1. Introduction
Esophageal squamous cell carcinoma (ESCC) is one of the most common and fatal cancers, ranking as the sixth leading cause of cancer-associated deaths globally. ESCC is the predominant histological subtype of esophageal cancer, constituting 90% of esophageal cancer worldwide[
1]. ESCC is a severe malignancy owing to its aggressive nature and very poor survival rate, with a 5-year survival rate of less than 30%[
2]. Although the screening and curative treatment techniques such as surgical resection, chemoradiotherapy and immune therapy have witnessed remarkable progress, the treatment for ESCC is still challenging, and the prognosis of patients with ESCC remains limited. There is an urgent need to identify promising biomarkers with higher prognostic accuracy and new drugs for ESCC therapies.
Traditional Chinese medicine (TCM) is a valuable resource for researching anti-tumor drug as it can effectively eliminate tumor cells from the body without causing side effects on normal organ function. Lycorine is a pharmacologically active alkaloid extracted from the traditional Chinese medicinal herb, Lycoris radiata, and has significant therapeutic potential. Recent evidence suggests that Lycorine and its derivatives have anticancer activities[
3], including significant inhibitory effects on various types of cancers such as leukemia, lymphoma, melanoma, breast cancer, ovarian cancer and prostate cancer etc.[
4]. Lycorine hydrochloride (Lyc. HCL), a derivative of Lycorine, is an isoquinoline alkaloid extracted from Lycoris. It shares similar properties with Lycorine, including anti-tumor effects and low toxicity to normal cells[
5], making it a promising candidate for an anticancer agent. Interestingly, Lyc. HCL exhibits a stronger therapeutic effect on tumor cells compared to Lycorine[
5]. However, the effects of Lyc. HCL on esophageal squamous cell carcinoma, including its mechanisms and targets, are still unclear. To deeper explore how Lyc. HCL works on ESCC, we focused on Tripartite motif-containing (TRIM) family, one of the critical regulators in human cellular function and their pathways, which could be the potential target of Lyc. HCL. Someone involved that TRIM family proteins are widely expressed and over 80 TRIM proteins were discovered in human[
6]. Many TRIM family proteins serve as critical regulators in gene regulation, post-transcriptional regulation, cell proliferation, cell signaling pathway, apoptosis, and tumorigenesis[
7]. It is important to note that different members of the TRIM family have diverse function in various cancers, including the regulation proliferation, migration and invasion. Several TRIM family genes have been found to be significantly under- or over-expressed, influencing cancer proliferation, metastases and invasion[
8,
9]. This suggests that the expression levels of TRIMs could serves as biomarkers and prognostic factors of cancer. Additionally, TRIM gene may play different roles in different types of cancers. For example, TRIM22, a member of the TRIM family, is expressed at low levels in melanoma, which is associated with cancer progression and poor prognosis[
10]. Knockout of TRIM22 effectively inhibited tumor proliferation and increased the sensitivity of glioblastoma cells to temozolomide in both in vivo and in vitro[
11]. Over-expression of TRIM22 inhibits osteosarcoma progression by destabilizing NRF2 and activating the ROS/AMPK/mTOR/autophagy signaling pathway[
12]. TRIM22 also acts as a negative regulator of MHCII expression, making it a potential target for checkpoint blockade immunotherapy in cancers treatment[
13]. However, the role of TRIM22 in esophageal squamous cell carcinoma has not been studied to date.
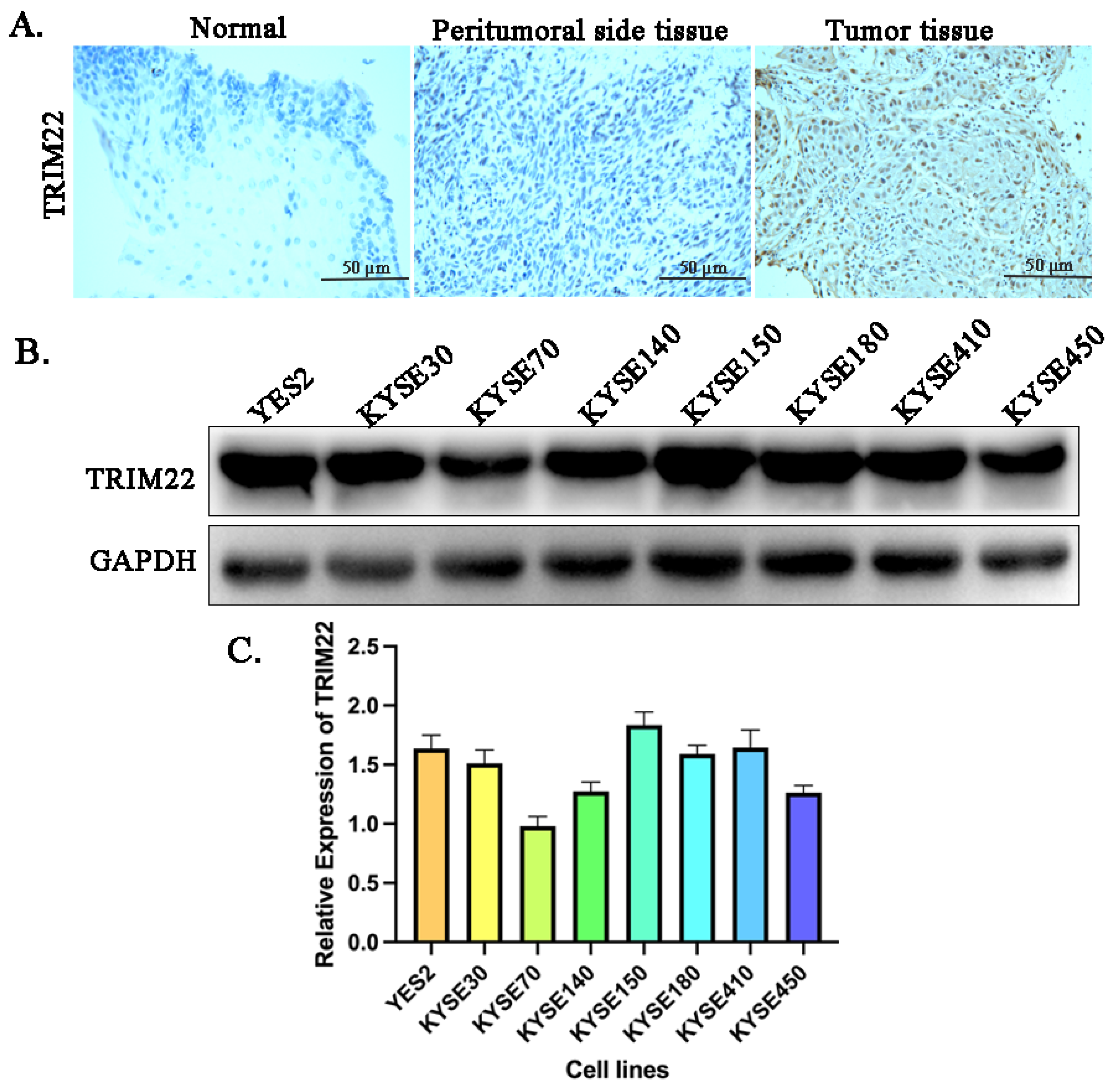
Our study revealed a high expression of TRIM22 in both ESCC patients and ESCC cell lines, such as YES2, KYSE70, KYSE140, KYSE150, KYSE180, KYSE410 and KYSE450. Furthermore, we confirmed that Lyc.HCL effectively suppressed the expression of TRIM22 in ESCC cell lines. Therefore, we propose the hypothesis that Lyc.HCL might exert an anti-esophageal squamous cell carcinoma effect by through its targeting of TRIM22.
2. Materials and Methods
2.1. Cell Culture, Reagents and Treatments
The human esophageal squamous cell carcinoma cell lines (YES2, KYSE30, KYSE70, KYSE140, KYSE150, KYSE180, KYSE410 and KYSE450) were generously provided by Dr. Y. Shimada (Kyoto University). All cells were cultured in RPMI 1640 medium (Gibco, USA), supplemented with 10% fetal bovine serum (Gibco, USA) and 1% penicillin/streptomycin (Gibco, USA). Both types of ESCC cells were cultured in a humidified incubator at 37 °C with 5% CO2. Lycorine hydrochloride (CAS No. 2188-68-3) was purchased from MedchemExpress. We dissolved Ly.HCL in DMSO and diluted it with RPMI 1640 medium (Gibo, USA).
2.2. Normal Esophageal Epithelial Tissue Samples, Patients and Esophageal Squamous Cancer Samples
The study received approval from the Pathology Department of Peking University Shenzhen Hospital (2021-111). From 2018 to 2021, we collected 35 cases of normal esophageal epithelial tissue, 35 cases of peritumoral side tissues and 35 cases of esophageal squamous cancer tissue from the Pathology Department of Peking University Shenzhen Hospital. All procedures conducted in studies involving human participants adhered to the ethical standards of the institution. Informed consent was obtained from all participants. All patients were initially diagnosed with untreated esophageal squamous cancer. To detect the protein expression level of TRIM22, we collected and stored all samples at -80 ℃ for immunohistochemistry assays.
2.3. Immunohistochemistry
The cancer tissue and normal tissue samples from patients were treated with xylene, followed by graded alcohol, Antigen retrieval was then performed in a 0.01 M citrate buffer. Hydrogen peroxide was used for blocking. Tissue sections were incubated with goat serum for 20 minutes. The slides were then incubated overnight at 4 ℃ with the TRIM22 antibody (1:300, Proteintech, USA). Immunostaining of TRIM22 was performed using the EliVision Super Kit from Maixin (Fuzhou, China). Two independent pathologists randomly examined all tumor slides. TRIM22 staining was observed in both the cytoplasmic and nuclear compartments of tumor cells. Nuclear staining was considered positive.
2.4. Cell Proliferation and Apoptosis Assay
Cells were seeded in 96-well plates and treated with Lyc.HCL at various concentrations. Cell proliferation was evaluated at 24h, 48h, and 72h after treatment using the MTS assay (Promega) following the manufacturer’s instructions. Apoptosis was measured after 48h of treatment using an Annexin V-FITC apoptosis detection kit (Beyotime, China) following the manufacturer’s instructions.
2.5. Colony Formation Assay
ESCC cells were seeded at a density of 5000 cells per well in 6-well plates. The cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum,1% penicillin/streptomycin, and specify concentrations of Lyc.HCL. After a 2-week incubation period, the cultures were washed with pre-cooled PBS, fixed with methanol, and stained with a 0.1% crystal violet solution for 30 minutes. The colonies were automatically examined and quantified using Image-Pro Plus.
2.6. Cell Cycle Assay
After treatment with different concentrations of Lyc.HCL for 48h, 1×106 cells were collected, trypsinized, and fixed in 70% ethanol overnight. Then the cells were washed 3 times with pre-cooled PBS and incubated with a PI-staining solution with RNase A (BD Biosciences, USA) for at least 15 minutes at room temperature before analysis. The cells were run on a FACS cytometer (BD Biosciences, USA) in accordance with the manufacturer’s guidelines.
2.7. Cell Migration Assays
The wound healing assay is one of the earliest methods developed to investigate directional cell migration in vitro. Cells were seeded in a 6-well plate and allowed to attach overnight until reaching 80% confluence. Subsequently, cell monolayers were wounded using 200 µL pipette tips and washed with 1× PBS 3 times to remove floating cells. Cells treated with different concentrations of Lyc.HCL were then incubated in RPMI 1640 medium. Cells migrated into the wound surface and the number of migrating cells was observed under an inverted microscope at 0, 12, and 24 hours. Three randomly chosen fields were observed for each well. The percentage of migration was calculated relative to untreated wells as 100%. All the independent experiments were conducted in triplicate. the results were analyzed using ImageJ and GraphPad prism 5.0.
2.8. Transwell Migration Assays
Migration of cells was evaluated in Transwell cell culture chambers with 6.5 mm diameter polycarbonate membrane filters containing 8 μm pore size (Neuro Probe, Gaithersburg, MD, United States). In total, the upper chamber was filled with 200 μL of serum-free medium with specify concentrations of Lyc.HCL and added 5×105 YES2 or KYSE150 cells, and the lower chamber was filled with 500 μL medium containing 20% FBS. After 24 hours of incubation at 37 °C, the non-migrated cells were removed from the upper surface of the membrane with a cotton swab. The filters were then fixed in methanol for 10 minutes, stained with crystal violet solution for 1 hour and counted. Five random microscopic fields (100×) were counted per well and analyzed with the mean value.
2.9. Cell Invasion Assay
A Matrigel invasion assay was conducted to examine tumor cell invasion. Initially, Transwell upper chambers were filled with 0.1 mL Matrigel (Becton Dickinson, Bedford, MA) and incubated at 37 °C for 1 hour. Subsequently, cells were treated with varying concentrations of Lyc.HCL medium for 24 hours. The cells were then trypsinized and suspended in serum free RPMI 1640 medium at a final concentration of 5 × 105 cells/mL. The. cell suspensions were then placed in the upper chamber, and medium with 20% fetal bovine serum was added to the lower chamber. After incubating at 37 °C with 5% CO2 for 24 hours, non-invading cells on the upper surface were removed using a cotton swab. All cells were stained with crystal violet and counted under an inverted microscope. Ten random views were selected for cell counting, and each independent experiments were repeated three times.
2.10. Lentiviral Infection, Transient Transfection and Cas9/sgRNA KO
The plasmids and siRNAs were transfected into cells using Lipofectamine 3000 reagent (Invitrogen, USA). The experimental operation was conducted strictly following the instructions provided by the transfection reagent manufacturer. The Leti-Cas9-puro and single-guide RNA (sgRNAs) lentiviruses were designed and constructed by Genechem (Shanghai, China). Lentivirus carrying Cas9 and sgRNAs infected YES2 and KYSE150 cells. After 48 hours of infection, the cells were cultured in fresh medium containing Puro for 15 days to screen cells with stable expression. All plasmids were designed and constructed by Hanbio (Shanghai, China).
2.11. Western Blotting
Protein extraction and western blotting were conducted as described previously[
14]. Briefly, proteins were isolated using RIPA lysis buffer (Beyotime, China). Then 24 μg protein was loaded and separated by 12% SDS-polyacrylamide gel electrophoresis and transferred onto a polyvinylidene difluoride membrane and incubated with following antibodies: JAK2 (#3230T), PI3K (#17366), mTOR (#2983), AKT (#4691), p-AKT (#4060T), p-MEK (#9127S), Erk (#4370), p-Erk (#4370T), and GAPDH (#51332), all of these antibodies were purchased from Cell Signaling Technology, and were diluted at a ratio of 1:1000 using a diluent purchased from Beyotime Biotechnology. Additionally, TRIM22 (#13744-1-AP) was purchased from Proteintech company and were diluted at a ratio of 1:500 using the same diluent. And STAT3 (#ab68153), p-STAT3 (#ab267373), p-p27 (#ab32034), p-p27 (#ab32034), p-Raf (#ab112053) were purchased from Abcam company and were diluted at a ratio of 1:1000 using the same diluent. The chemiluminescence signals were detected using the Amersham Imager 600 (GE, USA).
2.12. Rescue Assays
YES2 and KYSE150 cells were seeded at 20000 cells/well in 6-well plates and incubated overnight. Transient transfection of TRIM22siRNA and plasmid expressing TRIM22 into YES2 and KYSE150 cells for 24 hours, Add Lyc.HCL (4 μmol/mL) treated cells (YES2shTRIM22, YES2TRIM22, KYSE150shTRIM22, KYSE150TRIM22) for 48 hours. Then cells were collected and measured by colony formation assay, cell cycle assay, cell migration assay, cell invasion assay and western blotting assay as previously described.
2.13. Tumor Xenograft Experiments
Six-week-old female BALB/c nude mice were randomly divided into nine groups, with six mice in each. The groups were named as follows: KYSE150-NC, KYSE150-NC-Lyc.HCL (5 mg/kg), KYSE150-NC-Lyc.HCL (10 mg/kg), KYSE150-shTRIM22, KYSE150-shTRIM22-LycHCL (5 mg/kg), KYSE150-shTRIM22-Lyc.HCL (10 mg/kg), KYSE150-TRIM22, KYSE150-TRIM22-Lyc.HCL (5 mg/kg), KYSE150-TRIM22-Lyc.HCL (10 mg/kg). Every group received a subcutaneous injection of 5×106 cells on the right rear of the mice. After seven days of subcutaneous tumorigenesis, the mice in the groups KYSE150-NC-Lyc.HCL (5 mg/kg), KYSE150-NC-Lyc.HCL (10 mg/kg), KYSE150-shTRIM22-Lyc.HCL (5 mg/kg), KYSE150-shTRIM22-Lyc.HCL (10 mg/kg), KYSE150-TRIM22-Lyc.HCL (5 mg/kg), KYSE150-TRIM22-Lyc.HCL (10 mg/kg) were treated with intraperitoneal injection of Lyc.HCL (5 mg/kg/twice a day per mouse) and Lyc.HCL (10 mg/kg/twice a day per mouse) for 14 days respectively, while the other two groups mice were injected with DMSO. Tumor volumes (mm3) and mice weight were measured twice a day using a vernier caliper and calculated using the formula, tumor volume = L × W× 0.5 (L: the longest diameter of the tumor, W: the shortest diameter of the tumor).
Subsequently, mice were euthanized by intraperitoneal injection of 150 mg/kg sodium pentobarbital to halt their breathing, and cardiac arrest was confirmed. The tumor tissue was then dissected and photographed. Tumor tissue was embedded, cut into 4 µm thick slices, fixed in 4% paraformaldehyde for 15 min at room temperature, and then stained with hematoxylin for 10 min and eosin for 2 min at room temperature. The tissue was observed under an Olympus BX40 light microscope (Olympus Corporation). An immunohistochemistry assay was performed to analyze the expression of TIRM22, JAK2 and p-AKT. Western blot analysis was used to test the expression of TRIM22 and JAK2/STAT3/PI3K/AKT signaling pathways in tumor tissue.
2.14. Statistical Analysis
Each the experiments were conducted in duplicate and repeated three times. Statistical analysis of the in vitro data was performed using the student’s t-test and 1-way ANOVA. The student’s t-test was used to compare the different groups. *p < 0.05, **p < 0.01, and ***p < 0.001. A p-value of less than 0.05 was considered statistically significant for all data.
4. Discussion
Despite significant advancements in the treatment of ESCC, it continues to be a highly lethal cancer, with a survival rate of less than 30% at 5 years[
21]. Therefore, it is imperative to identify more effective therapeutic targets and develop new drugs to enhance the treatment efficiency of ESCC. Emerging evidence suggests that Lyc.HCL holds significant potential in the treatment of various human cancers[
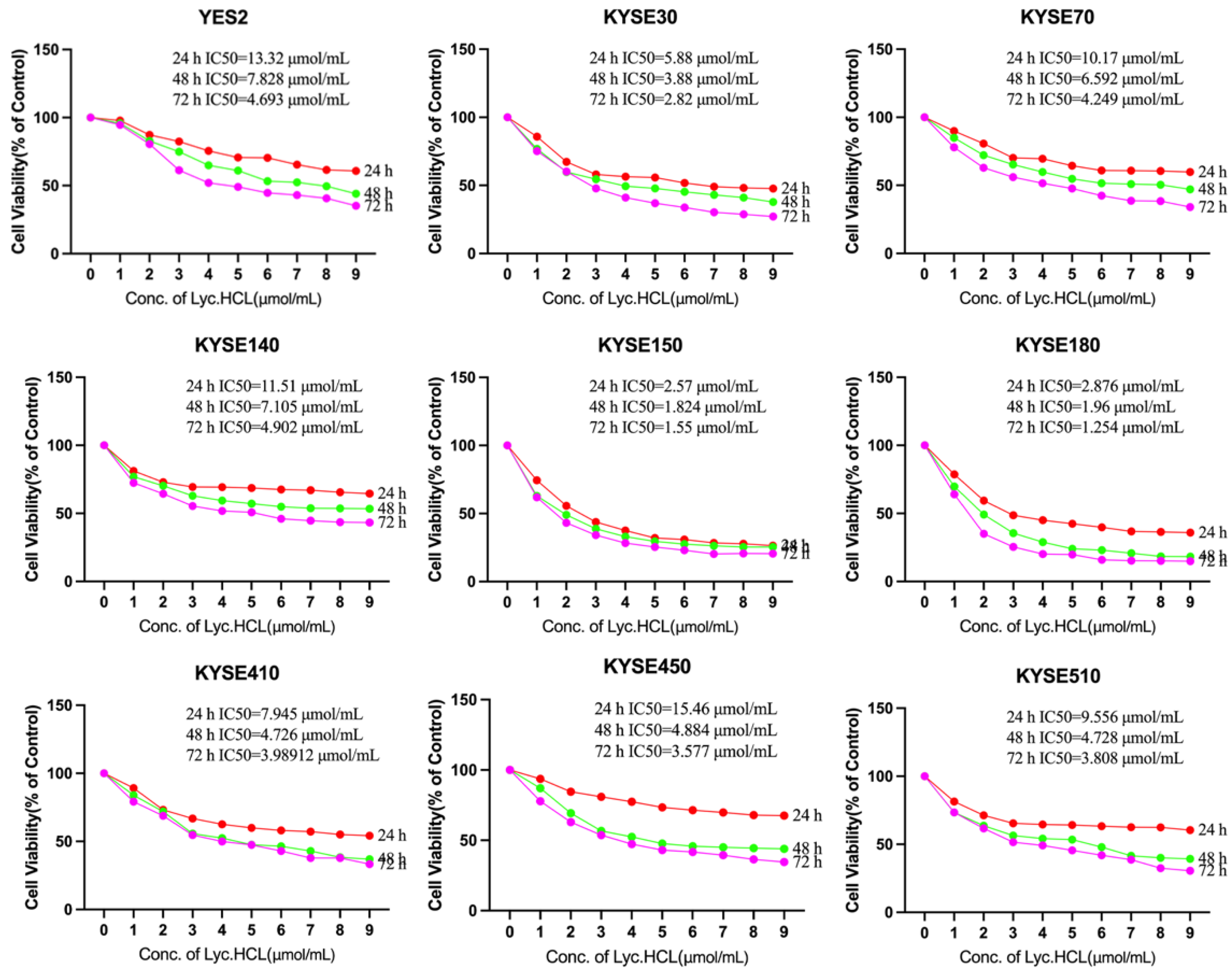
22]. Our study revealed that Lyc.HCL effectively suppressed the proliferation of ESCC cell lines in a dose- and time-dependent manner (
Figure 2 and
Figure 3A, B).
TRIM22 is a versatile protein that plays multiple roles in cellular differentiation and proliferation, inhibits viral replication, activates immune cells and regulates ubiquitination in vivo[
23,
24,
25,
26]. In addition, TRIM22 is involved in the regulation of cellular differentiation and proliferation[
27], which may have a crucial impact on various cancers. It is worth nothing that TRIM22 is downregulated in melanoma patients, who exhibit shorter survival compared to those with high expression of TRIM22[
10]. Conversely, TRIM22 is upregulated in non-small cell lung cancer, and the overexpression is associated with poor survival in lung cancer patients[
16]. Consequently, the functional role and status of TRIM22 in human cancers remain ambiguous, and its specific role in the progression of ESCC has yet to be elucidated. To determine the status of TRIM22 in ESCC, immunohistochemistry was performed to assess its protein expression in 35 cases of ESCC tissues, 35 cases of paracancerous tissues, and 35 cases of normal tissues. Our results demonstrate a significant upregulation of TRIM22 protein expression in ESCC patient specimens while it is not expressed in paracancerous tissues and normal esophagus tissues. The western blot data demonstrated that the level of TRIM22 was overexpressed in ESCC cell lines. It is noteworthy that Lyc.HCL significantly inhibited the expression of TRIM22 in vitro ESCC cell lines and in vivo xenograft tumors in a dose-dependent manner. Our results also showed that Lyc.HCL inhibited TRIM22 expression and inactivated JAK2/STAT3, PI3K/AKT, MAPK/ERK and FOXO pathways both in vitro and in vivo. Rescue assays demonstrated that overexpression of TRIM22 activated JAK2/STAT3, PI3K/AKT, MAPK/ERK and PI3K/AKT/FOXO pathways in vitro and in vivo. Conversely, TRIM22 knockout inactivated the same pathways both in vitro and in vivo. Lyc.HCL suppressed the expression of TIRM22 and the activation of pathways. These data suggest that TRIM22 could serve as a potential oncoprotein in ESCC and may be an excellent target for Lyc.HCL in the treatment of ESCC due to its control of muti-pathway expression in tumors. TRIM family genes are induced by IFN-γ stimulation and TRIM22 is one of the most strongly induced TRIM proteins by IFN-γ[
13]. IFN-γ induced activation of the JAK2/STAT and PI3K/AKT pathways[
28], both of which are responsible for cell growth and proliferation. The PI3K/AKT/mTOR pathway is hyperactivated or altered in many cancer types and regulates a broad range of cellular processes, including survival, proliferation, growth, metabolism, angiogenesis and metastasis[
29]. The PI3K/AKT pathway is regulated by a diverse array of upstream signaling proteins and collaborates with multiple compensatory signaling pathways, primarily the MAPK/ERK pathway, to regulate numerous downstream effectors[
29]. PI3K/AKT and MAPK/ERK signaling pathways are well-established oncogenic signaling pathways that play a crucial role in regulating tumorigenesis and development[
30]. Among all MAPK signal transduction pathways, the MAPK/ERK pathway stands out as the most vital signaling cascade, playing a pivotal role in the survival and development of tumor cells. The role of ERK in tumor proliferation, invasion and metastasis is prominently emphasized[
31]. FOXO1 is among the downstream targets of PI3K/AKT pathway[
32]. Therefore, we propose that Lyc.HCL has the potential to suppress ESCC cell proliferation, metastasis and invasion by targeting TRIM22 and modulating JAK2/STAT3, PI3K/AKT, MAPK and PI3K/AKT/FOXO signaling pathways. Nevertheless, the interplay among the JAK2/STAT, PI3K/AKT, MAPK and PI3K/AKT/FOXO signaling pathways, which holds promise for the advancement of novel therapeutic strategies for cancer, requires further investigation.
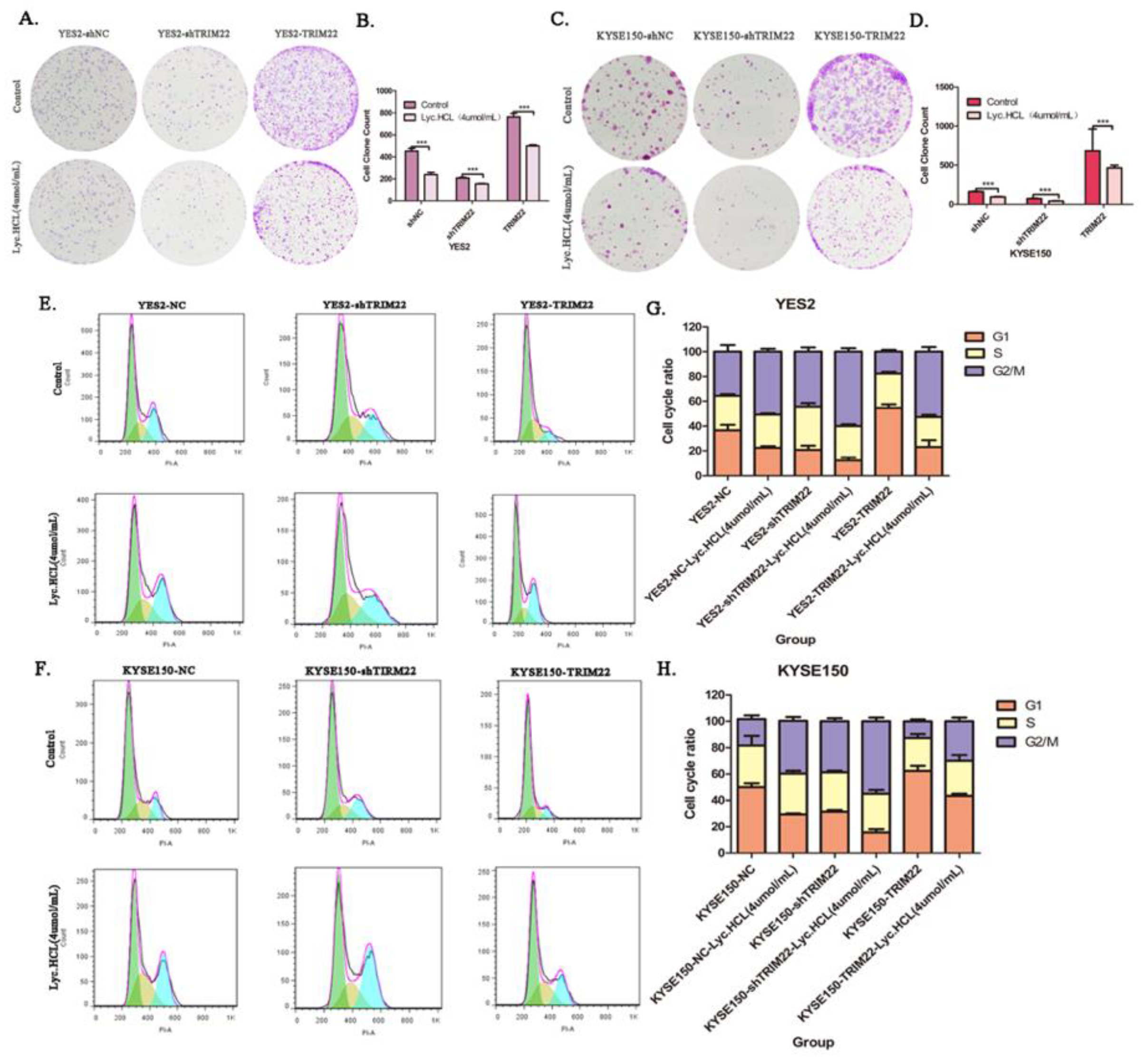
We conducted a colony formation assay to investigate the mechanism by which Ly.HCL mediates its anti-ESCC effect, the assay showed that Lyc.HCL suppressed cell growth in vitro. Additionally, we showed that Lyc.HCL treatment resulted in delaying tumor growth and reduced tumor cachexia in vivo. Both in vitro and in vivo rescue assays revealed that TRIM22 overexpression significantly promoted proliferation while silencing of TRIM22 reversed this effect. The ability of Lyc.HCL to reduce TRIM22-induced proliferation suggests that Lyc.HCL exerts its anti-proliferation effects by targeting TRIM22. Uncontrolled tumor growth has been primarily linked to aberrant cell cycle regulation[
33]. Cell cycle analysis revealed that Lyc. HCL can induce cell cycle arrest at the G2/M phase in YES2 and KYSE150 cells. Rescue assays showed that TRIM22 overexpression the significantly promoted cell cycle progression, while TRIM22 silencing reversed this effect. Lyc.HCL can induce cell cycle arrest at G2/M phase, which is triggered by TRIM22. Furthermore, we observed a significant inhibition of p-p27 in YES2 and KYSE150 cells treated with Lyc.HCL during cell cycle. p-p27 plays a crucial role in the activation of cyclin D/ CDKs complex or the cyclin E/ CDK2 complex, which promotes the cell cycle when cyclins bind to CDKs[
34].The phosphorylation of p27 at Thr157 is referred to as p-p27. Inhibition of p-p27 can hinder the formation of the cyclin D/ CDKs complex, leading to cell cycle arrest. p-p27 is downstream molecules of PI3K/AKT signaling pathway[
35]. Phosphorylation of AKT can upregulate cyclin D1 and downregulate the cyclin D kinase (CDK) inhibitors p21 and p27, promoting cell cycle progression[
36]. The present study indicated that inactivation of the PI3K/AKT pathway by Lyc.HCL in YES2 and KYSE150 cells led to the suppression of p-AKT and p-p27 ultimately causing cell cycle arrest. In addition, Fork head Box O (FOXO) is a subfamily of the fork-head transcription factor family that plays a crucial role in determining cell fate. This subfamily is also believed to function as a pivotal tumor suppressor in various cancers[
37]. FOXO proteins are translocated to the cytosol and subsequently degraded through the ubiquitin-proteasome pathway. In the absence of growth factors’ survival signals, FOXO proteins translocate to the nucleus and upregulate multiple target genes, promoting cell cycle arrest, stress resistance, and apoptosis[
38]. However, it is still not well understood whether the target genes of different transcription factors are distinct or similar. FOXO transcription factors may have different roles, and exhibit opposite functions under different conditions[
38]. Considering that FOXO1 is one of the gene associated with cell cycle transition[
39,
40]. In our study, we observed that Lyc.HCL suppressed the expression of p-FOXO1 in YES2 and KYSE150 cells, which may be linked to cell cycle arrest. Importantly, the PI3K/AKT pathway is the key pathway that interacts with FOXO in various types of cancers[
37]. FOXO is also a downstream molecular in the PI3K/AKT pathway[
41]. The PI3K/AKT pathway is strongly associated with tumor growth and metastasis[
42]. Our findings revealed that Lyc.HCL led to the downregulation of PI3K/AKT pathway, p-FOXO and p-p27. These data suggest that Lyc.HCL may inhibit the PI3K/AKT/FOXO pathway and Thr157 phosphorylation of p27 in a dose-dependent manner, ultimately leading to cell cycle arrest in the G2/M phase and inhibition of cell growth.
Previous studies have suggested that the reduction in tumor growth and cell viability can be attributed to the induction of cell cycle arrest and apoptosis[
43]. However, the cell apoptosis assay confirmed that Lyc.HCL induced apoptosis in YES2 and KYSE150 cells to some extent, although the effect was not obvious. Further investigation is required to explore alternative mechanism of cell death, including ferroptosis and autophagy.
Tumor metastasis and invasion are closely associated to prognosis and play a crucial role in tumor treatment[
44]. This study utilized the wood healing assay and Transwell assay to demonstrate that Lyc.HCL inhibited the metastatic ability of YES2 and KYSE150 cells. The Matrigel invasion assay confirmed that Lyc.HCL suppressed the invasive ability of YES2 and KYSE150 cells. This study is the first to demonstrate, at the cellular level, that Lyc.HCL inhibits the metastasis and invasion of ESCC YES2 and KYSE150 cells in a dose-dependent manner. Additionally, rescue assays demonstrated that TRIM22 overexpression significantly enhanced migration and invasion, whereas the results were significantly reversed upon TRIM22 depletion. Lyc.HCL inhibited migration and invasion induced by TRIM22, indicating its role in suppressing anti-ESCC metastasis and invasion by targeting TRIM22. Mechanically, previous studies have reported that TRIM22 knockout significantly inhibited the proliferation, migration and invasion of fibroblast-like synoviocytes in rheumatoid arthritis[
25]. Consistent with the afore mentioned studies, our study elucidated the role of TRIM22 in the ESCC cell lines and the effectiveness of Lyc.HCL in inhibiting ESCC metastasis and invasion. The results of the current study demonstrated that TRIM22 was expressed at significantly higher levels in ESCC cell lines and tumor tissues from ESCC patients. Lyc.HCL downregulated the expression of TRIM22 and significantly inhibited the proliferation, migration and invasion of YES2 and KYSE150 cell lines. The fork head family of transcription factors is associated with various biological processes, including proliferation, metastasis and invasion[
45]. It was reported that FOXC1 transcription factor has a binding site on the TRIM22 promoter. Overexpression of FOXC1 significantly increased the mRNA expression and protein expression of TRIM22[
25]. The current study suggested that p-FOXO1, a member of the fork head family of transcription factors, was reduced in YES2 and KYSE150 cells after the treatment with Lyc.HCL. This reduction may explain the mechanism of inhibition of migration and invasion in YES2 and KYSE150. In addition, rescue assays indicated a close relationship between the expression of p-FOXO1 and TRIM22, and Lyc.HCL suppressed p-FOXO1 by targeting TRIM22. However, further studies are needed to explore about the role of p-FOXO1 in reduction of migration and invasion by Lyc.HCL. The PI3K/AKT/mTOR pathway overtly regulates migration and invasion in various tumors[
46,
47,
48]. Additionally, it has been highlighted that activation of MAPK pathway promotes metastasis and invasion[
49,
50,
51,
52,
53,
54]. Recently, JAK2/STAT3 signaling axis was also reported to play a crucial role in metastasis and invasion of tumor cells[
55,
56], this indicates that potential drugs targeting the JAK2/STAT3 pathway may be developed for tumor therapy. The present study certificated that Lyc.HCL exerts its anti-migration and anti-invasion effects by targeting TRIM22 and simultaneously affecting the JAK/STAT3, PI3K/AKT, MAPK/ERK and FOXO pathways. This suggested that Lyc.HCL may be capable of controlling additional pathways and have one more effective function in inhibiting tumor metastasis and invasion, which is noteworthy. However, the role of downstream classical molecules in metastasis and invasion such as ECM and EMT, which are two critical steps[
57], needs to be further explored.
Meanwhile, it is important to critically consider the of our study. Firstly, our focus was solely on the expression of TRIM22 in ESCC patients without any treatment or drug interference. It is crucial to consider more detailed clinical information about ESCC patients, including tumor stages, metastasis, ages and gender. This will provide a more comprehensive understanding of potential effectiveness of TRIM22 as molecular marker for evaluating the prognosis of ESCC. Secondly, we findings indicated that Lyc.HCL inhibited proliferation, metastasis and invasion by targeting TRIM22 through JAK2/STAT3, PI3K/AKT, MAPK/ERK pathways. However, our in vivo experiments only involved the construction of a subcutaneous xenograft model to explore the inhibition of proliferation by Lyc.HCL. Additionally, we studied the role of TRIM22 in vivo. Although Lyc.HCL demonstrates significant anti-metastasis and anti-invasion effect in ESCC in vitro, we did not conduct further in vivo experiments to confirm these findings. Furthermore, our study utilized human ESCC cell line-derived xenograft models for in vivo research, rather than more diverse preclinical models such as ESCC patient-derived xenograft models or transgenic mice. These alternative models are necessary to fully unveil the anti-ESCC functions of Lyc.HCL and the role of TRIM22. Lastly, the crosstalk between different pathways that Lyc.HCL acted on to inhibit ESCC proliferation, metastasis and invasion has not been elucidated.
Figure 1.
High expression of TRIM22 in esophageal squamous cell carcinoma patients and human esophageal squamous cell carcinoma cell lines. (A) Immunohistochemistry assay shows the expression of TRIM22 in esophageal tissue, peritumoral side tissue and esophageal cancer tissue. (B) Western blot analysis of TRIM22 expression were performed on human esophageal squamous cell carcinoma cell lines. (C) The quantification of relative expression of TRIM22 in different cell lines by Image J.
Figure 1.
High expression of TRIM22 in esophageal squamous cell carcinoma patients and human esophageal squamous cell carcinoma cell lines. (A) Immunohistochemistry assay shows the expression of TRIM22 in esophageal tissue, peritumoral side tissue and esophageal cancer tissue. (B) Western blot analysis of TRIM22 expression were performed on human esophageal squamous cell carcinoma cell lines. (C) The quantification of relative expression of TRIM22 in different cell lines by Image J.
Figure 2.
Effect of Lyc.HCL on proliferation of human esophageal squamous cell carcinoma cell lines (ESCC). YES2, KYSE30, KYSE70, KYSE140, KYSE150, KYSE180, KYSE410 and KYSE450 cells were treated with the indicated concentrations of Lyc.HCL for 24h, 48h and 72 h, cell viability was assessed using MTS assay. IC50 values were calculated using the GraphPad Prism 5.0 software. Data are presented as mean±SD.
Figure 2.
Effect of Lyc.HCL on proliferation of human esophageal squamous cell carcinoma cell lines (ESCC). YES2, KYSE30, KYSE70, KYSE140, KYSE150, KYSE180, KYSE410 and KYSE450 cells were treated with the indicated concentrations of Lyc.HCL for 24h, 48h and 72 h, cell viability was assessed using MTS assay. IC50 values were calculated using the GraphPad Prism 5.0 software. Data are presented as mean±SD.
Figure 3.
Effect of Lyc.HCL on cell colony formation, cell cycle and apoptosis of ESCC cells. (A) Results of colony formation assays for YES2 and KYSE150 cells. ESCC cells were treated with the indicated concentrations of Lyc.HCL for 14 days. (B) The quantification of the cell colonies in (A) is presented as the mean percentage of viable cells (mean±SD), averaged from 3 independent experiments, each with 3 replicates per condition.(C) YES2 and KYSE150 cells were treated with vehicle, 2, 4, and 6µmol/mL of Lyc.HCL for 48 h, and then stained with PI and subjected by FACS. (D and E) cell cycle was analyzed by Mod Fit 5.0. All data are presented as the mean±SD. A 1-way analysis of variance, followed by a Tukey’s post-hoc test, was used to compare the different groups. *P<.05, **P<.01, ***P<.001 versus vehicle. (F) YES2 and KYSE150 cells were treated with vehicle, 2, 4, and 6 µmol/mL of Lyc.HCL for 48 h. Flow cytometry analyzed apoptotic cells stained by Annexin V and PI. (G) data are presented as the mean±SD. A 1-way analysis of variance, followed by a Tukey’s post-hoc test, was used to compare the different groups. *P<.05, **P<.01, ***P<.001 versus vehicle.
Figure 3.
Effect of Lyc.HCL on cell colony formation, cell cycle and apoptosis of ESCC cells. (A) Results of colony formation assays for YES2 and KYSE150 cells. ESCC cells were treated with the indicated concentrations of Lyc.HCL for 14 days. (B) The quantification of the cell colonies in (A) is presented as the mean percentage of viable cells (mean±SD), averaged from 3 independent experiments, each with 3 replicates per condition.(C) YES2 and KYSE150 cells were treated with vehicle, 2, 4, and 6µmol/mL of Lyc.HCL for 48 h, and then stained with PI and subjected by FACS. (D and E) cell cycle was analyzed by Mod Fit 5.0. All data are presented as the mean±SD. A 1-way analysis of variance, followed by a Tukey’s post-hoc test, was used to compare the different groups. *P<.05, **P<.01, ***P<.001 versus vehicle. (F) YES2 and KYSE150 cells were treated with vehicle, 2, 4, and 6 µmol/mL of Lyc.HCL for 48 h. Flow cytometry analyzed apoptotic cells stained by Annexin V and PI. (G) data are presented as the mean±SD. A 1-way analysis of variance, followed by a Tukey’s post-hoc test, was used to compare the different groups. *P<.05, **P<.01, ***P<.001 versus vehicle.
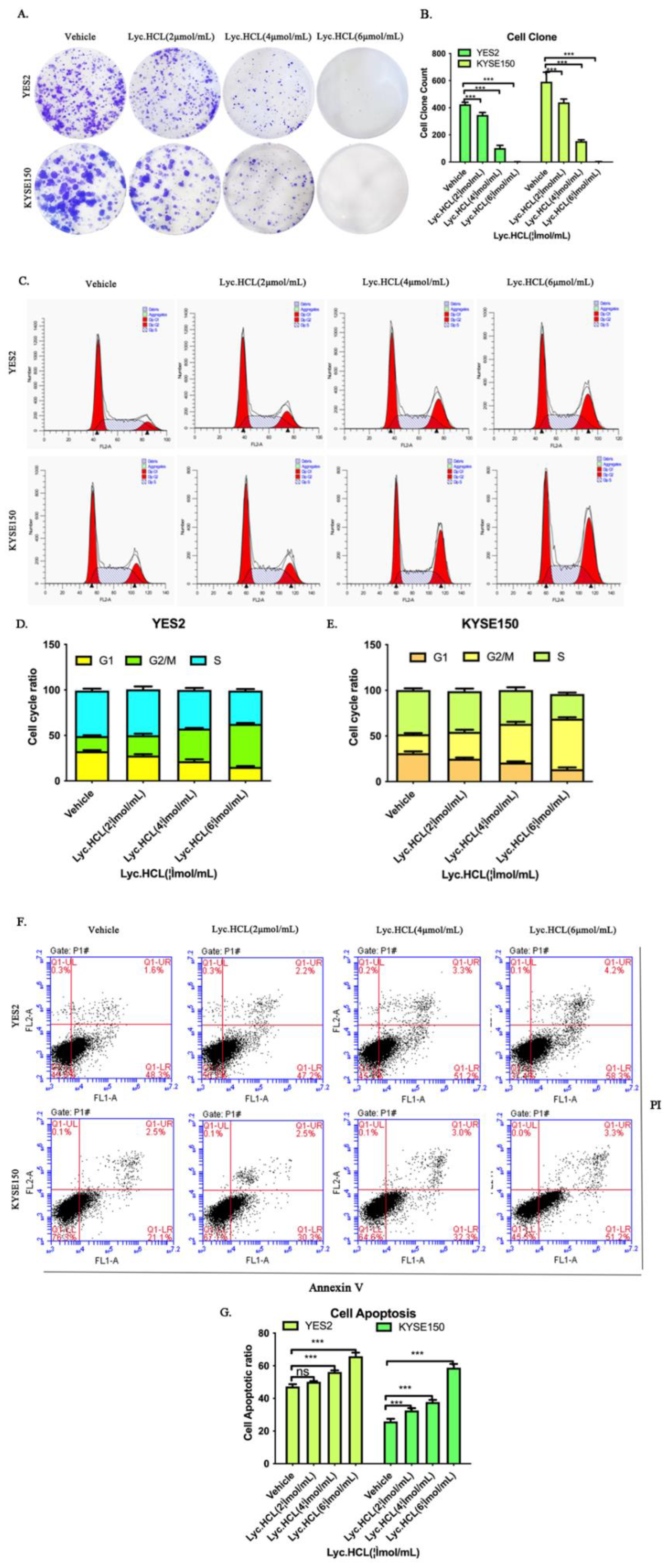
Figure 4.
Lyc.HCL inhibited migration and invasion in YES2 and KYSE150 cells. (A and C) YES2 and KYSE150 cells were treated with vehicle, 2, 4, and 6 µmol/mL of Lyc.HCL for 48 h. Cell migration was evaluated by the wound healing assay. Scale bar, 200µm. (B and D) Gap Width data were presented as the mean±SD. A 1-way analysis of variance, followed by a Tukey’s post-hoc test, was used to compare the different groups. *P<.05, **P<.01, ***P<.001 versus vehicle. (E) YES2 and KYSE150 cells, pretreated with vehicle, 2, 4, and 6µmol/mL of Lyc.HCL for 12 h, were plated onto the apical side of filters in serum free medium containing either vehicle or Lyc.HCL, medium containing 20% FBS was placed in the basolateral chamber to act as a chemoattractant for 24h. Cells on the bottom of the filter were stained by 0.5% crystal violet and then counted. (F) Quantification of the migrated cells in (E) was displayed on the right. The results were displayed as the mean±SD. A 1-way analysis of variance, followed by a Tukey’s post-hoc test, was used to compare the different groups. *P<.05, **P<.01, ***P<.001 versus vehicle. (G) Before experiment, transwell chamber was covered with matrix glue. YES2 and KYSE150 cells, pretreated with vehicle, 2, 4, and 6µmol/mL of Lyc.HCL for 12h, were plated onto the apical side of filters in serum free medium containing either vehicle or Lyc.HCL, medium containing 20% FBS was placed in the basolateral chamber to act as a chemoattractant for 24hours. Cells on the bottom of the filter were stained by 0.5% crystal violet and then counted. (H) Quantification of the invasive cells in (G) was displayed on the right. The results were displayed as the mean±SD. A 1-way analysis of variance, followed by a Tukey’s post-hoc test, was used to compare the different groups. *P<.05, **P<.01, ***P<.001 versus vehicle.
Figure 4.
Lyc.HCL inhibited migration and invasion in YES2 and KYSE150 cells. (A and C) YES2 and KYSE150 cells were treated with vehicle, 2, 4, and 6 µmol/mL of Lyc.HCL for 48 h. Cell migration was evaluated by the wound healing assay. Scale bar, 200µm. (B and D) Gap Width data were presented as the mean±SD. A 1-way analysis of variance, followed by a Tukey’s post-hoc test, was used to compare the different groups. *P<.05, **P<.01, ***P<.001 versus vehicle. (E) YES2 and KYSE150 cells, pretreated with vehicle, 2, 4, and 6µmol/mL of Lyc.HCL for 12 h, were plated onto the apical side of filters in serum free medium containing either vehicle or Lyc.HCL, medium containing 20% FBS was placed in the basolateral chamber to act as a chemoattractant for 24h. Cells on the bottom of the filter were stained by 0.5% crystal violet and then counted. (F) Quantification of the migrated cells in (E) was displayed on the right. The results were displayed as the mean±SD. A 1-way analysis of variance, followed by a Tukey’s post-hoc test, was used to compare the different groups. *P<.05, **P<.01, ***P<.001 versus vehicle. (G) Before experiment, transwell chamber was covered with matrix glue. YES2 and KYSE150 cells, pretreated with vehicle, 2, 4, and 6µmol/mL of Lyc.HCL for 12h, were plated onto the apical side of filters in serum free medium containing either vehicle or Lyc.HCL, medium containing 20% FBS was placed in the basolateral chamber to act as a chemoattractant for 24hours. Cells on the bottom of the filter were stained by 0.5% crystal violet and then counted. (H) Quantification of the invasive cells in (G) was displayed on the right. The results were displayed as the mean±SD. A 1-way analysis of variance, followed by a Tukey’s post-hoc test, was used to compare the different groups. *P<.05, **P<.01, ***P<.001 versus vehicle.
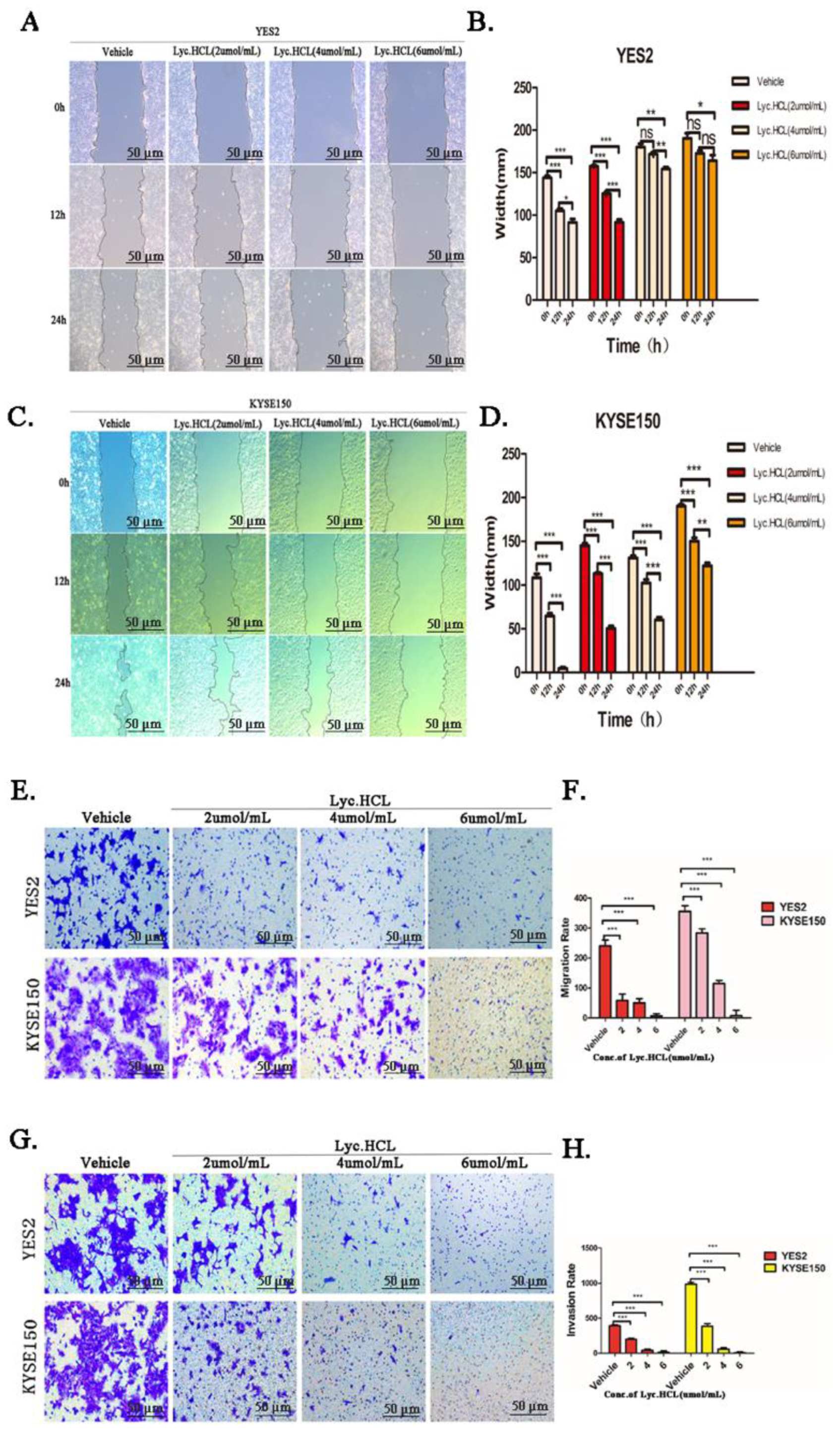
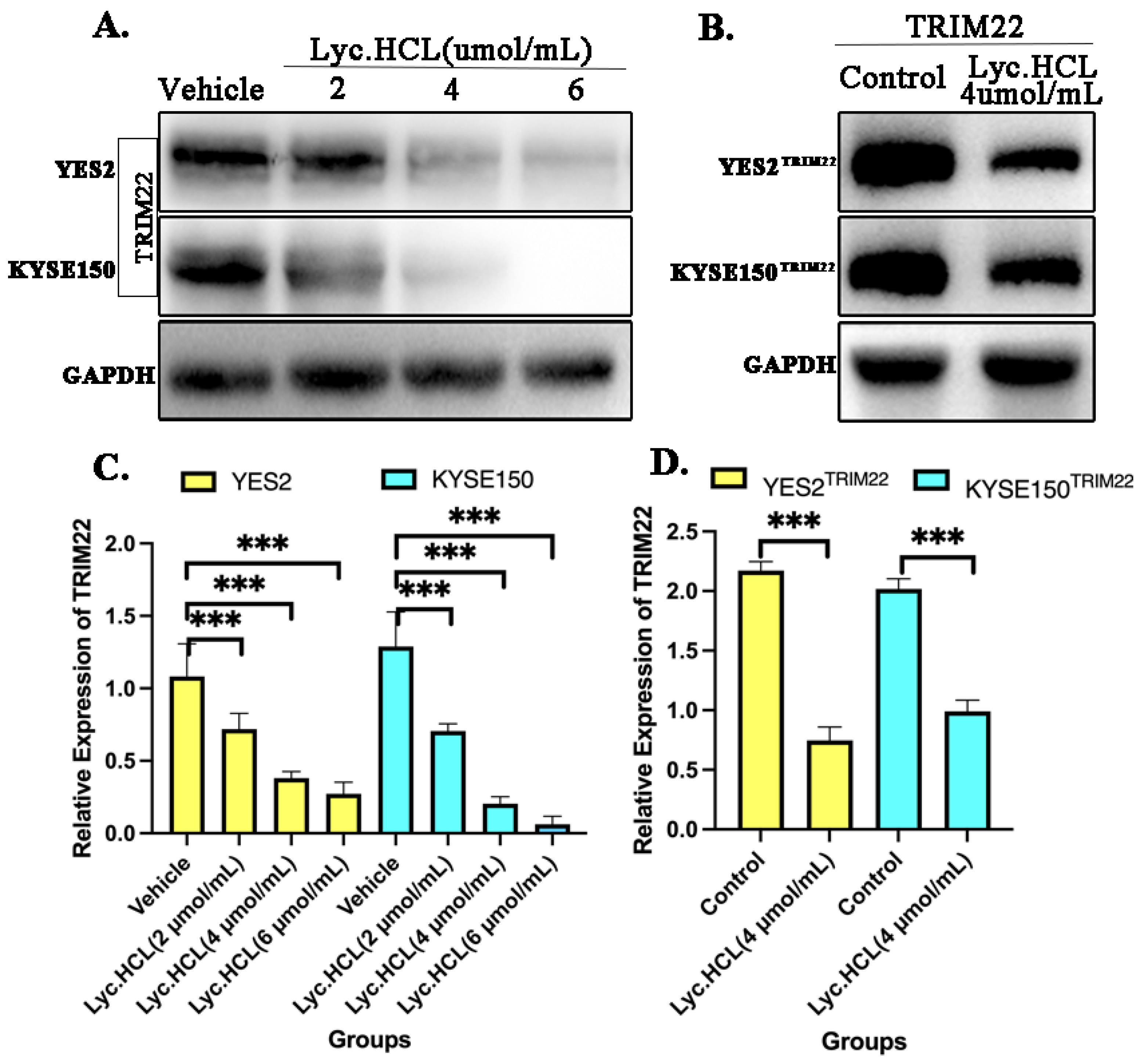
Figure 5.
TRIM22 was inhibited in YES2 and KYSE150 treated by Lyc.HCL. (A)Western blotting analysis of TRIM22 was performed when YES2 and KYSE150 were treated with vehicle, 2, 4, and 6µmol/mL of Lyc.HCL for 48h. (B) Both YES2 and KYSE150 cells were transfected with TRIM22, constructing over-expressing TRIM22 YES2 and KYSE150 cells. Treatment with Lyc.HCL reduced both the over-expression levels of TRIM22 in YES2TRIM22 and KYSE150TRIM22 cells. (C) The quantification of relative expression of TRIM22 in YES2 and KYSE150 cell by Image J. (D) The quantification of relative expression of TRIM22 in YES2TRIM22 and KYSE150TRIM22 by Image J.
Figure 5.
TRIM22 was inhibited in YES2 and KYSE150 treated by Lyc.HCL. (A)Western blotting analysis of TRIM22 was performed when YES2 and KYSE150 were treated with vehicle, 2, 4, and 6µmol/mL of Lyc.HCL for 48h. (B) Both YES2 and KYSE150 cells were transfected with TRIM22, constructing over-expressing TRIM22 YES2 and KYSE150 cells. Treatment with Lyc.HCL reduced both the over-expression levels of TRIM22 in YES2TRIM22 and KYSE150TRIM22 cells. (C) The quantification of relative expression of TRIM22 in YES2 and KYSE150 cell by Image J. (D) The quantification of relative expression of TRIM22 in YES2TRIM22 and KYSE150TRIM22 by Image J.
Figure 6.
By regulating cell cycle, over-expression of TRIM22 alleviates the inhibition of cell growth induced by Lyc.HCL, while knockout of TRIM22 promotes the inhibition of cell proliferation mediated by Lyc.HCL. (A and C) Representative images showed that TRIM22 alleviates Lyc.HCL-induced cell colony formation inhibition; shTRIM22 promoted Lyc.HCL-induced cell colony formation inhibition. (B and D) The quantification of the cell colonies in (A and C) is presented as the mean percentage of viable cells (mean±SD), averaged from 3 independent experiments, each with 3 replicates per condition. (E and F) Cell cycle assay showed that TRIM22 reduces Lyc.HCL-induced cell cycle arrest while shTRIM22 increased Lyc.HCL-induced cell cycle arrest. (G and H) cell cycle was analyzed by FlowJo 10.9.0. All data are presented as the mean±SD. A 1-way analysis of variance, followed by a Tukey’s post-hoc test, was used to compare the different groups. *P<.05, **P<.01, ***P<.001 versus vehicle.
Figure 6.
By regulating cell cycle, over-expression of TRIM22 alleviates the inhibition of cell growth induced by Lyc.HCL, while knockout of TRIM22 promotes the inhibition of cell proliferation mediated by Lyc.HCL. (A and C) Representative images showed that TRIM22 alleviates Lyc.HCL-induced cell colony formation inhibition; shTRIM22 promoted Lyc.HCL-induced cell colony formation inhibition. (B and D) The quantification of the cell colonies in (A and C) is presented as the mean percentage of viable cells (mean±SD), averaged from 3 independent experiments, each with 3 replicates per condition. (E and F) Cell cycle assay showed that TRIM22 reduces Lyc.HCL-induced cell cycle arrest while shTRIM22 increased Lyc.HCL-induced cell cycle arrest. (G and H) cell cycle was analyzed by FlowJo 10.9.0. All data are presented as the mean±SD. A 1-way analysis of variance, followed by a Tukey’s post-hoc test, was used to compare the different groups. *P<.05, **P<.01, ***P<.001 versus vehicle.
Figure 7.
Over-expression of TRIM22 promotes cell migration and invasion while shTRIM22 reduces cell migration and invasion. (A and C) Wound healing assay was performed to detected cell migration in ESCCTRIM22 cells or ESCCshTRIM22 cells treated with Lyc.HCL. ( B and D) Gap Width data were presented as the mean±SD. A 1-way analysis of variance, followed by a Tukey’s post-hoc test, was used to compare the different groups. *P<.05, **P<.01, ***P<.001 versus vehicle. (E and G) Transwell assay was performed to measure cell migration ability in ESCCTRIM22 and ESCCshTRIM22 cells, the migration ability changes of these cells after being treated with Lyc.HCL. (F and H) Quantification of the migrated cells in (E and G) was displayed on the right. The results were displayed as the mean ± SD. A 1-way analysis of variance, followed by a Tukey’s post-hoc test, was used to compare the different groups. *P<.05, **P<.01, ***P<.001 versus vehicle. (I and K) Before experiment, transwell chamber was covered with matrix glue. Transwell assay was performed to measure the cell invasive ability in ESCCTRIM22 cells, ESCCshTRIM22 cells and the invasion ability changes of these cells after being treated with Lyc.HCL. Cells on the bottom of the filter were stained by 0.5% crystal violet and then counted. (J and L) Quantification of the invasive cells in (I and K) was displayed on the right. The results were displayed as the mean±SD. A 1-way analysis of variance, followed by a Tukey’s post-hoc test, was used to compare the different groups. *P<.05, **P<.01, ***P<.001 versus vehicle.
Figure 7.
Over-expression of TRIM22 promotes cell migration and invasion while shTRIM22 reduces cell migration and invasion. (A and C) Wound healing assay was performed to detected cell migration in ESCCTRIM22 cells or ESCCshTRIM22 cells treated with Lyc.HCL. ( B and D) Gap Width data were presented as the mean±SD. A 1-way analysis of variance, followed by a Tukey’s post-hoc test, was used to compare the different groups. *P<.05, **P<.01, ***P<.001 versus vehicle. (E and G) Transwell assay was performed to measure cell migration ability in ESCCTRIM22 and ESCCshTRIM22 cells, the migration ability changes of these cells after being treated with Lyc.HCL. (F and H) Quantification of the migrated cells in (E and G) was displayed on the right. The results were displayed as the mean ± SD. A 1-way analysis of variance, followed by a Tukey’s post-hoc test, was used to compare the different groups. *P<.05, **P<.01, ***P<.001 versus vehicle. (I and K) Before experiment, transwell chamber was covered with matrix glue. Transwell assay was performed to measure the cell invasive ability in ESCCTRIM22 cells, ESCCshTRIM22 cells and the invasion ability changes of these cells after being treated with Lyc.HCL. Cells on the bottom of the filter were stained by 0.5% crystal violet and then counted. (J and L) Quantification of the invasive cells in (I and K) was displayed on the right. The results were displayed as the mean±SD. A 1-way analysis of variance, followed by a Tukey’s post-hoc test, was used to compare the different groups. *P<.05, **P<.01, ***P<.001 versus vehicle.
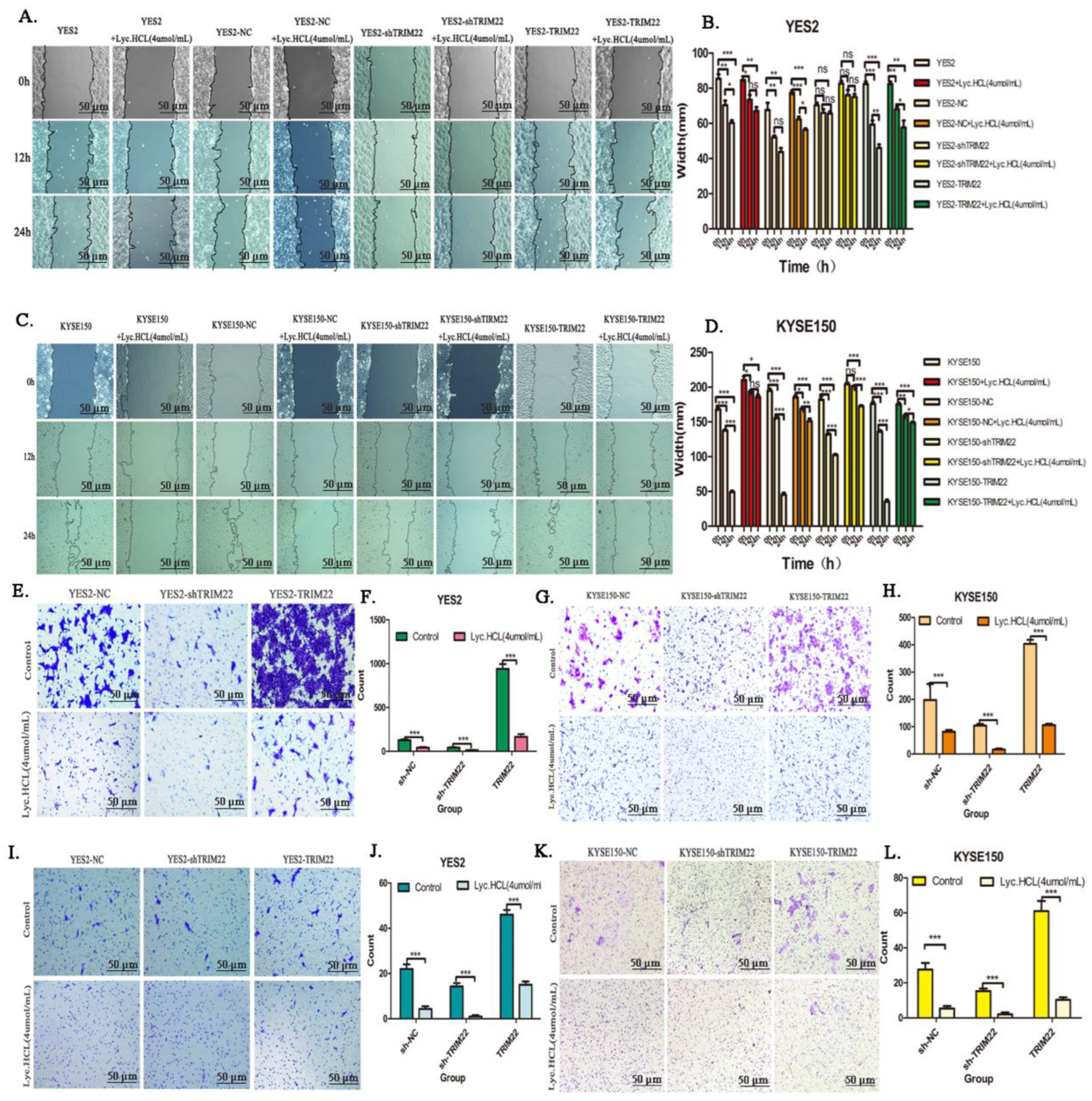
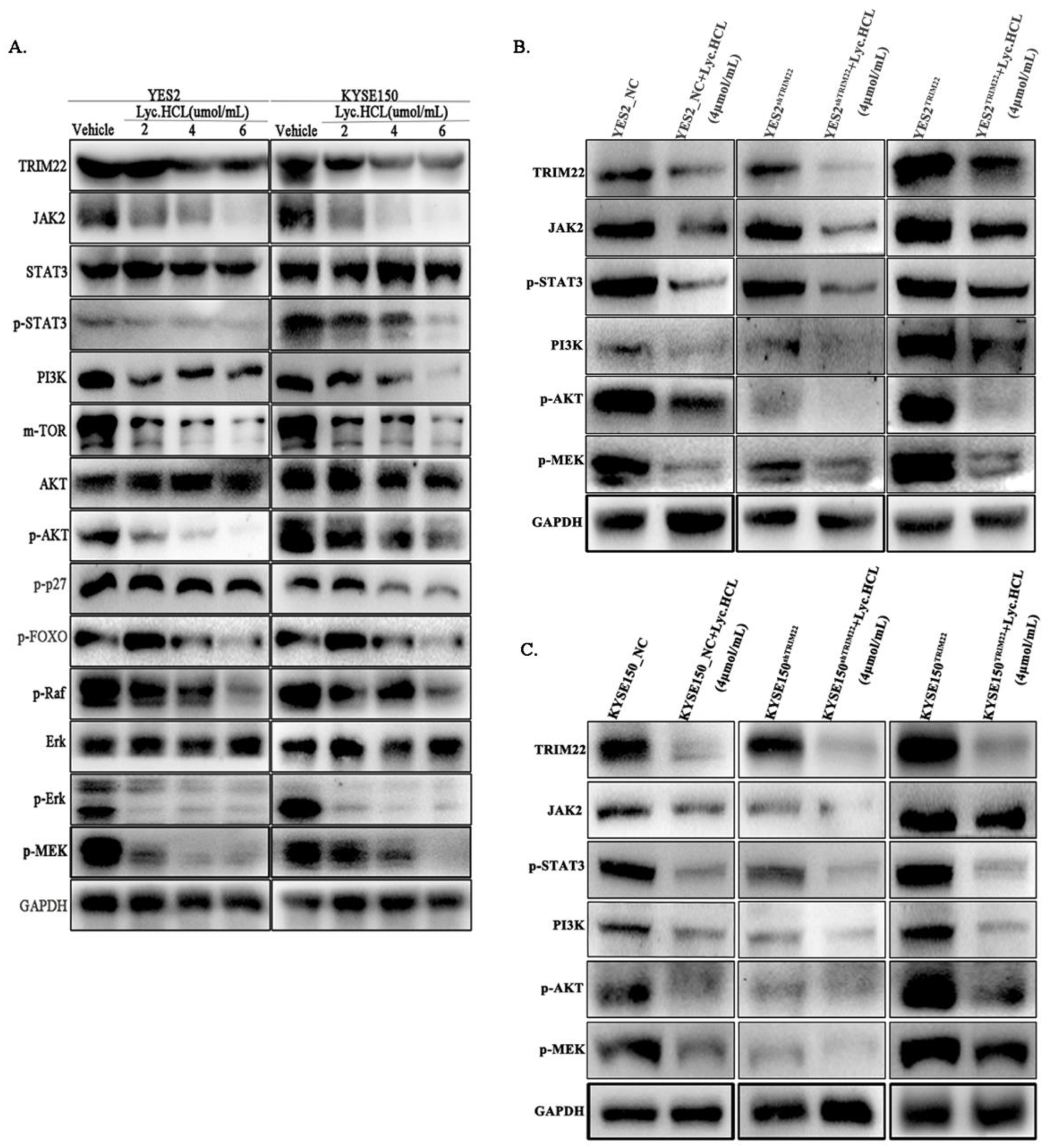
Figure 8.
Lyc.HCL suppressed JAK2/STAT3, PI3K/AKT and MAPK/ERK signaling pathway via down-regulation of TRIM22 in esophageal squamous cell carcinoma cells. The expression of JAK2, STAT3, p-STAT3, PI3K, AKT, p-AKT, Erk, p-Erk, p-MEK were assessed using the corresponding antibodies via Western blotting. The results were repeated with at least three independent experiments. (A) YES2 and KYSE150 were treated with indicated concentrations of Lyc.HCL for 48h, proteins in signal pathway were assessed using western blot. (B and C) Western blotting results showed the expression changes of TRIM22, JAK2, p-STAT3, PI3K, p-AKT, p-MEK in YES2, KYSE150, YES2TRIM22, YES2shTRIM22, KYSE150TRIM22, KYSE150shTRIM22 cells and these cells were treated with Lyc.HCL.
Figure 8D, 8E and 8F are quantification of relative expression of proteins in
Figure 8A,
Figure 8B and
Figure 8C by Image J, respectively. .
Figure 8.
Lyc.HCL suppressed JAK2/STAT3, PI3K/AKT and MAPK/ERK signaling pathway via down-regulation of TRIM22 in esophageal squamous cell carcinoma cells. The expression of JAK2, STAT3, p-STAT3, PI3K, AKT, p-AKT, Erk, p-Erk, p-MEK were assessed using the corresponding antibodies via Western blotting. The results were repeated with at least three independent experiments. (A) YES2 and KYSE150 were treated with indicated concentrations of Lyc.HCL for 48h, proteins in signal pathway were assessed using western blot. (B and C) Western blotting results showed the expression changes of TRIM22, JAK2, p-STAT3, PI3K, p-AKT, p-MEK in YES2, KYSE150, YES2TRIM22, YES2shTRIM22, KYSE150TRIM22, KYSE150shTRIM22 cells and these cells were treated with Lyc.HCL.
Figure 8D, 8E and 8F are quantification of relative expression of proteins in
Figure 8A,
Figure 8B and
Figure 8C by Image J, respectively. .
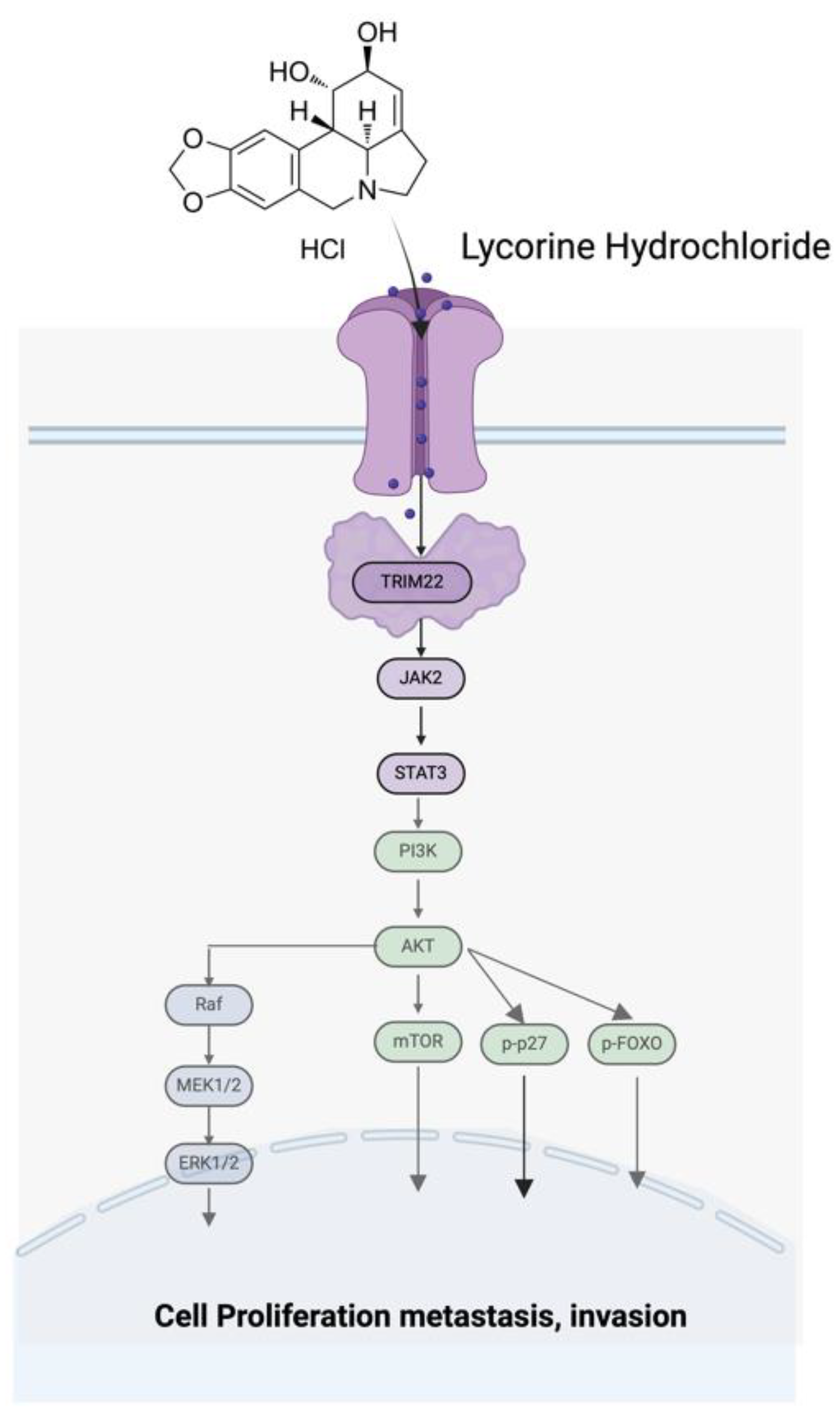
Figure 9.
Schematic diagram of the TRIM22-JAK2/STAT3-PI3K/AKT-MAPK/ERK signal pathway axis.
Figure 9.
Schematic diagram of the TRIM22-JAK2/STAT3-PI3K/AKT-MAPK/ERK signal pathway axis.
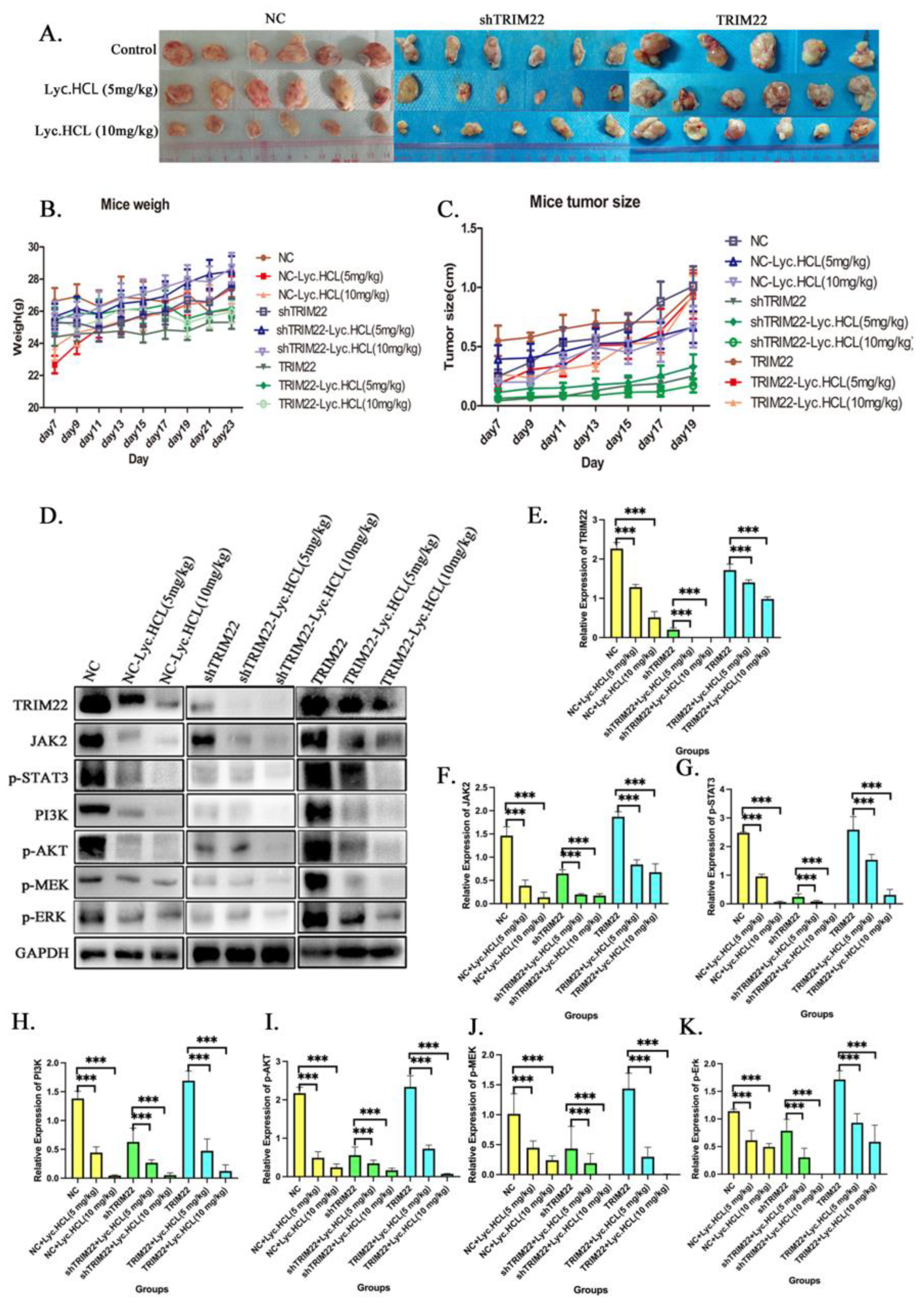
Figure 10.
Over-expression TRIM22 rescues Lyc.HCL-induced tumor inhibition in vivo and knockout TRIM22 promotes Lyc.HCL-induced tumor inhibition in vivo. (A) KYSE150-Control, KYSE150-shTRIM22 and KYSE150-TRIM22 cells were injected subcutaneously into the nude mice. When the average tumor volume achieved a volume of about 100 mm3, the mice were constructed by intra-peritoneal injections of either DMSO or Lyc.HCL (5 mg/kg/twice a day per mouse) or Lyc.HCL (10mg/kg/twice a day per mouse) from day7 to day23. (B)The body weights of the nude mice over the indicated time. (C) In vivo tumor growth was measured by Vernier caliper over the indicated time. (D)Western blot analysis of the TRIM22, JAK2, pSTAT3, PI3K, p-AKT, p-MEK expression levels in the dissected tumor tissues from different groups. (E-K) Quantification of relative expression of proteins in Figure10D.
Figure 10.
Over-expression TRIM22 rescues Lyc.HCL-induced tumor inhibition in vivo and knockout TRIM22 promotes Lyc.HCL-induced tumor inhibition in vivo. (A) KYSE150-Control, KYSE150-shTRIM22 and KYSE150-TRIM22 cells were injected subcutaneously into the nude mice. When the average tumor volume achieved a volume of about 100 mm3, the mice were constructed by intra-peritoneal injections of either DMSO or Lyc.HCL (5 mg/kg/twice a day per mouse) or Lyc.HCL (10mg/kg/twice a day per mouse) from day7 to day23. (B)The body weights of the nude mice over the indicated time. (C) In vivo tumor growth was measured by Vernier caliper over the indicated time. (D)Western blot analysis of the TRIM22, JAK2, pSTAT3, PI3K, p-AKT, p-MEK expression levels in the dissected tumor tissues from different groups. (E-K) Quantification of relative expression of proteins in Figure10D.
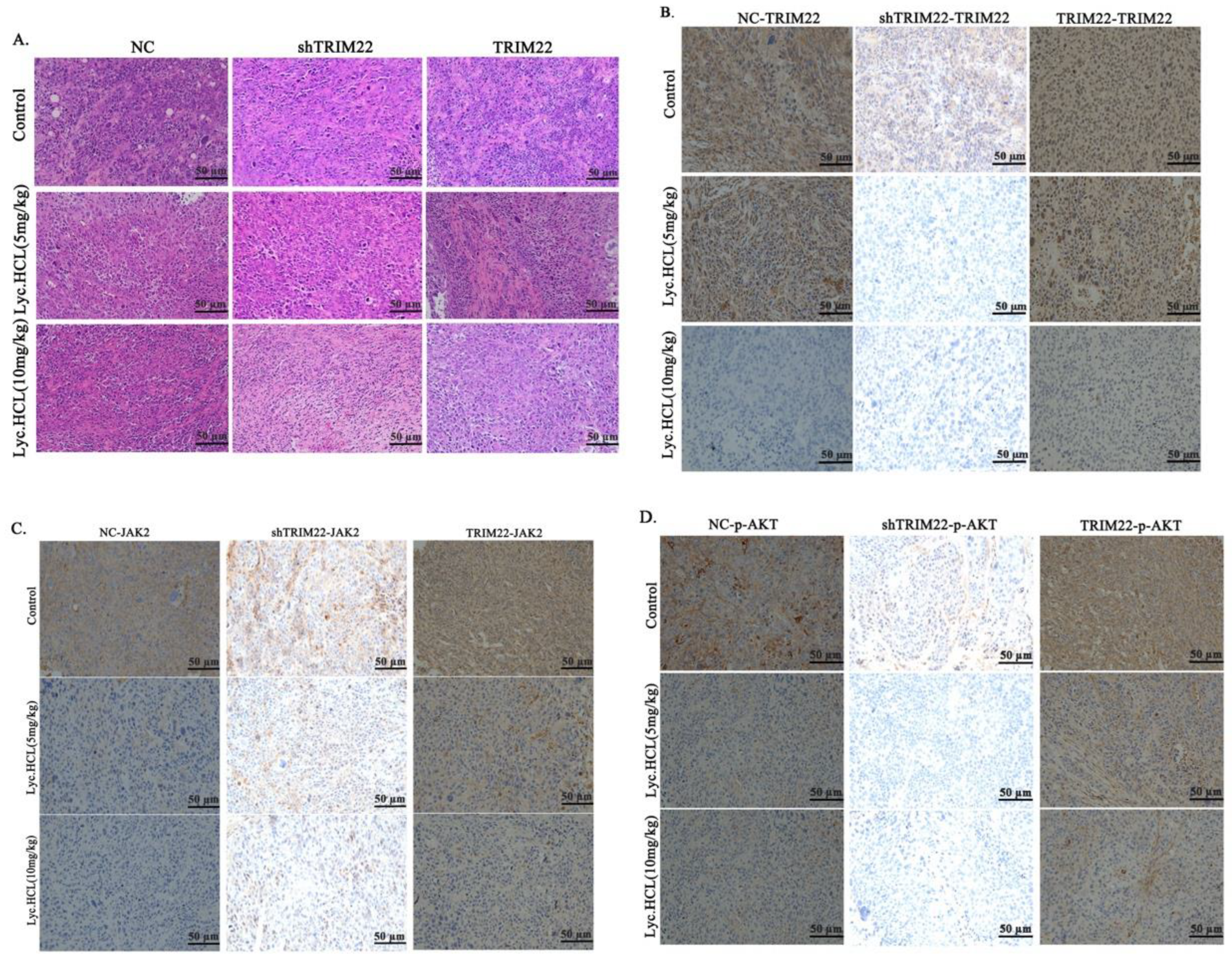
Figure 11.
HE and IHC of tumor tissues from different groups. (A) Representative HE stained of dissected tumor tissues from different groups. Scale bar 50μm. (B) IHC analyzed the expression of TRIM22 of tumors tissues from different groups; Scale bar 50μm. (C) IHC analyzed the expression of JAK2 of tumors tissues from different groups; Scale bar 50μm. (D) IHC analyzed the expression of p-AKT of tumors tissues from different groups; Scale bar 50μm.
Figure 11.
HE and IHC of tumor tissues from different groups. (A) Representative HE stained of dissected tumor tissues from different groups. Scale bar 50μm. (B) IHC analyzed the expression of TRIM22 of tumors tissues from different groups; Scale bar 50μm. (C) IHC analyzed the expression of JAK2 of tumors tissues from different groups; Scale bar 50μm. (D) IHC analyzed the expression of p-AKT of tumors tissues from different groups; Scale bar 50μm.
Table 1.
The cells count that expressed TRIM22 in tissues.
Table 1.
The cells count that expressed TRIM22 in tissues.
| Tissue No. |
Normal Tissue |
Peritumoral Tissue |
ESCC Tissue |
| 1 |
0 |
0 |
1556 |
| 2 |
0 |
0 |
1507 |
| 3 |
0 |
0 |
1687 |
| 4 |
0 |
0 |
3657 |
| 5 |
0 |
0 |
3676 |
| 6 |
0 |
0 |
2031 |
| 7 |
0 |
0 |
2240 |
| 8 |
0 |
0 |
1786 |
| 9 |
0 |
0 |
1099 |
| 10 |
0 |
0 |
2345 |
| 11 |
0 |
0 |
1455 |
| 12 |
0 |
0 |
2345 |
| 13 |
0 |
0 |
1596 |
| 14 |
0 |
0 |
2329 |
| 15 |
0 |
0 |
2020 |
| 16 |
0 |
0 |
1945 |
| 17 |
0 |
0 |
1425 |
| 18 |
0 |
0 |
1709 |
| 19 |
0 |
0 |
2063 |
| 20 |
0 |
0 |
1113 |
| 21 |
0 |
0 |
1162 |
| 22 |
0 |
0 |
1092 |
| 23 |
0 |
0 |
1927 |
| 24 |
0 |
0 |
1634 |
| 25 |
0 |
0 |
1552 |
| 26 |
0 |
0 |
3374 |
| 27 |
0 |
0 |
3068 |
| 28 |
0 |
0 |
2813 |
| 29 |
0 |
0 |
1552 |
| 30 |
0 |
0 |
1142 |
| 31 |
0 |
0 |
1750 |
| 32 |
0 |
0 |
1414 |
| 33 |
0 |
9 |
1213 |
| 34 |
0 |
0 |
1709 |
| 35 |
0 |
0 |
1786 |