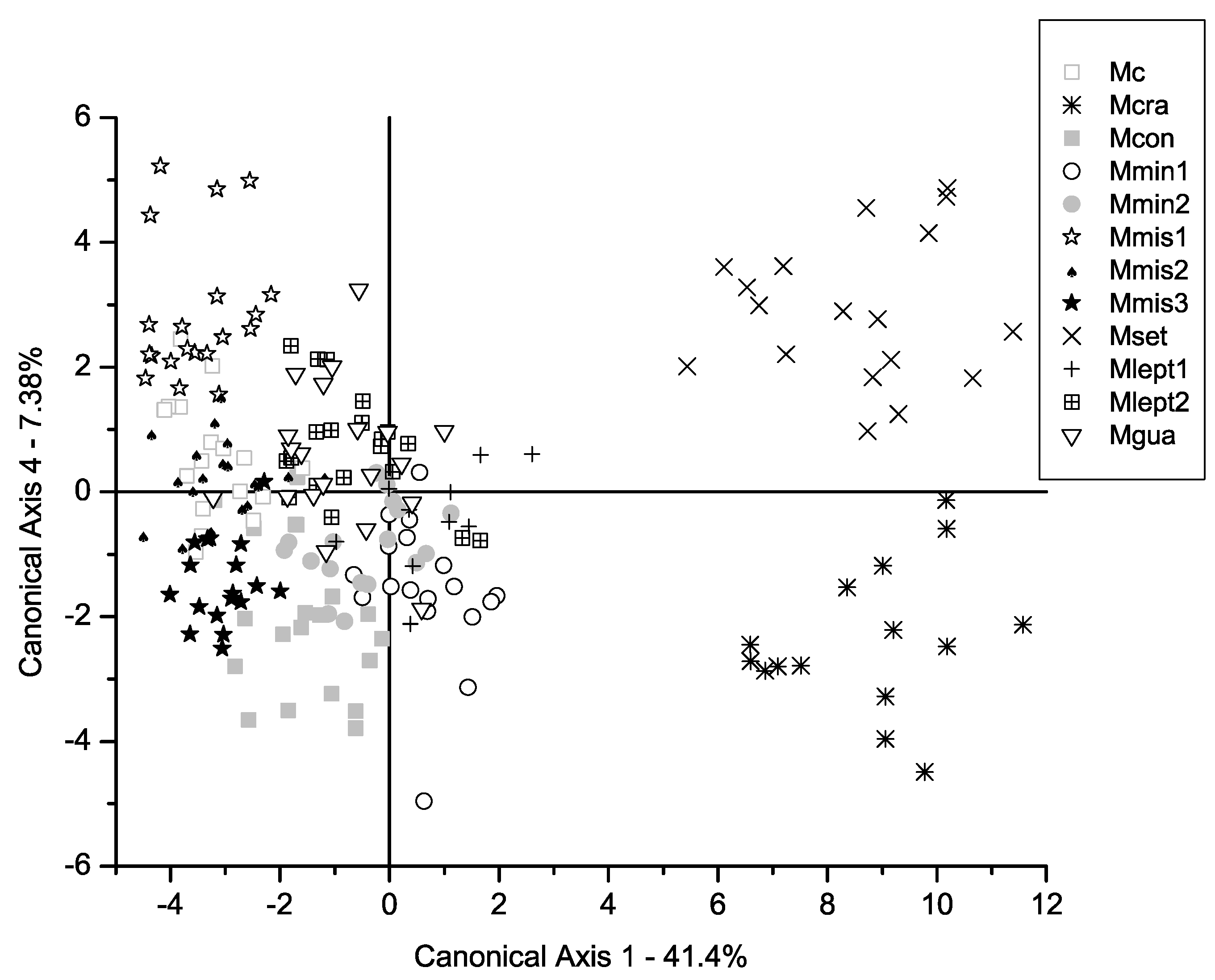
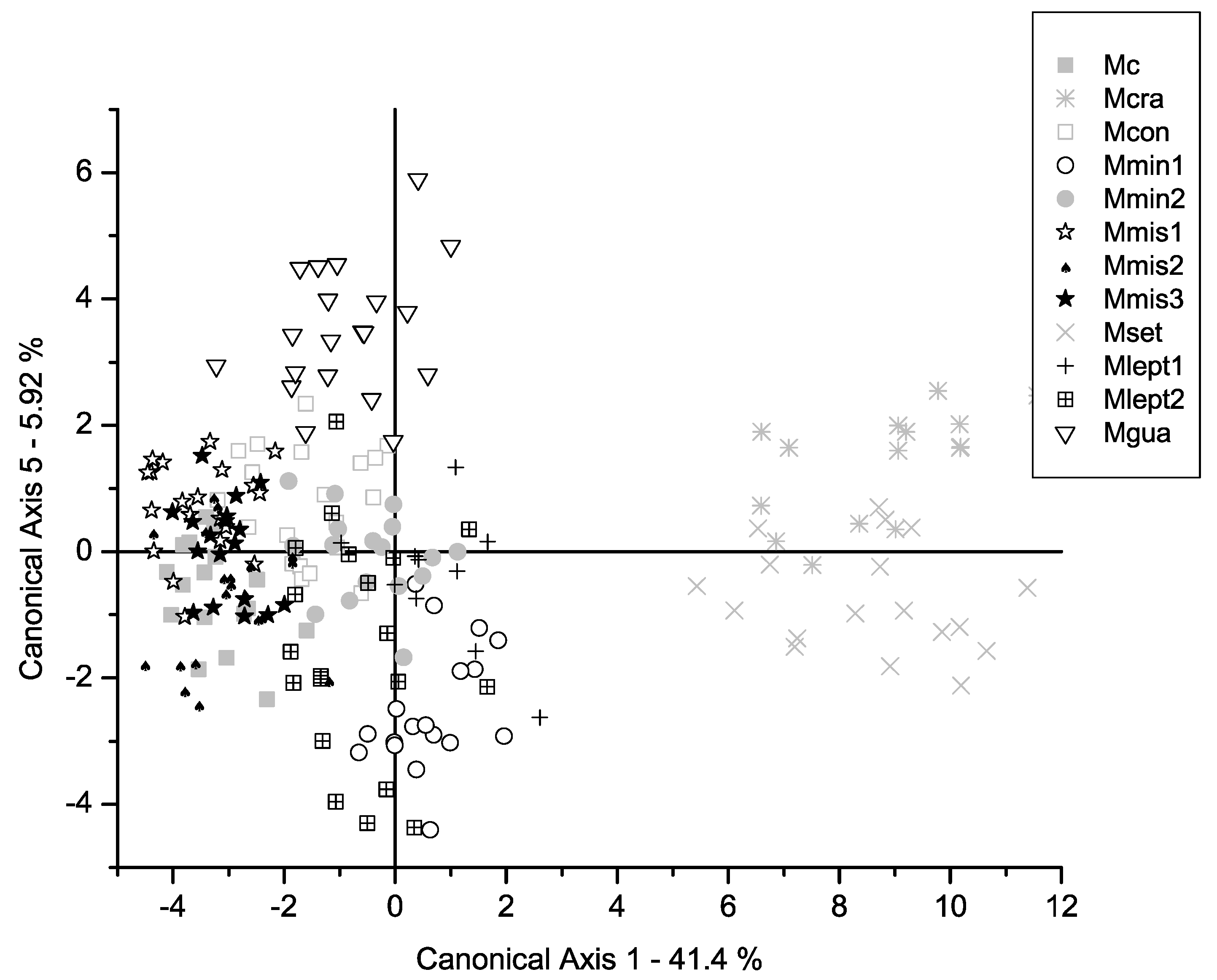
2. Results
3. Discussion
3.1. Comparison Between Morphometric Patterns and Traditional Taxonomy of the M. misera Species Complex
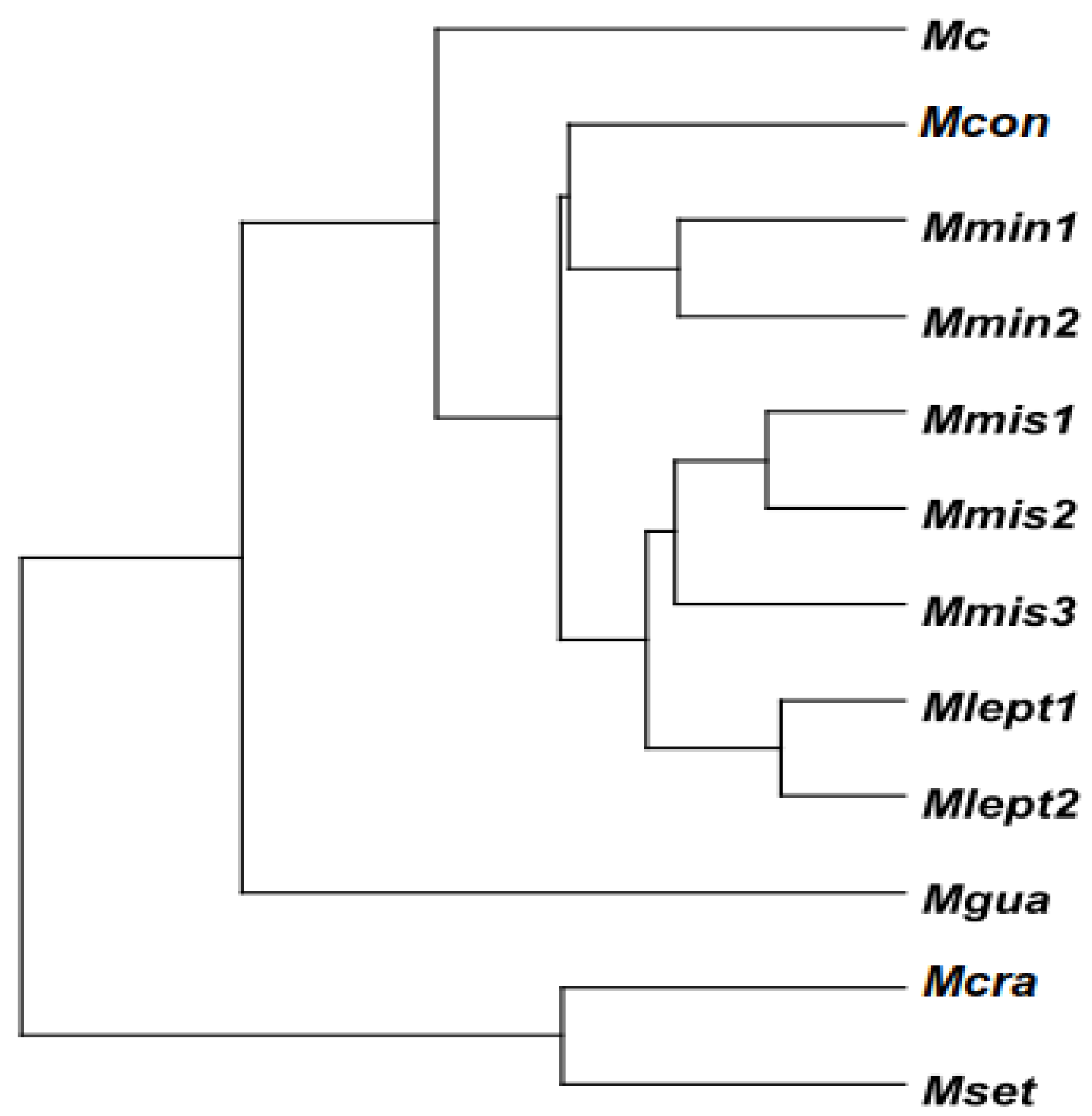
3.2. Overall Comparison Between Morphometric Clusters and Phylogenetic Relationships
3.3. Usefulness and Perspectives for the Use of Multivariate Morphometrics in Leguminosae
4. Materials and Methods
4.1. Data Collection
4.2. Data Analysis
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Simon, M.F.; Grether, R.; Queiroz, L.P.; Särkinen, T.E.; Dutra, V.F.; Hughes, C.E. The evolutionary history of Mimosa (Leguminosae): toward a phylogeny of sensitive plants. Amer. J. Bot. 2011, 98, 1201–1221. [Google Scholar] [CrossRef] [PubMed]
- Lewis, G.P.; Schrire, B.; Mackinder, B.; Lock, M. Legumes of the world; Royal Botanic Gardens, Kew: Richmond, United Kingdom, 2005. [Google Scholar]
- Barneby, R.C. Sensitivae Censitae: a description of the genus Mimosa Linnaeus (Mimosaceae) in New World. Mem. New York Bot. Gard. 1991, 65, 1–835. [Google Scholar]
- Dutra, V.F.; Garcia, F.C.P. Three new species of Mimosa (Leguminosae) from Minas Gerais, Brazil. Syst. Bot. 2013, 38, 398–405. [Google Scholar] [CrossRef]
- Nascimento, J.G.A.; Rocha,L. ; Dutra, V.F.; Queiroz, L.P.; van den Berg, C. Phytotaxa 2023, 599, 265–279. [Google Scholar]
- Barneby, R.C. The genus Mimosa (Mimosaceae) in Bahia, Brazil: new taxa and nomenclatural adjustments. Brittonia 1985, 37, 125–153. [Google Scholar] [CrossRef]
- Bentham, G. Notes on Mimoseae, with a short synopsis of species. J. Bot. (Hooker) 1842, 4, 323–418. [Google Scholar]
- Borba. E.L.; Shepherd, G.J.; van den Berg, C.; Semir. J. Floral and vegetative morphometrics of five Pleurothalis (Orchidaceae) species: correlation with taxonomy, phylogeny, genetic, variability and pollination systems. Ann. Bot. 2002, 90, 219–230. [Google Scholar] [CrossRef]
- Goldman, D.H.; van den Berg, C.; Griffith, M.P. Morphometric circumscription of species and infraspecific taxa in Calopogon R.Br. (Orchidaceae). Pl. Syst. Evol. 2004, 274, 37–60.
- Schmalzel, R.J.; Nixon, R.T.; Best, A.L. Morphometric variation in Coryphantha robustipina (Cactaceae). Syst. Bot. 2004, 29, 553–568. [Google Scholar] [CrossRef]
- Oliveira, R.P.; Borba. E.L.; Longhi-Wagner, H.M. Morphometrics of herbaceous bamboos of the Raddia brasiliensis complex (Poaceae–Bambusoideae): implications for the taxonomy of the genus and new species from Brazil. Pl. Syst. Evol. 2007, 270, 159–182. [Google Scholar] [CrossRef]
- Ribeiro, P.L.; Borba, E.L.; Smidt, E.C.; Lambert, S.M.; Schnadelbach, A.S.; van den Berg, C. Genetic and morphological variation in the Bulbophyllum exaltatum (Orchidaceae) complex occurring in the Brazilian “campos rupestres”: implications for taxonomy and biogeography. Pl. Syst. Evol. 2008, 270, 109–137. [Google Scholar] [CrossRef]
- Venhuis, C.; Venhuis, P.; Oostermeijer, J.G.B.; van Tienderen, P.H. Morphological systematics of Serapias L. (Orchidaceae). Pl. Syst. Evol. 2007, 265, 165–177. [Google Scholar] [CrossRef]
- Andrade. M.; Mayo, S.J.; Kirkup, D.; van den Berg, C. Comparative morphology of populations of Monstera Adans. (Araceae) from natural forest fragments in Northeast Brazil using Elliptic Fourier Analysis of leaf outlines. Kew Bull. 2008, 63, 193–211. [Google Scholar] [CrossRef]
- Marcussen, T.; Borgen, L. Species delimitation in the Ponto-Caucasian Viola sieheana complex, based on evidence from allozymes, morphology, ploidy levels, and crossing experiments. Pl. Syst. Evol. 2011, 291, 183–196. [Google Scholar] [CrossRef]
- Sosa, M.M.; Panseri, A.; Damateis, M. Morphometric analysis of Stemodia hyptoides and S. stricta (Plantaginaceae). Pl. Syst. Evol. 2012, 298, 1315–1326.
- Doğan, M.O.; Kence, A.; Tigin, C. Numerical taxonomic study on the genus Lathyrus (Leguminoseae). Edinburgh J. Bot. 1992, 49, 333–341. [Google Scholar] [CrossRef]
- Rahman, M.Z.; Rahman, M.O. Morphometric analysis of Desmodium Desv. in Bangladesh. Bangladesh J. Bot. 2012, 41, 143–148. [Google Scholar] [CrossRef]
- Rahman, M.O.; Rhaman, M.Z.; Begum, A. Numerical taxonomy of the genus Senna Mill. from Bangladesh. Bangladesh J. Pl. Taxon. 2013, 20, 77–83. [Google Scholar] [CrossRef]
- Casiva, P.V.; Saidman, B.O.; Vilardijc, J.C.; Cialdella, A.M. First comparative phenetic studies of Argentinean species of Acacia (Fabaceae), using morphometric, isozymal, and RAPD approaches. Amer. J. Bot. 2002, 89, 843–853. [Google Scholar] [CrossRef]
- Souza, I.M.; Funch, L.S.; Queiroz, L.P. Morphological analyses suggest a new taxonomic circumscription for Hymenaea courbaril L. (Leguminosae, Caesalpinioideae). PhytoKeys 2014, 38, 101–118. [Google Scholar] [CrossRef]
- Knaus, B.J. Morphometric architecture of the most taxon-rich species in the U.S. Flora: Astragalus lentiginosus (Fabaceae). Amer. J. Bot. 2010, 97, 1816–1826. [Google Scholar] [CrossRef]
- Boonkerd, T.; Saengmanee, S.; Baum, B.R. The varieties of Bauhinia pottsii G. Don in Thailand (Leguminosae-Caesalpinioideae). Pl. Syst. Evol. 2002, 232, 51–62. [Google Scholar] [CrossRef]
- Conceição, A.S.; Queiroz, L.P.; Lambert, S.M.; Pereira, A.C.S.; ,Borba. E.L. Biosystematics of Chamaecrista sect. Absus subsect. Baseophyllum (Leguminosae-Caesalpinioideae) based on allozyme and morphometric analyses. Pl. Syst. Evol. 2008, 270, 183–207. [Google Scholar]
- Estrella, M.; Aedo, C. ; Velayos. M. A morphometric analysis of Daniellia (Fabaceae–Caesalpinioideae). Bot. J. Linn. Soc. 2009, 159, 268–279.
- Chandler, G.T.; Crisp, M.D. Morphometric and phylogenetic analysis of the Daviesia ulicifolia complex (Fabaceae, Mirbelieae). Pl. Syst. Evol. 1998, 209, 93–122. [Google Scholar] [CrossRef]
- Hoffman, D.L.; Muehlbauer. F.J.; Ladizinsky. G. Morphological variation in Lens (Leguminosae). Syst. Bot. 1988, 13, 89–96. [Google Scholar] [CrossRef]
- Riggins, R.; Pimentel. R.A.; Walters, D.R. Morphometrics of Lupinus nanus (Leguminosae). I. Variation in natural populations. Syst. Bot. 1977, 2, 317–326. [Google Scholar] [CrossRef]
- Agulló, J.C.; Juan, A.; Alonso. M.; Terrones, A.; Crespo, M.B. Taxonomic status of Ononis tridentata (Fabaceae) from Morocco, resolved by multivariate morphometric analyses. Pl. Biosyst. 2013, 147, 645–653. [Google Scholar] [CrossRef]
- Ahmad, M.; McNeil, D.L.; Sedcole, J.R. Phylogenetic relationships in Lens species and their interspecific hybrids as measured by morphological characters. Euphytica 1997, 94, 101–111. [Google Scholar] [CrossRef]
- Maxted, N. A phenetic analysis of Vicia section Atossa series Truncatulae (Leguminosae: Vicieae). Kew Bull. 1993, 48, 739–753. [Google Scholar] [CrossRef]
- Kirchner, F.; Bullock, J.M. Taxonomic separation of Ulex minor Roth. and U. gallii Planch.: morphometrics and chromosome counts. Watsonia 1999, 22, 365–376. [Google Scholar]
- Sheidai, M.; Yazdanbakhsh, Z.; Assadi, M.; Moussavi, M. Cytology and morphometry study of Alhagi (Leguminosae) species in Iran. Nordic J. Bot. 2001, 21, 83–91. [Google Scholar] [CrossRef]
- Ceolin, B.G.; Miotto, S.T.S. Combining ecological and morphometrical approaches to increase the resolution within the Galactia neesii (Leguminosae) complex. Pl. Syst. Evol. 2012, 298, 645–652. [Google Scholar] [CrossRef]
- Fisher, R.A. The use of multiple measurements in taxonomic problems. Ann. Eugenics 1936, 7, 179–188. [Google Scholar] [CrossRef]
- Rencher, A.C.; Christensen, W.F. Methods of multivariate statistics, 3rd ed.; John Wiley & Sons: Hoboken, U.S.A, 2012; pp. 281–308. [Google Scholar]
- Zelditch, M.L.; Swiderski, D.L.; Sheets, H.D. Geometric morphometrics for biologists: a primer., 2nd ed.; Academic Press: San Diego, U.S.A, 2012; pp. 151–167. [Google Scholar]
- Bateman. R.M. Evolution and classification of European orchids: insights from molecular and morphological characters. J. Eur. Orch. 2001, 33, 501–568. [Google Scholar]
- Kropf, M. Intraspecific patterns of European mountain plants: a morphometric analysis confirms molecular results in the submediterranean oreophyte. Taxon 2008, 57, 511–524. [Google Scholar]
- Sousa, F.P.S.T.; Lewis, G.P.; Hawkins, J.A. A revision of the South American genus Apuleia (Leguminosae, Cassieae). Kew Bull. 2010, 65, 225–232. [Google Scholar] [CrossRef]
- Bartlett, M.S. Multivariate analysis. J. Royal Stat. Soc. 1947, 9, 176–197. [Google Scholar] [CrossRef]
- Sneath, P.H.A.; Sokal, R.R. Numerical taxonomy; W.H.Freeman: San Francisco, U.S.A.; 1973. [Google Scholar]

| Axis removed | Eigenvalue | Canonical R | Wilks' lambda | Х2 | d.f.* | p** |
|---|---|---|---|---|---|---|
| 0 | 16.49 | 0.971 | 0.000001 | 2671.032 | 418 | 0.000000 |
| 1 | 8.67 | 0.947 | 0.000010 | 2138.772 | 370 | 0.000000 |
| 2 | 6.74 | 0.933 | 0.000098 | 1716.765 | 324 | 0.000000 |
| 3 | 4.59 | 0.906 | 0.000760 | 1336.009 | 280 | 0.000000 |
| 4 | 3.00 | 0.866 | 0.004248 | 1015.796 | 238 | 0.000000 |
| 5 | 2.14 | 0.825 | 0.016978 | 758.105 | 198 | 0.000000 |
| 6 | 1.60 | 0.785 | 0.053239 | 545.530 | 160 | 0.000000 |
| 7 | 1.21 | 0.741 | 0.138549 | 367.634 | 124 | 0.000000 |
| 8 | 0.84 | 0.677 | 0.306866 | 219.730 | 90 | 0.000000 |
| 9 | 0.36 | 0.516 | 0.566029 | 105.855 | 58 | 0.000127 |
| 10 | 0.30 | 0.478 | 0.771238 | 48.315 | 28 | 0.009909 |
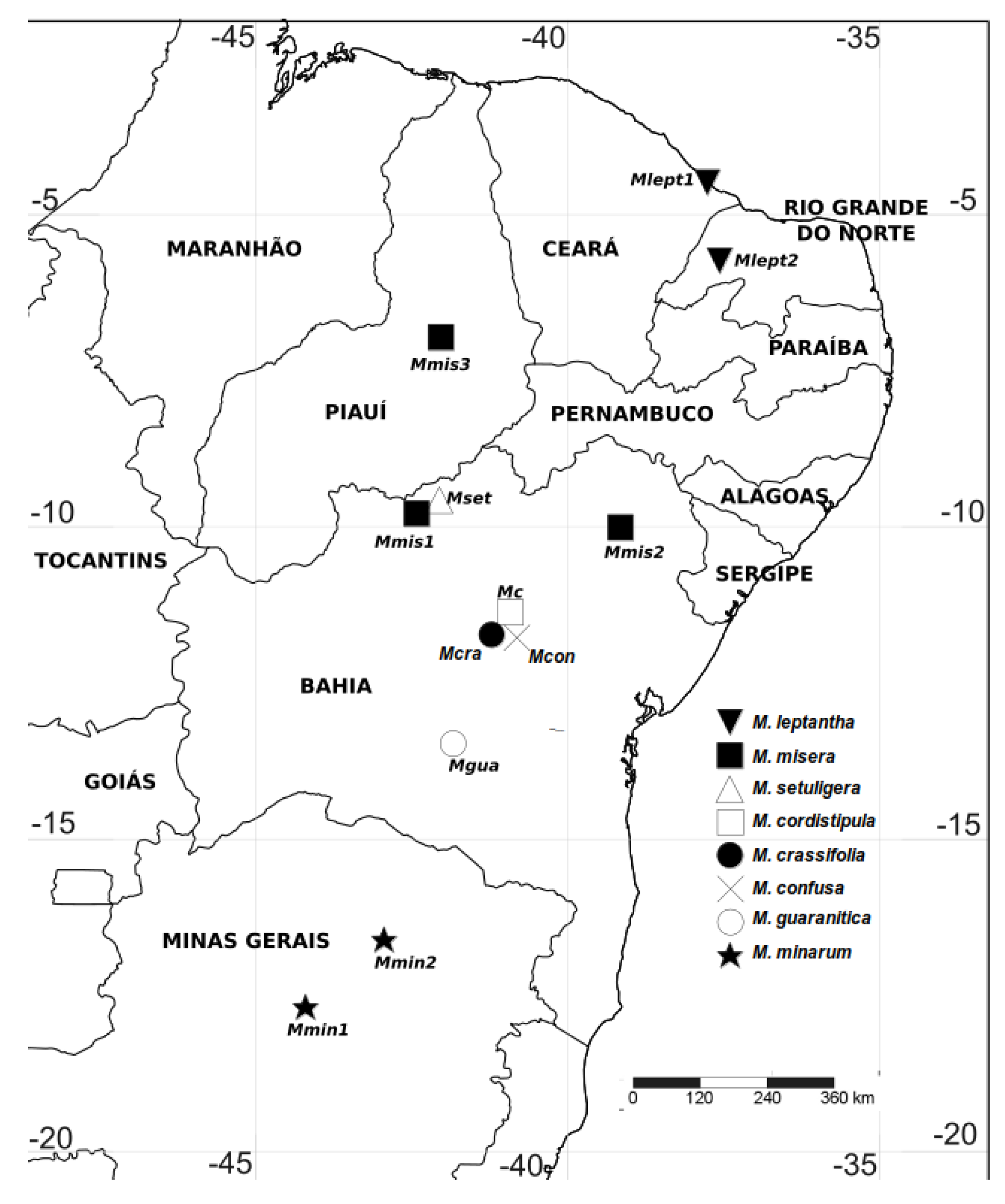
| Taxon | Voucher/sample size | Population Code | Locality data (state, municipality, site) | Geographical coordinates |
|---|---|---|---|---|
| M. confusa | Queiroz et al. 7733 (20) | Mcon | Bahia, Morro do Chapéu, Lajes in the road to Irecê | 11º36’58”S, 41º00’18”W |
| M. cordistipula | Nascimento 149 (19) | Mc | Bahia, Morro do Chapéu | 11º36’4.7”S, 41º9’46.6”W |
| M. crassifolia | Santos et al. 355 (15) | Mcra | Bahia, Morro do Chapéu, Tabuleiro dos Tigres | 11º36’4.7”S, 41º9’46.6”W |
| M. guaranitica | Santos 807 (20) | Mgua | Bahia, Rio de Contas, Estrada para o Pico das Almas | 13º28’12”S, 41º50’25”W |
| M. leptantha | Nascimento 453 (19) | Mlept1 | Ceará, Aracati, Cumbi | 4º29’34”S, 37’45”33.2W |
| M. leptantha | Nascimento 473 (19) | Mlept2 | Rio Grande do Norte, Caraúbas, road to Governador Rosado | 5º44’46.9”S, 37º33’38.7”W |
| M. minarum | Nascimento 495 (18) | Mmin1 | Minas Gerais, Joaquim Felício, Serra do Cabral | 17º41’34”S, 44º11’56”W |
| M. minarum | Nascimento 520 (17) | Mmin2 | Minas Gerais, Grão Mogol, 3km ao N de Grão-Mogol | 16º36’47.3”S, 42º56’S 27.6”W |
| M misera | Nascimento 359 (20) | Mmis1 | Bahia, Remanso, road to São Raimundo Nonato | 9º45’S, 42º17’W |
| M. misera | Lima 177 (20) | Mmis2 | Bahia, Canudos, Estação Biológica de Canudos, Base II | 10°1'S, 39°9'W |
| M. misera | Nascimento 381 (19) | Mmis3 | Piauí, Oeiras, road to Gaturiana | 6º58’18.4”S, 42º1’37”W |
| M. setuligera | Nascimento 338 (19) | Mset | Bahia, Remanso, road to São Raimundo Nonato | 9º45’S, 42º18’W |
| Variable | Character |
|---|---|
| 1 | Stipule length (mm) |
| 2 | Stipule width at ¾ of the length from the base (mm) |
| 3 | Stipule width at ½ of the length from the base (mm) |
| 4 | Stipule width at ¼ of the length from the base (mm) |
| 5 | Rachis length (mm) |
| 6 | Petiole length (including pulvinus) (mm) |
| 7 | First interpinal segment length (mm) |
| 8 | Last interpinal segment length (mm) |
| 9 | Length of the rachis in the pinna of first pair (mm) |
| 10 | Length of the rachis in the pinna of last pair (mm) |
| 11 | Minimum number of pinnae pairs |
| 12 | Maximum number of pinnae pairs |
| 13 | Number of leaflet pairs in the first pinna |
| 14 | Number of leaflet pairs in the last pinna |
| 15 | First leaflet pair length (mm) |
| 16 | First leaflet pair width in the first pinna at 3/4 of the length from the base (mm) |
| 17 | First leaflet pair width in the first pinna at ½ of the length from the base (mm) |
| 18 | First leaflet pair width in the first pinna at ½ of the length from the base (mm) |
| 19 | Length of the third pair of leaflets (mm) |
| 20 | Third leaflet pair width in the first pinna at ¾ of the length from the base (mm) |
| 21 | Third leaflet pair width in the first pinna at ½ of the length from the base (mm) |
| 22 | Third leaflet pair width in the first pinna at ¼ of the length from the base (mm) |
| 23 | Length of the last pair of leaflets (mm) |
| 24 | Last leaflet pair width in the first pinna at ¾ of the length from the base (mm) |
| 25 | Last leaflet pair width in the first pinna at ½ of the length from the base (mm) |
| 26 | Last leaflet pair width in the first pinna at¾ of the length from the base (mm) |
| 27 | Peduncle length (mm) |
| 28 | Number of flowers per head |
| 29 | Bracteole length (mm) |
| 30 | Bracteole width at ¾ of the length from the base (mm) |
| 31 | Bracteole width at½ of the length from the base (mm) |
| 32 | Bracteole width at ¼ of the length from the base (mm) |
| 33 | Calyx length (mm) |
| 34 | Corolla tube length (mm) |
| 35 | Corolla lobes length (mm) |
| 36 | Corolla lobes width at the base (mm) |
| 37 | Gynoecium length (mm) |
| 38 | Androecium length (mm) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




