1. Introduction
Orius nagaii, a predatory natural enemy insect, is widely distributed in both northern and southern regions of China [
1,
2]. This species is known for its ability of nymphs and adults to feed on a multiple of pests, including thrips, mites, aphids, whiteflies, and other minute pests [
3,
4,
5,
6]. Additionally,
O. nagaii has shown efficient control over the eggs and neonates of various Lepidoptera pests [
7,
8,
9]. It plays a key role in controlling pests in greenhouse vegetables, flowers, and field cash crops, making it a natural enemy insect with high potential for development [
10,
11]. Semi-field cage experiments demonstrated that the efficacy of
O. nagaii in controlling
Megalurothrips usitatus was notably superior to that achieved through conventional chemical control measures [
2], which highlighted its important value in pest control. However, the molecular mechanisms underlying the interactions between
O. nagaii and its host pests remains unclear. Therefore, conducting gene expression analysis is of great importance in exploring its gene functions and further understanding its interaction mechanisms with host pests.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) is a widely employed rapid detection technique of gene quantification for accurately measuring the expression levels of target genes in various biological conditions [
12], which is crucial for the subsequent examination of the expression patterns of target genes in
O. nagaii. However, when working with different experimental samples, errors may occur in the detection results of target genes due to factors such as the quality of RNA in the starting material, concentration of template cDNA, and experimental manipulation errors [
13,
14]. Hence, it is crucial to meticulously choose and validate suitable reference genes for normalization as the first step in ensuring the success of RT-qPCR analysis. Previous research has shown that many commonly used reference genes may not exhibit consistent expression under complex experimental conditions [
15]. In
Drosophila melanogaster, the
ribosomal protein L18 (RPL18) was found to be the most stable in methanol-treated flies, while its expression was the least stable in ethyl acetate (ETOAC)-treated flies [
16,
17]. Meanwhile, the transcriptional level of
glyceraldehyde-3-phosphate dehydrogenase (
GAPDH) gene from
Spodoptera litura was found to be stable in different developmental periods, but unstable in different tissues and geographical populations [
18]. Hence, it is essential to identify the best optimal reference gene with a stable expression under various research conditions for accurate target gene quantitative analysis.
Due to the lack of stability evaluation studies for reference genes in O. nagaii, ten frequently employed reference genes, including β-Actin (Act), glyceraldehyde 3-phosphate dehydrogenase (GAPDH), β-tubulin (β-Tub), elongation factor-1-alpha (EF1-α), ribosomal protein S10 (RPS10), ribosomal protein S15 (RPS15), ribosomal protein L6 (RPL6), ribosomal protein L13 (RPL13), ribosomal protein L32 (RPL32), and heat shock protein 90 (HSP90) of O. nagaii, were chosen as candidate reference genes. RefFinder, which integrates four analysis methods (NormFinder, geNorm, the ΔCt method, and BestKeeper), was used to assess the expression stability of these genes under four experimental conditions (developmental period, tissue, host, and temperature). Meanwhile, T-box transcription factor (TBX1) gene was used to verify the dependability of the evaluation results. This research will offer reliable reference genes for the quantitative analysis of key gene expression in the interaction between O. nagaii and host pests.
2. Materials and Methods
2.1. Preparation of Insects
Orius nagaii samples were collected from the plantation base of Shandong Academy of Agricultural Sciences (Jinan, China), classified, and identified in the Key Laboratory of Natural Enemies Insects, Ministry of Agriculture and Rural Areas, P.R. China, and have been continuously propagated under the following conditions: 26 ± 1 °C, 70 ± 5% RH, and a 16/8 h (L/D) cycle [
19].
Phaseolus vulgaris was used as the oviposition substrate for females, while nymphs and adults were fed
Sitotroga cerealella eggs [
19].
2.2. Sample Handling and Retrieval
Developmental stage: All phases of development were collected, including eggs, nymphs of the 1st to 5th instar, and adults. Samples were collected on the first day of each instar, with three biological replicates per sample. The quantity of individuals gathered for each repetition at various developmental phases is as follows: 90 eggs for the egg stage; 60 individuals in each of the 1st and 2nd instar; 30 individuals in each of the 3rd and 4th instar; 15 individuals in each of the 5th instar and adult stage.
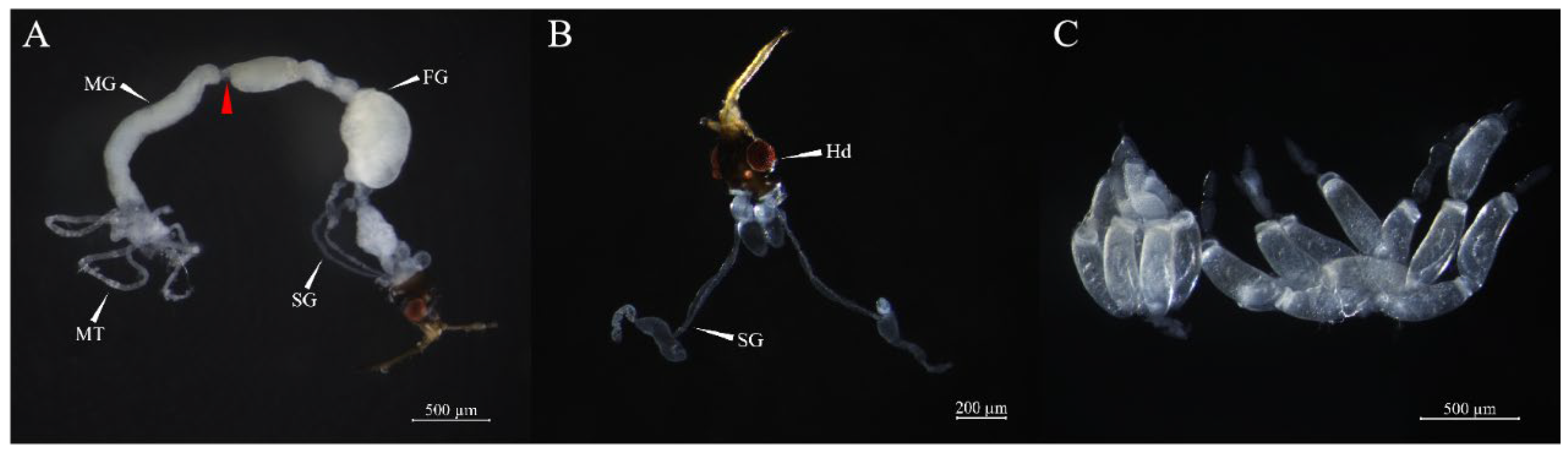
Tissue: One-day-old female adults were dissected under a compact stereo microscope Stemi 508 (ZEISS, Germany), and different body tissues were identified and collected (
Figure 1), specifically the head (Hd), salivary gland (SG), foregut (FG), midgut (MG), ovary (Ov), Malpighian tubule (MT), and residual body (RB). Three biological replicates were set per sample, with 30 insects dissected per replicate.
Host: Diet-induced stress was utilized to assess the stability of candidate reference gene (CRG) expression during feeding on different hosts (S. cerealella egg, Corcyra cephalonica egg, Frankliniella occidentalis nymph and Megalurothrips usitatus nymph). Newly hatched nymphs were fed on different hosts until adult emergence, and the newly emerged adults were collected. Three biological replicates were set per sample, with 15 adults in each replicate.
Temperature: Three distinct temperatures, 8°C, 25°C, and 35°C, were used as temperature-induced stress [
20]. Newly emerged adults were exposed to these temperatures for 3 h, and then collected. Each sample had three biological replicates, with 15 adults per replicate.
All samples were transferred in RNase-free tubes, snap-frozen in liquid nitrogen for 2-3 min, and gathered at -80°C for future utilization.
2.3. Production of cDNA Template
Sample RNA was isolated of multiple experimental conditions by following the manufacturer’s instructions for Trizol (Invitrogen, Carlsbad, CA, USA). The RNA concentration was assessed with a NanoDrop One spectrophotometer (Thermo Scientific, Waltham, MA, USA) and 1% agarose gel electrophoresis was used to check its integrity. First-strand complementary DNA (cDNA) was initially synthesized from 1 μg of sample RNA in two steps via the PrimeScript™ RT reagent Kit with the gDNA Eraser (Takara, Tokyo, Japan). The gDNA Eraser within the kit exhibits robust DNA degradation capabilities, which can effectively eliminate genomic DNA at 42°C for 2 min. Additionally, the reverse transcription reagent includes elements that suppress the function of the DNA decomposition enzyme, so the sample treated with gDNA Eraser can directly synthesize cDNA through the reverse transcription reaction at 37℃, 15 min, and 85℃, 5 s.
2.4. Primer Design and Verification
In our research, frequently utilized CRGs were screened for RT-qPCR analysis (
Table 1). Utilizing conserved sequences as a foundation, precise primers were crafted through the NCBI Primer-BLAST web tool (
https://www.ncbi.nlm.nih.gov/tools/primer-blast/) and the uniqueness of the primers was confirmed through 1% agarose gel electrophoresis. Amplified fragments of 80 to 200 bp of a single band were used to further assess the stability of CRGs.
2.5. RT-qPCR
RT-qPCR experiments were conducted using ChamQ Universal SYBR qPCR Master Mix (Vazyme, Nanjing, China) in a 20 µL reaction mixture. The reaction program was as follows: Hold stage at 95℃ for 30 s, PCR stage (40 cycles) at 95℃ for 10 s and 60℃ for 30 s, and melt curve stage using the default system program. Melting and standard curves of reference genes were derived using Applied Biosystem™ QuantStudio5™ Real-Time PCR System (Thermo Scientific, Waltham, MA, USA). The amplification efficiency (E) of RT-qPCR was calculated using the threshold-cycle (
Ct value) obtained from the 5-fold dilution of the template, as follows: E = (10
[−1/slope] − 1) × 100% [
21,
22].
2.6. Stability Assessment
The stability of CRGs was assessed through RefFinder (
http://blooge.cn/RefFinder/) [
23], which incorporates four algorithms (
NormFinder,
geNorm, the
ΔCt method, and
BestKeeper) to offer a thorough evaluation of genetic stability [
24,
25,
26,
27]. The pairwise variation (V
n/n+1) value, computed with geNorm, was employed to ascertain the ideal number of reference genes needed to normalize the target gene. A value of V
n/n+1 < 0.15 indicates that there is no need to add another reference gene, meaning that the initial n reference genes are sufficient to achieve the standardization of the target gene.
2.7. Validation of Optimal Reference Genes in Various Tissues
The T-box transcription factor (
TBX1) gene was picked as the target gene to confirm the dependability of the CRGs. Prior research has demonstrated that
TBX1 participates in the initial phase of heart development during insect embryonic development [
28,
29]. The primer sequence for the target gene is as listed:
TBX1, F: (5’- TGGGAAGAATCACAGCATCA-3’) and R: (5’- ATCGGTATCGTTTATCGTCAAG-3’). The two most- and two least-stable selected reference genes were utilized to standardize the
Ct values of
TBX1 across various tissues. The average
TBX1 transcript level was then calculated using the 2
−∆∆Ct method. The obtained data were statistically analyzed using one-way ANOVA in GraphPad Prism 9.0.
3. Results
3.1. Dependability of Primers for CRGs
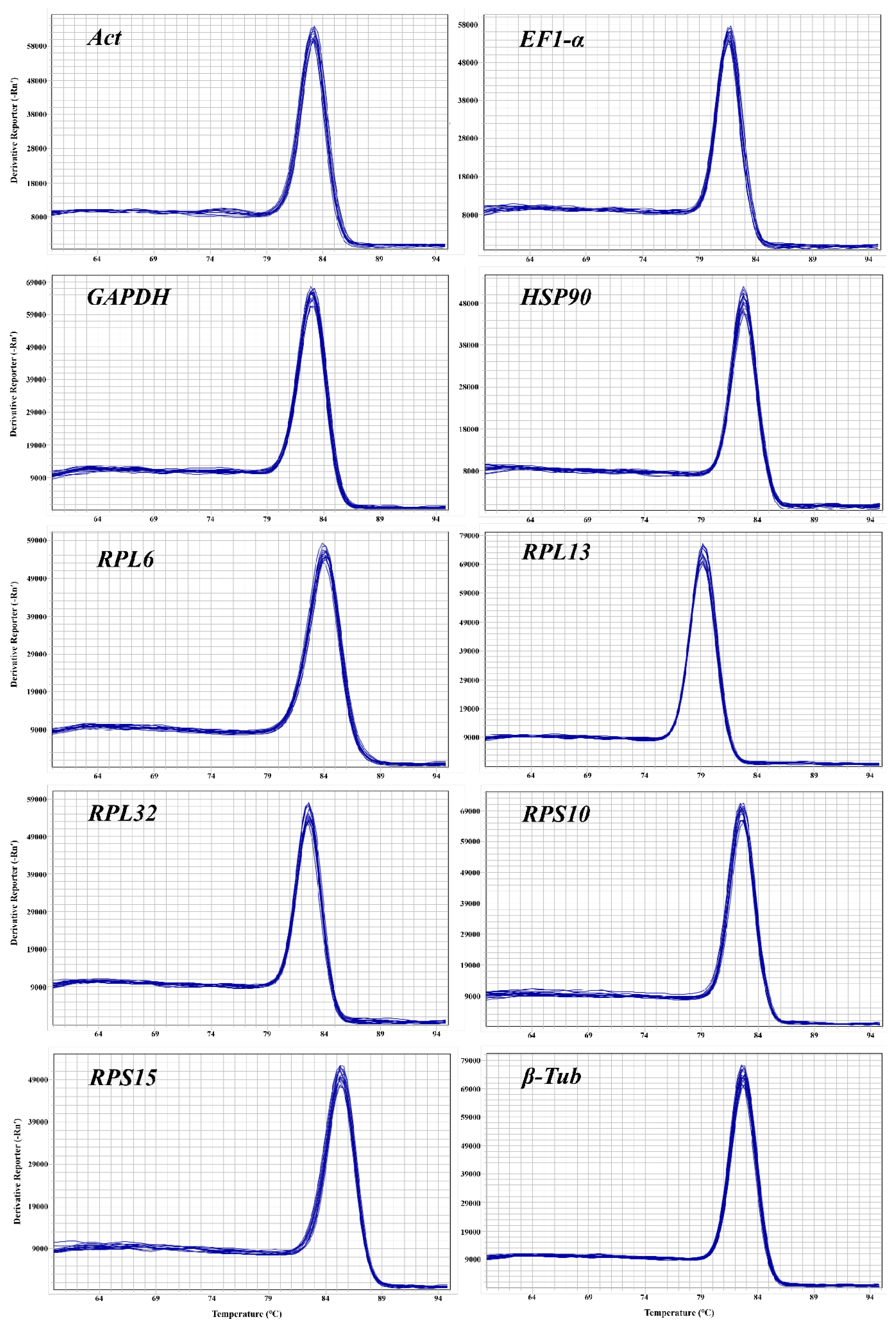
Before assessing the stability of CRGs, it is essential to confirm the dependability of primers, containing specificity and amplification efficiency. 1% agarose gel electrophoresis of all CRGs showed a singular band of amplified fragments (
Figure S1), and the melting curves of their RT-qPCR analysis also showed a single peak (
Figure 2). These results indicate that the designed primers have excellent specificity. This study provided standard curves (
Figure S2) for all CRGs and calculated the primer amplification efficiency via the slope of the curve. The primer amplification efficiency ranged from 102.63-109.02% (
Table 1), and the correlation coefficient varied between 0.9956-0.9999 (
Table 1), indicating high quality of the standard curve and reliable quantitative results.
3.2. Expression Profile of CRGs
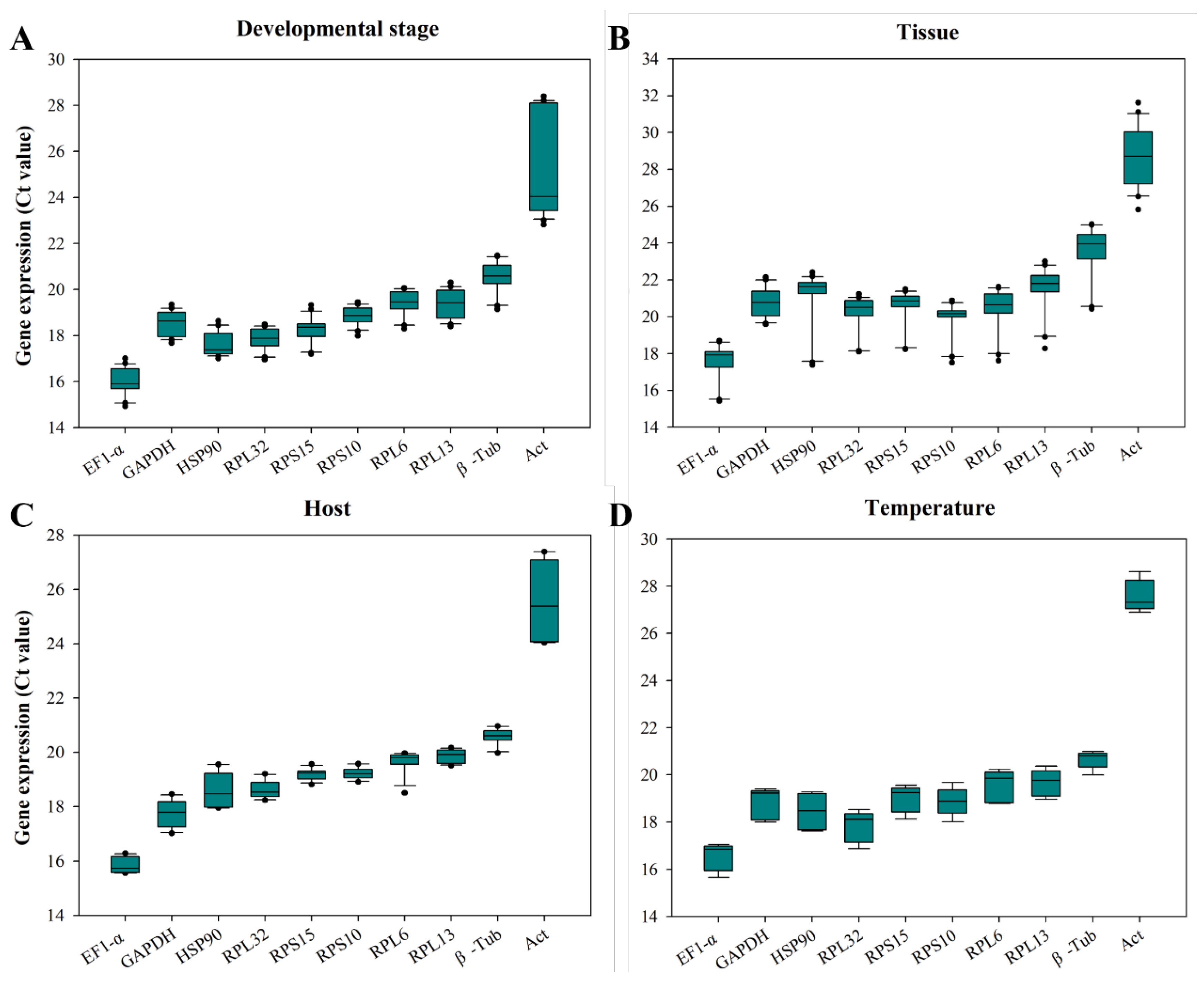
The
Ct values of all reference genes under four experimental conditions ranged from 15.4 to 31.6.
EF1-α was the best richness reference gene, while
β-Tub and
Act were the least abundant reference genes (
Figure 3). At multi-stages of development, the
Ct value of
RPS10 fluctuates in the smallest range, with a difference value of 1.45 (
Figure 3A); In different tissues, the
Ct value of
GAPDH had the smallest fluctuation range, with a difference value of 2.55 (
Figure 3B); When feeding on different hosts, the
Ct value of
RPS10 had the smallest fluctuation range, with a difference value of 0.66 (
Figure 3C); The
Ct value of
β-Tub has the smallest fluctuation range under temperature-induced stress, with a difference value of 1.0 (
Figure 3D). The
Ct value of
Act exhibited the largest fluctuations among all experimental conditions (
Figure 3).
3.3. Assess the Expression Stability of CRGs
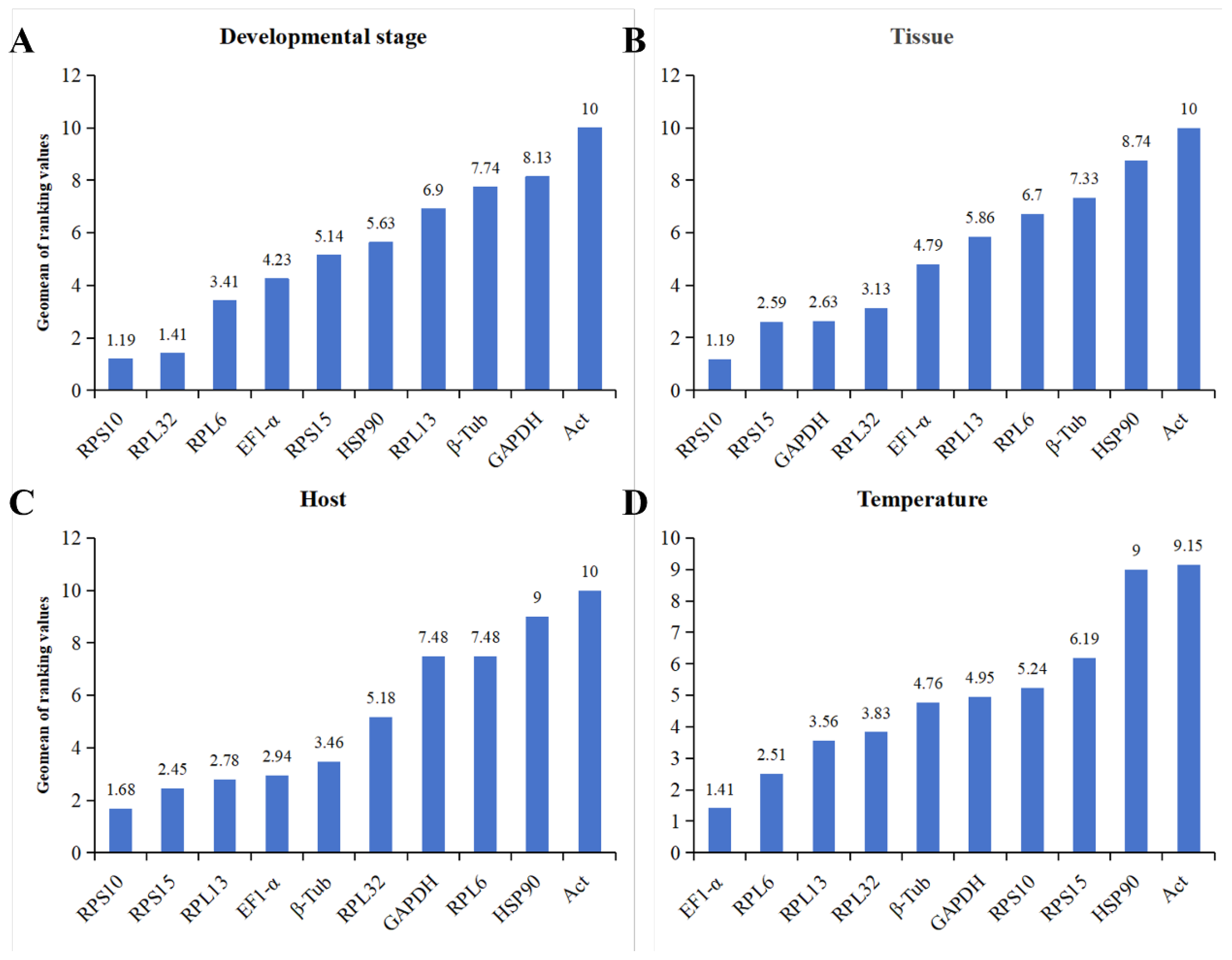
Based on comprehensive evaluation of
RefFinder, the best to the least stable CRGs at multi-stages of development were ranked as:
RPS10 >
RPL32 >
RPL6 >
EF1-α >
RPS15 >
HSP90 >
RPL13 >
β-Tub >
GAPDH >
Act (
Figure 4A). Within different tissues, the ranking:
RPS10 >
RPS15 >
GAPDH >
RPL32 >
EF1-α >
RPL13 >
RPL6 >
β-Tub >
HSP90 >
Act (
Figure 4B). The ranking of expression stability of CRGs in
O. nagaii fed with different hosts was:
RPS10 >
RPS15 >
RPL13 >
EF1-α >
β-Tub >
RPL32 >
GAPDH >
RPL6 >
HSP90 >
Act (
Figure 4C). Under different temperature stresses, the ranking:
EF1-α >
RPL6 >
RPL13 >
RPL32 >
β-Tub >
GAPDH >
RPS10 >
RPS15 >
HSP90 >
Act (
Figure 4D). The expression stability values and rankings of all CRGs under various experimental conditions were obtained by
Normfinder,
geNorm, the
ΔCt method and
BestKeeper, which were shown in this study (
Table 2).
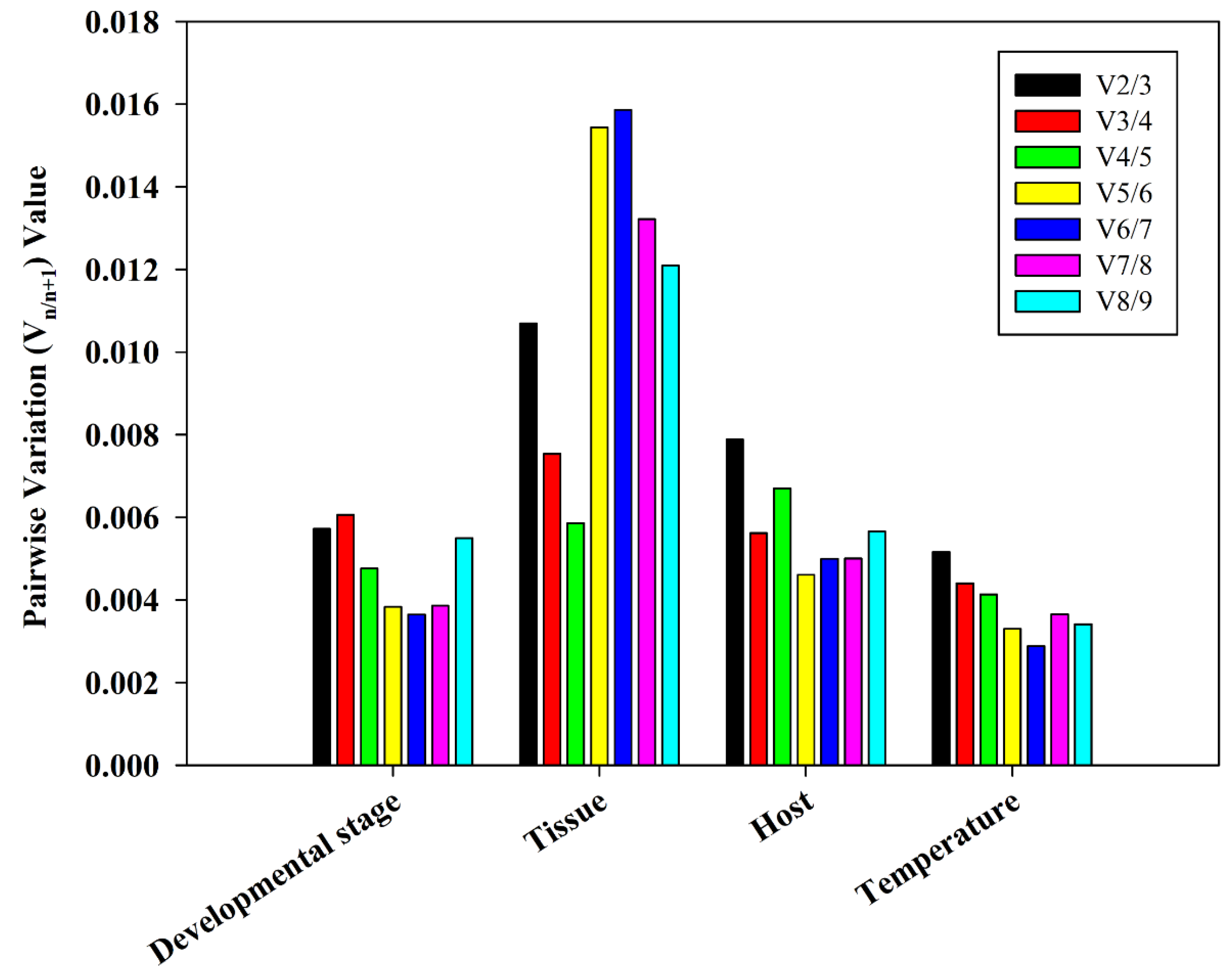
3.4. Ideal Number of Reference Genes Across Various Experimental Conditions
The
geNorm is designed to provide guidance on determining the ideal number of reference genes. A V
n/n+1 value less than 0.15, as determined by
geNorm, was used as our criterion for determining the ideal number of reference genes [
30]. In multiple experimental conditions, the values of V
2/3 were found to be less than 0.15, suggesting that only the two most stable reference genes are needed for normalization of the target genes (
Figure 5). Correlation analysis with the stability (
Table 2) showed that the ideal reference genes for different developmental periods were
RPS10 and
RPL32. In different tissues,
RPS10 and
RPS15 were revealed to be the ideal reference genes. Additionally,
RPS10 and
RPS15 were the ideal housekeeping genes when feeding on different hosts. Under temperature-induced stress,
EF1-α and
RPL6 were identified as the best suitable reference genes.
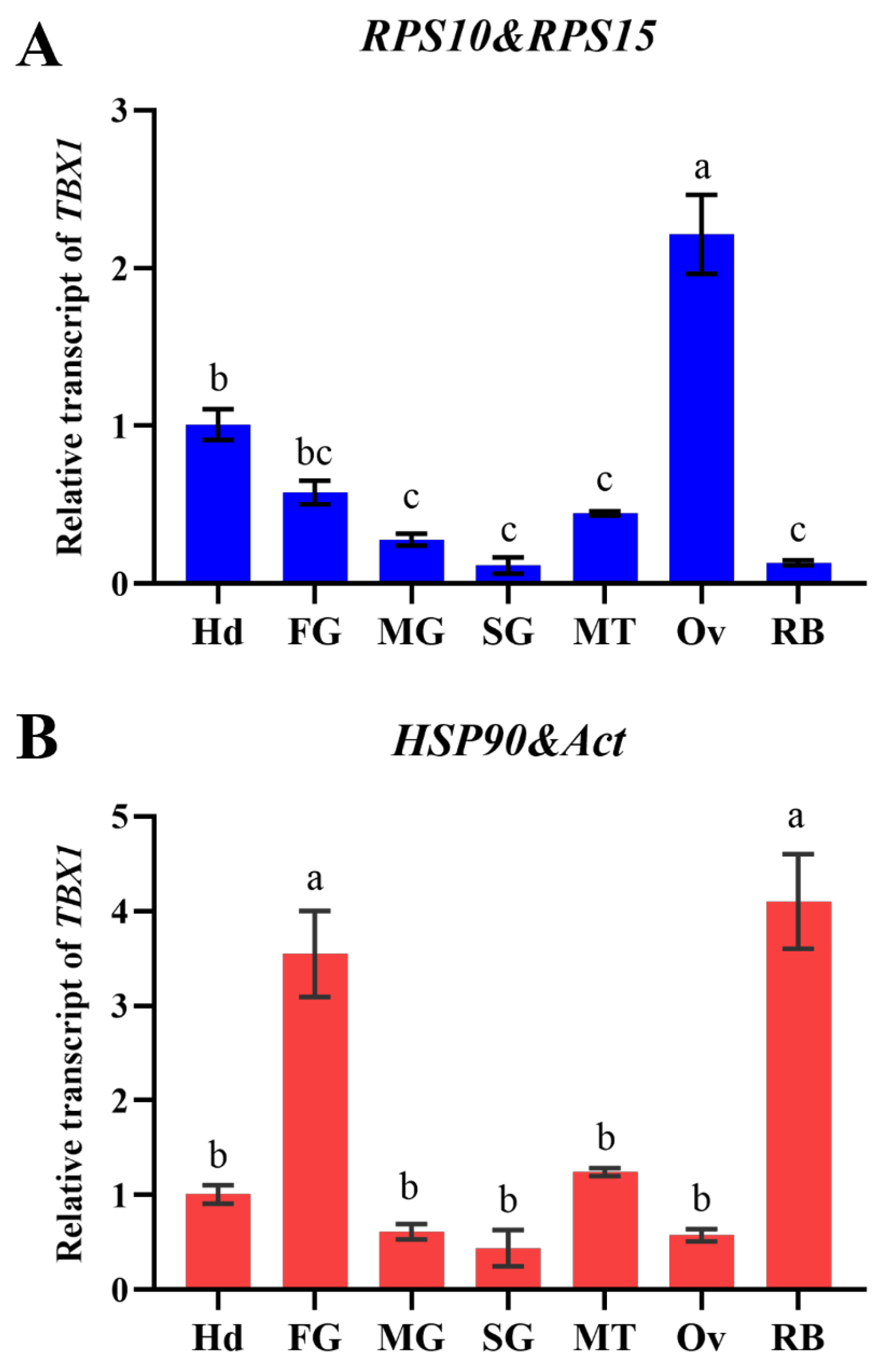
3.5. Confirmation of Optimal Reference Genes
To verify the dependability of the selected reference gene in different tissues, the relative expression of the
TBX1 gene was examined. It was discovered that the
Ct values of
TBX1 were normalized using the two best-stable reference genes (
RPS10 and
RPS15) and the two least-stable reference genes (
HSP90 and
Act), and their expression patterns in different tissues varied significantly (
Figure 6). Using
RPS10 and
RPS15 as reference genes (F = 46.87,
P < 0.0001),
TBX1 was found to be expressed at the highest level in the ovary (Ov), significantly higher than in other tissues. However, using
HSP90 and
Act as reference genes (F = 31.50,
P < 0.0001),
TBX1 expression levels were higher in the foregut (FG) and residual body (RB), and significantly higher than in the Ov and other tissues.
4. Discussion
RT-qPCR is a generally used quantitative technique for exploring the expression patterns and relative expression levels of target genes under different biological processes and abiotic conditions [
12,
31]. In the times ahead, we plan to employ RNA interference (RNAi) to investigate the roles of pivotal genes engaged in the interaction between
O. nagaii and host pests, and RT-qPCR will be widely used to assess the expression fluctuation of genes. The selection and verification of appropriate reference genes is crucial for ensuring reliable analysis results. However, there has been no report on the reference genes of
O. nagaii. Prior research has found that no single gene is consistently expressed under complex experimental conditions [
15]. Blindly selecting reference genes may result in difficulty detecting expression differences in target genes and could even lead to incorrect conclusions [
32,
33].
Here, we chose ten frequently employed reference genes and investigated their stability using various methods across four distinct experimental conditions. The results indicate that accurate normalization of target genes can be achieved by using only two of the best stable internal reference genes under various experimental conditions.
RPS10 and
RPL32 were recognized as the best stable reference genes at multi-stages of development. These genes belong to ribosomal protein-coding genes involved in protein synthesis within the cell [
34,
35]. Our findings were consistent with previous research where
RPL32 was found to have stable expression at multiple developmental periods of
Bactrocera minax [
36]. However, in contrast,
RPL32 was identified to be stably expressed in
Holotrichia parallela under different sex and photoperiodic conditions, but was unstable in different developmental stages [
37]. Currently, there is limited research on the stability evaluation of
RPS10. Additionally,
RPS10 and
RPS15 were determined to be the best appropriate reference genes across various tissues and while feeding on different hosts. Previous research has also demonstrated that
RPS15 is a suitable reference gene for tissue studies of
Nilaparvata lugens [
15] and
Helicoverpa armigera [
38], as well as for studying temperature stress in
H. armigera [
39]. We found that
EF1-α and
RPL6 were stably expressed in
O. nagaii under temperature-induced stress, which differed from
EF1-α being stably expressed in different tissues of
Rhodnius prolixu [
40] and
Agrilus planipennis [
41]. Similarly,
RPL6 is the best appropriate reference gene for
Mylabris sibirica under temperature-induced stress [
22]. In summary, there is no “universal” reference gene that can be applied to various experimental conditions.
To further confirm the internal reference genes of O. nagaii screened in this study, we assessed the relative expression levels of TBX1 gene in various tissues. The findings indicated that the best appropriate reference genes (RPS10 and RPS15) were used to normalize TBX1 gene, which had the highest expression in the ovary. However, the least stable internal reference genes (HSP90 and Act) were used for normalization, and the expression level of TBX1 was found to be significantly higher in the residual body and foregut compared to other tissues, and the average expression level of TBX1 in the ovary was only 1/4 of that when using the best reference gene. Additionally, the expression pattern of TBX1 was also found to be significantly altered in different tissues. These results highlight the importance of using appropriate reference genes to obtain reliable quantitative results for target genes. Therefore, it is necessary to screen and verify the optimal reference genes, and our study provides reliable reference genes for the quantitative analysis of functional genes involved in the interaction between O. nagaii and its host pests under strict screening conditions.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org.
Author Contributions
Conceptualization, C.W., Y.Z. and L.Z.; Data curation, Z.Y. and C.W.; Funding acquisition, Y.Z. and L.Z.; Investigation, Y.W., and C.W.; Methodology, C.W., Y.L., S.Z., X.D. and R. W.; Software, L.S. and H.C.; Writing-original draft, C.W. and Y.Z.; writing-review and editing, C.W. and Y.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by National Key R&D Program of China (2023YFE0123000), Taishan Scholars program of Shandong Province (tsqn202312293) and Agricultural Science and Technology Innovation Project of Shandong Academy of Agricultural Sciences (CXGC2024G01).
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no competing interests.
References
- Hu, C.; Li, Y.; Chen, G.; Duan, P.; Wu, D.; Liu, Q.; Yin, H.; Xu, T.; Zhang, X. Population dynamics of Frankliniella occidentalis Pergrande and its predator Orius similis Zheng on common crops and surrounding plants. J. Asia-Pac. Entomol. 2021, 24, 555–563. [Google Scholar] [CrossRef]
- Dai, X.; Wang, R.; Liu, Y.; Su, L.; Yin, Z.; Wu, M.; Chen, H.; Zheng, L.; Zhai, Y. Control effect and field application of four predatory Orius species on Megalurothrips usitatus (Thysanoptera: Thripidae). J. Econ. Entomol. 2024, 117, 448–456. [Google Scholar] [CrossRef] [PubMed]
- Fathi, S.A.A.; Nouri-Ganbalani, G. Assessing the potential for biological control of potato field pests in Ardabil, Iran: functional responses of Orius niger (Wolf.) and O. minutus (L.) (Hemiptera: Anthocoridae). J. Pest Sci. 2010, 83, 47–52. [Google Scholar] [CrossRef]
- Ge, Y.; Camara, I.; Wang, Y.; Liu, P.; Zhang, L.; Xing, Y.; Li, A.; Shi, W. Predation of Aphis craccivora (Hemiptera: Aphididae) by Orius sauteri (Hemiptera: Anthocoridae) under different temperatures. J. Econ. Entomol. 2018, 111, 2599–2604. [Google Scholar] [CrossRef]
- Sharifi, M.; Malkeshi, S.H.; Madahi, K.; Mobasheri, M.T.; Malek Shahkoei, S.; Ghaderi, K.; Rajaei, A.; Khamar, E. Evaluation of predator and prey preference of Orius niger (Wolff) (Hemiptera: Anthocoridae) in the control of important sucking pests of oilseeds. J. Plant Protect. Res. 2021, 8, 107–118. [Google Scholar]
- Silva, L.P.; Souza, I.L.; Marucci, R.C.; Guzman-Martinez, M. Doru luteipes (Dermaptera: Forficulidae) and Orius insidiosus (Hemiptera: Anthocoridae) as nocturnal and diurnal predators of thrips. Neotrop. Entomol. 2023, 52, 263–272. [Google Scholar] [CrossRef]
- Lins Jr, J.; Bueno, V.; Silva, D.; van Lenteren, J.; Calixto, A.; Sidney, L. Tuta absoluta egg predation by Orius insidiosus. IOBC WPRS Bulletin. 2011, 68, 101–104. [Google Scholar]
- Salehi, Z.; Yarahmadi, F.; Rasekh, A.; Sohani, N. Functional responses of Orius albidipennis Reuter (Hemiptera, Anthocoridae) to Tuta absoluta Meyrick (Lepidoptera, Gelechiidae) on two tomato cultivars with different leaf morphological characteristics. Entomol. Gen. 2016, 36, 127–136. [Google Scholar] [CrossRef]
- Desneux, N.; Wajnberg, E.; Wyckhuys, K.A.G.; Burgio, G.; Arpaia, S.; Narváez-Vasquez, C.A.; González-Cabrera, J.; Catalán Ruescas, D.; Tabone, E.; Frandon, J.; et al. Biological invasion of European tomato crops by Tuta absoluta: ecology, geographic expansion and prospects for biological control. J. Pest Sci. 2010, 83, 197–215. [Google Scholar] [CrossRef]
- Yue-Li, J.; Yu-Qing, W.U.; Yun, D.; Xin-Guo, G. Control efficiencies of releasing Orius sauteri (Heteroptera:Anthocoridae) on some pests in greenhouse pepper. Chin. J. Biol. Control 2011, 27, 414–417. [Google Scholar]
- Jian, Y.; Xinguo, G.; Yuqing, W.U.; Yuli, J.; Shuntong, L.; Aiju, D.; Ziqi, Z.; Changying, L. Thrips control on the greenhouse eggplant by releasing Orius sauteri (Heteroptera: Anthocoridae). Chin. J. Biol. Control 2013, 29, 459–462. [Google Scholar]
- Gomez, R.; Sendín, L. Relative expression analysis of target genes by using reverse transcription-quantitative PCR, 2020, 2072, 51–63.
- Bustin, S.A.; Benes, V.; Nolan, T.; Pfaffl, M.W. Quantitative real-time RT-PCR - a perspective. J. Mol. Endocrinol. 2005, 34, 597–601. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Higgins, P.J.; Crawford, D.R. Control selection for RNA quantitation. Biotechniques. 2000, 29, 332–337. [Google Scholar] [CrossRef]
- Yuan, M.; Lu, Y.; Zhu, X.; Wan, H.; Shakeel, M.; Zhan, S.; Jin, B.; Li, J. Selection and evaluation of potential reference genes for gene expression analysis in the brown planthopper, Nilaparvata lugens (Hemiptera: Delphacidae) using reverse-transcription quantitative PCR. PLoS One 2014, 9, e86503. [Google Scholar] [CrossRef]
- Dong, R.; Cao, F.; Yu, J.; Yuan, Y.; Wang, J.; Li, Z.; Zhu, C.; Li, S.; Li, N. Selection and validation of reference genes for quantitative real-time PCR analysis in cockroach parasitoid Tetrastichus hagenowii (Ratzeburg). Insects 2024, 15, 668. [Google Scholar] [CrossRef]
- Sagri, E.; Koskinioti, P.; Gregoriou, M.; Tsoumani, K.T.; Bassiakos, Y.C.; Mathiopoulos, K.D. Housekeeping in Tephritid insects: the best gene choice for expression analyses in the medfly and the olive fly. Sci. Rep. 2017, 7, 45634. [Google Scholar] [CrossRef]
- Lu, Y.; Yuan, M.; Gao, X.; Kang, T.; Zhan, S.; Wan, H.; Li, J. Identification and validation of reference genes for gene expression analysis using quantitative PCR in Spodoptera litura (Lepidoptera: Noctuidae). PLoS One 2013, 8, e68059. [Google Scholar] [CrossRef]
- Du, H.; Wang, R.; Dai, X.; Yin, Z.; Liu, Y.; Su, L.; Chen, H.; Zhao, S.; Zheng, L.; Dong, X.; et al. Effect of guanylate cyclase-22-like on ovarian development of Orius nagaii (Hemiptera: Anthocoridae). Insects 2024, 15, 110. [Google Scholar] [CrossRef]
- Lü, J.; Chen, S.; Guo, M.; Ye, C.; Qiu, B.; Wu, J.; Yang, C.; Pan, H. Selection and validation of reference genes for RT-qPCR analysis of the ladybird beetle Henosepilachna vigintioctomaculata. Front. Physiol. 2018, 9, 1614. [Google Scholar] [CrossRef]
- Shen, X.; Zhang, G.; Zhao, Y.; Zhu, X.; Yu, X.; Yang, M.; Zhang, F. Selection and validation of optimal reference genes for RT-qPCR analyses in Aphidoletes aphidimyza Rondani (Diptera: Cecidomyiidae). Front. Physiol. 2023, 14, 1277942. [Google Scholar] [CrossRef]
- Shen, C.; Tang, M.; Li, X.; Zhu, L.; Li, W.; Deng, P.; Zhai, Q.; Wu, G.; Yan, X. Evaluation of reference genes for quantitative expression analysis in Mylabris sibirica (Coleoptera, Meloidae). Front. Physiol. 2024, 15, 1345836. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Wang, J.; Zhang, B. RefFinder: a web-based tool for comprehensively analyzing and identifying reference genes. Funct. Integr. Genomics 2023, 23, 125. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, h31–h34. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef]
- Plageman Jr, T.F.; Yutzey, K.E. T-box genes and heart development: Putting the “T” in heart. Dev. Dyn. 2005, 232, 11–20. [Google Scholar] [CrossRef]
- Porsch, M.; Hofmeyer, K.; Bausenwein, B.S.; Grimm, S.; Weber, B.H.F.; Miassod, R.; Pflugfelder, G.O. Isolation of a Drosophila T-box gene closely related to human TBX1. Gene 1998, 212, 237–248. [Google Scholar] [CrossRef]
- Yang, G.; Yu, X.; Zhang, Y.; Luo, J.; Li, X.; Zhu, L.; Zhang, H.; Jin, L.; Wu, G.; Yan, X.; et al. Screening and validation of stable reference genes for qRT-PCR analysis in Epicauta gorhami (Coleoptera: Meloidae). Preprints 2024, 2024102173. [Google Scholar] [CrossRef]
- Zhou, L.; Meng, J.; Ruan, H.; Yang, C.; Zhang, C. Expression stability of candidate RT-qPCR housekeeping genes in Spodoptera frugiperda (Lepidoptera: Noctuidae). Arch. Insect Biochem. Physiol. 2021, 108, e21831. [Google Scholar] [CrossRef]
- Huggett, J.; Dheda, K.; Bustin, S.; Zumla, A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005, 6, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Liang, P.; Guo, Y.; Zhou, X.; Gao, X. Expression profiling in Bemisia tabaci under insecticide treatment: Indicating the necessity for custom reference gene selection. PLoS One 2014, 9, e87514. [Google Scholar] [CrossRef] [PubMed]
- Anger, A.M.; Armache, J.; Berninghausen, O.; Habeck, M.; Subklewe, M.; Wilson, D.N.; Beckmann, R. Structures of the human and Drosophila 80S ribosome. Nature 2013, 497, 80–85. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Zuo, M.; Zhang, Y.; Li, N.; Ma, C.; Dong, M.; Gao, N. Structural snapshots of human pre-60S ribosomal particles before and after nuclear export. Nat. Commun. 2020, 11, 3542. [Google Scholar] [CrossRef]
- Zhi-Chuang, L.; Liu-Hao, W.; Rui-Lin, D.; Gui-Fen, Z.; Jian-Ying, G.; Fang-Hao, W. Evaluation of endogenous reference genes of Bactrocera (Tetradacus) minax by gene expression profiling under various experimental conditions. Fla. Entomol. 2014, 97, 597–604. [Google Scholar]
- Gong, Z.; Zhang, J.; Chen, Q.; Li, H.; Zhang, Z.; Duan, Y.; Jiang, Y.; Li, T.; Miao, J.; Wu, Y. Comprehensive screening and validation of stable internal reference genes for accurate qRT-PCR analysis in Holotrichia parallela under diverse biological conditions and environmental stresses. Insects 2024, 15, 661. [Google Scholar] [CrossRef]
- Zhang, S.; An, S.; Li, Z.; Wu, F.; Yang, Q.; Liu, Y.; Cao, J.; Zhang, H.; Zhang, Q.; Liu, X. Identification and validation of reference genes for normalization of gene expression analysis using qRT-PCR in Helicoverpa armigera (Lepidoptera: Noctuidae). Gene 2015, 555, 393–402. [Google Scholar] [CrossRef]
- Shakeel, M.; Zhu, X.; Kang, T.; Wan, H.; Li, J. Selection and evaluation of reference genes for quantitative gene expression studies in cotton bollworm, Helicoverpa armigera (Lepidoptera: Noctuidae). J. Asia-Pac. Entomol. 2015, 18, 123–130. [Google Scholar] [CrossRef]
- Majerowicz, D.; Alves-Bezerra, M.; Logullo, R.; Fonseca-de-Souza, A.L.; Meyer-Fernandes, J.R.; Braz, G.R.C.; Gondim, K.C. Looking for reference genes for real-time quantitative PCR experiments in Rhodnius prolixus (Hemiptera: Reduviidae). Insect Mol. Biol. 2011, 20, 713–722. [Google Scholar] [CrossRef]
- Rajarapu, S.P.; Mamidala, P.; Mittapalli, O. Validation of reference genes for gene expression studies in the emerald ash borer (Agrilus planipennis). Insect Sci. 2012, 19, 41–46. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).