Submitted:
11 November 2024
Posted:
12 November 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Sample Collection
2.3. Detection of O. tsutsugamushi by Nested PCR
2.4. PCR Amplification of 56 kDA VDI, VII and VIII
2.5. DNA Sequencing and Phylogenetic Analysis
3. Results
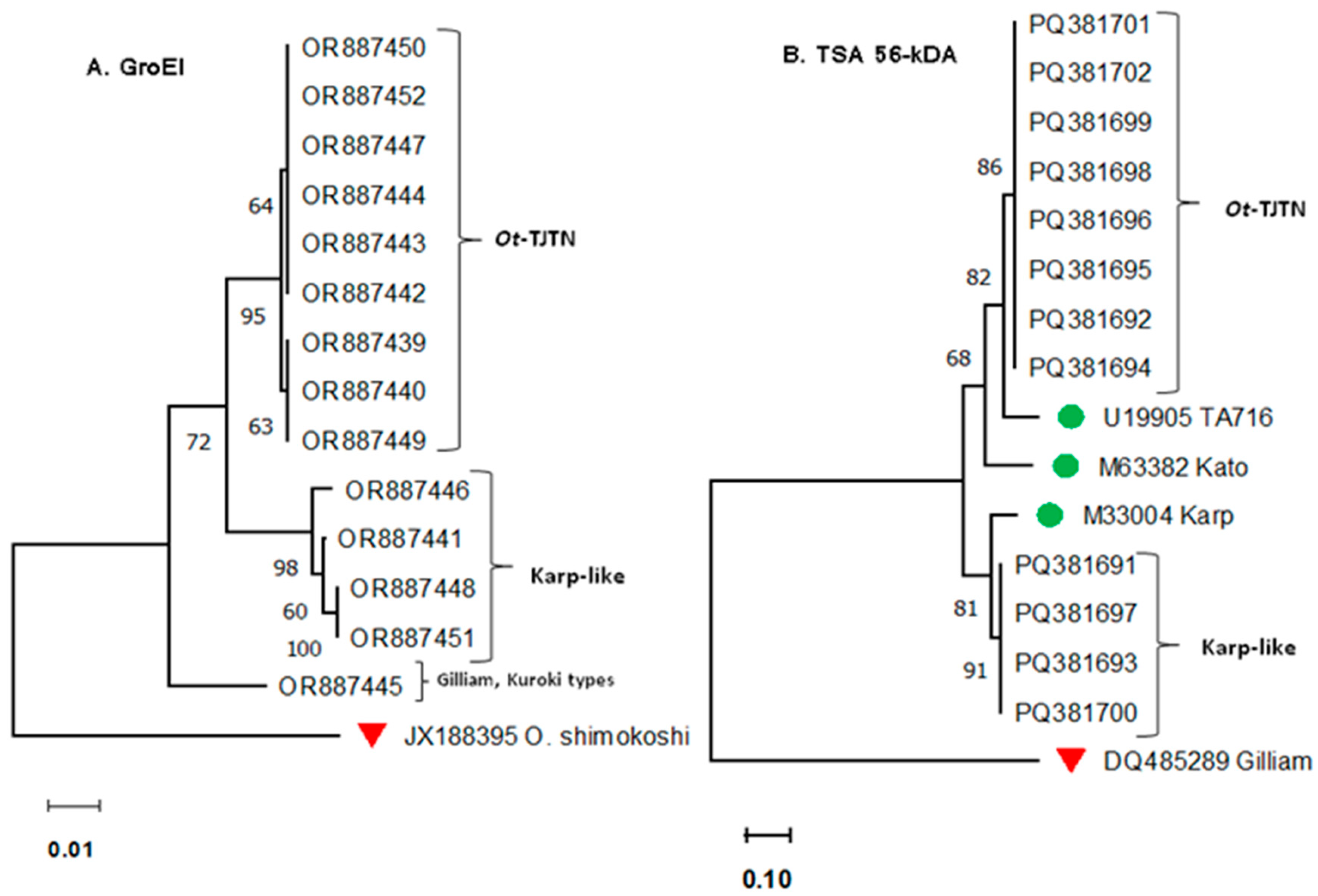
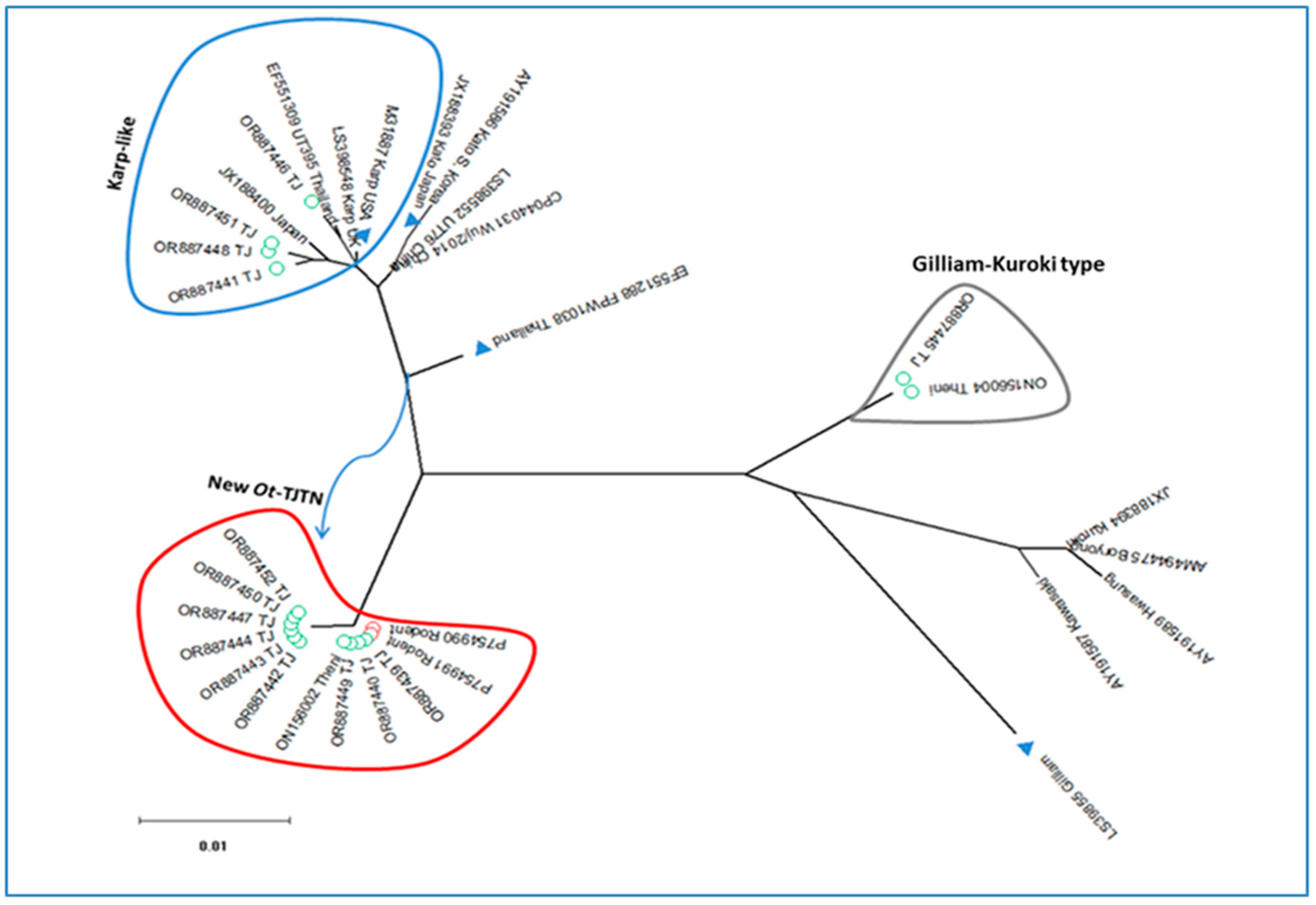
3.1. GroEL Gene Sequence Analysis
3.2. TSA 56 kDA Gene VDI-III Sequence Analysis
3.3. TSA 56 kDA Gene VDI-III Similarity Plot Analysis
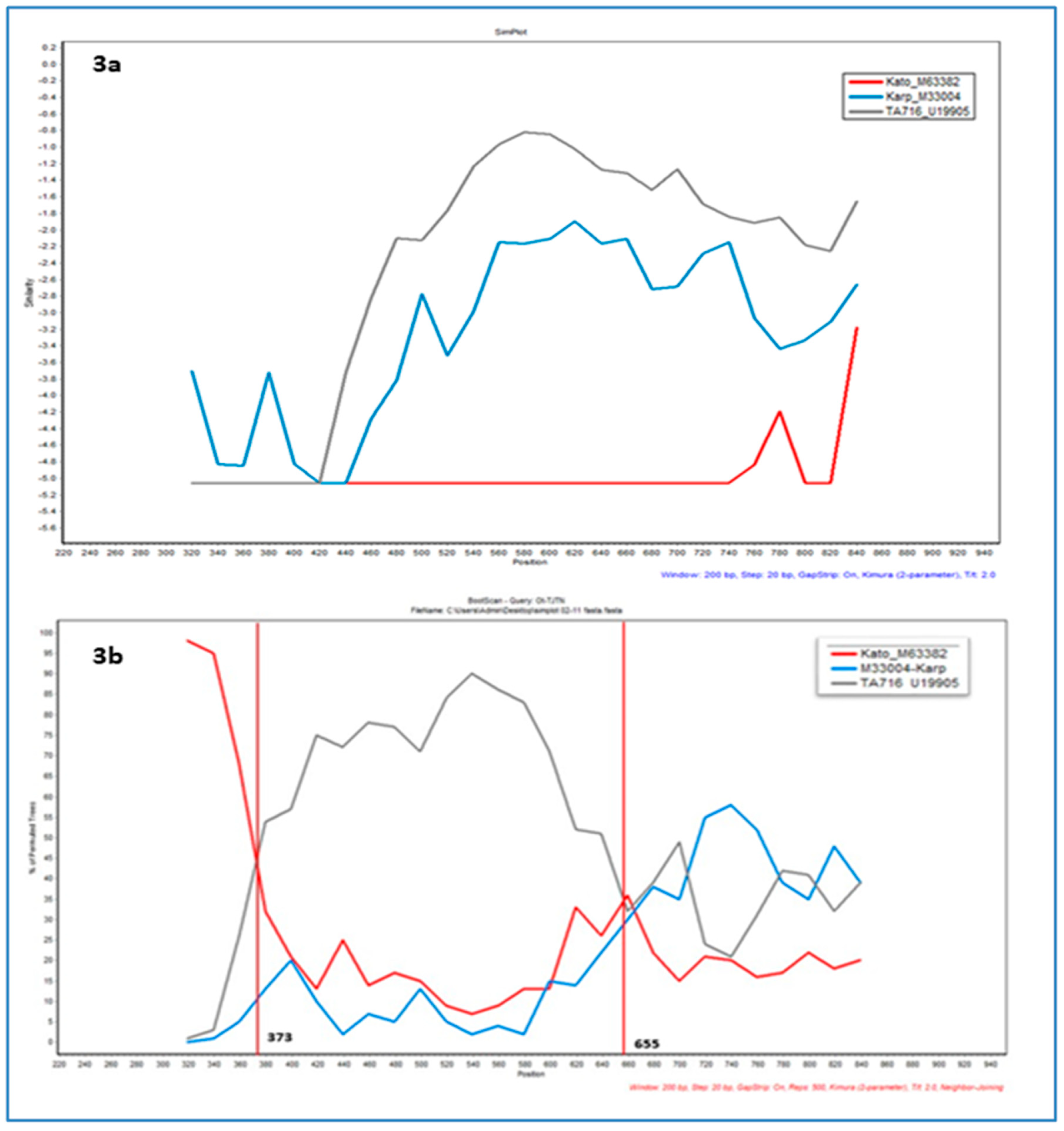
4. Discussions
5. Conclusions
Author Contributions
Funding
Ethics Statement
Data Availability Statement
Acknowledgments
Conflict of Interest
References
- Tilak, R.; Kunwar, R.; Wankhade, U.; Tilak, V. Emergence of Schoengastiella ligula as the vector of scrub typhus outbreak in Darjeeling: Has Leptotrombidium deliense been replaced? Indian. J Pub. Hlth. 2011, 55,92. [CrossRef]
- Candasamy, S.; Ayyanar, E.; Paily, K.; Karthikeyan, P.; Sundararajan, A.; Purushothaman, J. Abundance & distribution of trombiculid mites & Orientia tsutsugamushi, the vectors & pathogen of scrub typhus in rodents & shrews collected from Puducherry & Tamil Nadu, India. Ind. J Med. Res. 2016, 144, 893. [CrossRef]
- Bhate, R.; Pansare, N.; Chaudhari, S.P.; Barbuddhe, S.B.; Choudhary, V.K.; Kurkure, N.V.; Kolte, S.W. Prevalence and phylogenetic analysis of Orientia tsutsugamushi in rodents and mites from central India. Vec. Borne Zoon. Dis. 2017, 17(11), 749-54. [CrossRef]
- Zaman, K. Scrub typhus, a salient threat: Needs attention. PLoS Negl. Trop Dis. 2023,17,e0011427. [CrossRef]
- Kala, D.; Gupta, S.; Nagraik.; R, Verma, V.; Thakur, A.; Kaushal, A. Diagnosis of scrub typhus: recent advancements and challenges. 3 Biotech, 2020,10,396. [CrossRef]
- Bonell, A.; Lubell, Y.; Newton, P.N.; Crump, J.A.; Paris, D.H. Estimating the burden of scrub typhus: A systematic review. PLoS Negl. Trop. Dis. 2017,11,e0005838. [CrossRef]
- Banerjee, A.; Kulkarni, S. Orientia tsutsugamushi: The dangerous yet neglected foe from the East. Int. J Med. Microbiol. 2021, 311,151467. [CrossRef]
- https://cdn.who.int/media/docs/default-source/documents/publications/who-recommended-surveillance-standards17363eff-9860-48c1-9f5f-3c0c3a4f955d.pdf?sfvrsn=ee2893e4_1.
- Kelly, D.J.; Fuerst, P.A.; Ching, W.; Richards, A.L. Scrub Typhus: The Geographic Distribution of Phenotypic and Genotypic Variants of Orientia tsutsugamushi. Clin. Infec. Dis. 2009, 48, S203–30. [CrossRef]
- Izzard, L.; Fuller, A.; Blacksell, S.D,; Paris, D.H.; Richards, A.L.; Aukkanit, N.; Nguyen, C.; Jiang, J.; Fenwick, S.; Day, N.P. et. al. Isolation of a novel Orientia species (O. chuto sp. nov.) from a patient infected in Dubai. J Clin. Microbiol. 2010, 48(12),4404-9. [CrossRef]
- Abarca Villaseca, K.; Martinez-Valdebenito, C.; Angulo, J.; Jiang, J.; Farris, C.M.; Richards, A.L. Acosta-Jamett, G.; Weitzel, T. Molecular Description of a Novel Orientia Species Causing Scrub Typhus in Chile. Emerg. Infect. Dis. 2020, 26, 2148–56. [CrossRef]
- Ohashi, N.; Koyama, Y.; Urakami, H.; Fukuhara, M.; Tamura, A.; Kawamori, F.; Yamamoto, S.; Kasuya, S.; Yoshimura, K. Demonstration of antigenic and genotypic variation in Orientia tsutsugamushi which were isolated in Japan, and their classification into type and subtype. Microbiol. Immunol. 1996, 40(9), 627-38. PMID: 8908607. [CrossRef]
- Devasagayam, E.; Dayanand, D.; Kundu, D.; Kamath, M.S.; Kirubakaran, R.; Varghese, G.M. The burden of scrub typhus in India: A systematic review. PLoS Negl. Trop. Dis. 2021,15 e0009619. [CrossRef]
- Varghese, G.M.; Janardhanan, J.; Mahajan, S.K.; Tariang, D.; Trowbridge, P.; Prakash, J.A.; David, T.; Sathendra, S.; Abraham, O.C. Molecular epidemiology and genetic diversity of Orientia tsutsugamushi from patients with scrub typhus in 3 regions of India. Emerg. Infect. dis. 2015, 21(1), 64-9. [CrossRef]
- Prakash, J.A.; Kamarasu, K.; Samuel, P.P.; Govindarajan, R.; Govindasamy, P.; Johnson, L.A.; Ramalingam, P.; Nirmalson, J.; Seran, K.C. Detection of Orientia tsutsugamushi in novel trombiculid mite species in northern Tamil Nadu, India: Use of targeting the multicopy traD gene. J Med. Entomol. 2022, 59(2), 693-9. [CrossRef]
- Murali, R.; Kalpana, S.; Satheeshkumar, P.; Dhandapani, P. Seroprevalence and Genotypic Characterization of Orientia tsutsugamushi in Febrile Paediatric Patients Admitted in Tertiary Care Hospital of Chennai, South India. J Pure. Appl. Microbiol. 2023, 17, 2232–42. [CrossRef]
- Kumaraswamy, J.; Govindasamy, P.; Nagarajan, L.S.; Gunasekaran, K.; Abhilash, K.P.P.; Prakash, J.A.J. Genotyping of Orientia tsutsugamushi circulating in and around Vellore (South India) using TSA 56 gene. Ind. J Med. Microbiol. 2024, 47, 100483. Epub 2023 Oct 25. PMID: 37890413. [CrossRef]
- Devamani, C.S.; Prakash, J.A.J.; Alexander, N.; Stenos, J.; Schmidt, W.P. The incidence of Orientia tsutsugamushi infection in rural South India. Epidemiol. Infect. 2022,150,e132. [CrossRef]
- Fournier, P.E.; Siritantikorn, S.; Rolain, J.M.; Suputtamongkol, Y.; Hoontrakul, S.; Charoenwat, S.; Losuwanaluk, K.; Parola, P.; Raoult, D. Detection of new genotypes of Orientia tsutsugamushi infecting humans in Thailand. Clin. Microbiol. Infect. 2008,14(2),168-73. Epub 2007 Dec 10. PMID: 18076670. [CrossRef]
- Sonthayanon, P.; Peacock, S.J.; Chierakul, W.; Wuthiekanun, V.; Blacksell, S.D.; Holden, M.T.; Bentley, S.D.; Feil, E.J.; Day, N.P. High rates of homologous recombination in the mite endosymbiont and opportunistic human pathogen Orientia tsutsugamushi. PLoS Neg. Trop. Dis. 2010, 20, 4(7):e752. [CrossRef]
- Arai, S.; Tabara, K.; Yamamoto, N.; Fujita, H.; Itagaki, A.; Kon, M.; Satoh, H,; Araki, K.; Tanaka,T.K.; Takada, N.; et. al. Molecular phylogenetic analysis of Orientia tsutsugamushi based on the groES and groEL genes. Vec. Borne Zoon. Dis. 2013,13(11), 825-9. [CrossRef]
- Ruang-areerate, T.; Jeamwattanalert, P.; Rodkvamtook, W.; Richards, A.L. Sunyakumthorn, P.; Gaywee, J. Genotype Diversity and Distribution of Orientia tsutsugamushi Causing Scrub Typhus in Thailand . J Clin. Microbiol. 2011, 49. [CrossRef]
- https://thanjavur.nic.in/agriculture-2/.
- Li, W.; Dou, X.; Zhang, L.; Lyu, Y.; Du, Z.; Tian, L.; Zhang, X.; Sun, Y.; Guan, Z.; Chen, L.; Li, X. Laboratory diagnosis and genotype identification of scrub typhus from Pinggu district, Beijing, 2008 and 2010. Am. J Trop. Med. Hyg. 2013, 89(1), 123-9. [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–7. [CrossRef]
- Lole, K.S.; Bollinger, R.C.; Paranjape, R.S.; Gadkari, D.; Kulkarni, S.S.; Novak, N.G.; Ingersoll, R. Sheppard, H.W.; Ray, S.C. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J virol. 1999, 73(1),152-60. [CrossRef]
- Minahan, N.T.; Yen, T.Y.; Guo, Y.L.L.; Shu, P.Y.; Tsai, K.H. Concatenated ScaA and TSA56 Surface Antigen Sequences Reflect Genome-Scale Phylogeny of Orientia tsutsugamushi: An Analysis Including Two Genomes from Taiwan. Pathogens, 2024, 13, 299. [CrossRef]
- Paulraj, P.; Renu, G.; Ranganathan, K.; Leo, V.; Veeramanoharan, R. First seroprevalence report of scrub typhus from the tribal belts of the Nilgiris district, Tamil Nadu, India. Ind. J Med. Res. 2021,153, 503. [CrossRef]
- Seong, S.Y.; Kim, H.R.; Huh, M.S.; Park, S.G.; Kang, J.S.; Han, T.H.; Choi, M.S.; Chang, W.H.; Kim, I.S. Induction of neutralizing antibody in mice by immunization with recombinant 56 kDa protein of Orientia tsutsugamushi. Vaccine, 1997, 15(16), 1741-1747.
- Nallan, K.; Rajan, G.; Sivathanu, L.; Devaraju, P.; Thiruppathi, B.; Kumar, A.; Rajaiah, P. Molecular detection of multiple genotypes of Orientia tsutsugamushi causing scrub typhus in febrile patients from Theni District, South India. Trop.Med.Infect.Dis. 2023, 16:8(3), 174. [CrossRef]
- Long, J.; Wei, Y.; Tao, X.; He, P.; Xu, J.; Wu, X.; Zhu, W.; Chen, K.; Yang, Z. Representative genotyping, recombination and evolutionary dynamics analysis of TSA56 gene segment of Orientia tsutsugamushi. Front. Cell. Infect. Microbiol. 2020, 10, 383. [CrossRef]
- Takhampunya R, Korkusol A, Promsathaporn S, Tippayachai B, Leepitakrat S, Richards AL, Davidson SA. Heterogeneity of Orientia tsutsugamushi genotypes in field-collected trombiculid mites from wild-caught small mammals in Thailand. PLoS Negl Trop Dis. 2018 Jul 16;12(7):e0006632. PMID: 30011267; PMCID: PMC6062101. [CrossRef]
- Shirai, A.; Wisseman, Jr C.L. Serologic classification of scrub typhus isolates from Pakistan. Am. J Trop. Med. Hyg. 1975, 24(1), 145-53.
- Basu, S.; Chakravarty, A. Neurological Manifestations of Scrub Typhus. Curr. Neurol. Neurosci. Rep. 2022, 22(8), 491-498. Epub 2022 Jun 21. PMID: 35727462. [CrossRef]
- Lu, HY.; Tsai, K.H.; Yu, S.K.; Cheng, C.H.; Yang, J.S.; Su, C.L.; Hu, H.C.; Wang, H.C.; Huang, J.H.; Shu, P.Y Phylogenetic analysis of 56-kDa type-specific antigen gene of Orientia tsutsugamushi isolates in Taiwan. Am J Trop. Med. Hyg. 2010, 83(3), 658-63. PMID: 20810835; PMCID: PMC2929066. [CrossRef]


| Sl. No. | DNA code | Age/ Gender |
Districts | GenBank Nos. (Present study) | Related Strains | Closest GenBank Acc No. |
Percentage Identity |
Country |
|---|---|---|---|---|---|---|---|---|
| O. tsutsugamushi-New genotype (Ot-TJTN, 9/14) | ||||||||
| 1 | TJ09 | 13/M | Mayiladuthurai | OR887439 | Common New Ot-TJTN | |||
| 2 | TJ11 | 54/F | Nagapattinam | OR887440 | ||||
| 3 | TJ13 | 56/F | Thanjavur | OR887442 | Theni | ON156002 | 100.00 | India |
| 4 | TJ24 | 60/F | Pudukkottai | OR887443 | Vellore | CP166954 | 100.00 | India |
| 5 | TJ25 | 05/F | Pudukkottai | OR887444 | TA716 | EF551288 | 97.71 | Thailand |
| 6 | TJ35 | 37/F | Thanjavur | OR887447 | Wuj/2014 | CP044031 | 97.43 | China |
| 7 | TJ39 | 03/F | Pudukkottai | OR887449 | UT76 | LS398552 | 97.43 | China |
| 8 | TJ42 | 06/M | Pudukkottai | OR887450 | ||||
| 9 | TJ147 | 67/M | Thanjavur | OR887452 | ||||
| Karp-like (4/14) | ||||||||
| 10 | TJ12 | 35/M | Thanjavur | OR887441 | HSBI 2049 Karp |
JX188400 EF551291 LS398548 |
99.43 99.43 99.14 |
Japan Thailand UK |
| 11 | TJ38 | 65/F | Nagapattinam | OR887448 | Karp-like HSB1 Karp |
ON156000 JX188400 LS398548 |
99.71 99.71 99.43 |
India Japan UK |
| 12 | TJ46 | 35/F | Thanjavur | OR887451 | ||||
| 13 | TJ33 | 42/F | Thanjavur | OR887446 | UT395 Karp-like Karp |
EF551309 ON155999 M31887 |
99.71 99.43 99.43 |
Thailand India USA |
| Kuroki-Gilliam type (1/14) | ||||||||
| 14 | TJ26 | 50/F | Thanjavur | OR887445 | Kuroki Kuroki Boryong Kawasaki Hwasung Gilliam |
ON156004 LX188394 AM494475 AY191587 AY191589 LS398551 |
100.00 96.86 96.86 96.86 96.57 96.29 |
India Japan S. Korea |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





