Submitted:
07 November 2024
Posted:
11 November 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Site Description and Soil Sampling
2.2. Microcosm Experiments
2.3. Methane Oxidation Measurements
2.4. Soil DNA Extraction, Amplification and Metagenome Sequencing
2.5. 16S rRNA Gene Amplicon Analyses
2.6. Statistical Analysis
3. Results
3.1. Basic Parameters of Soil Under Investigation
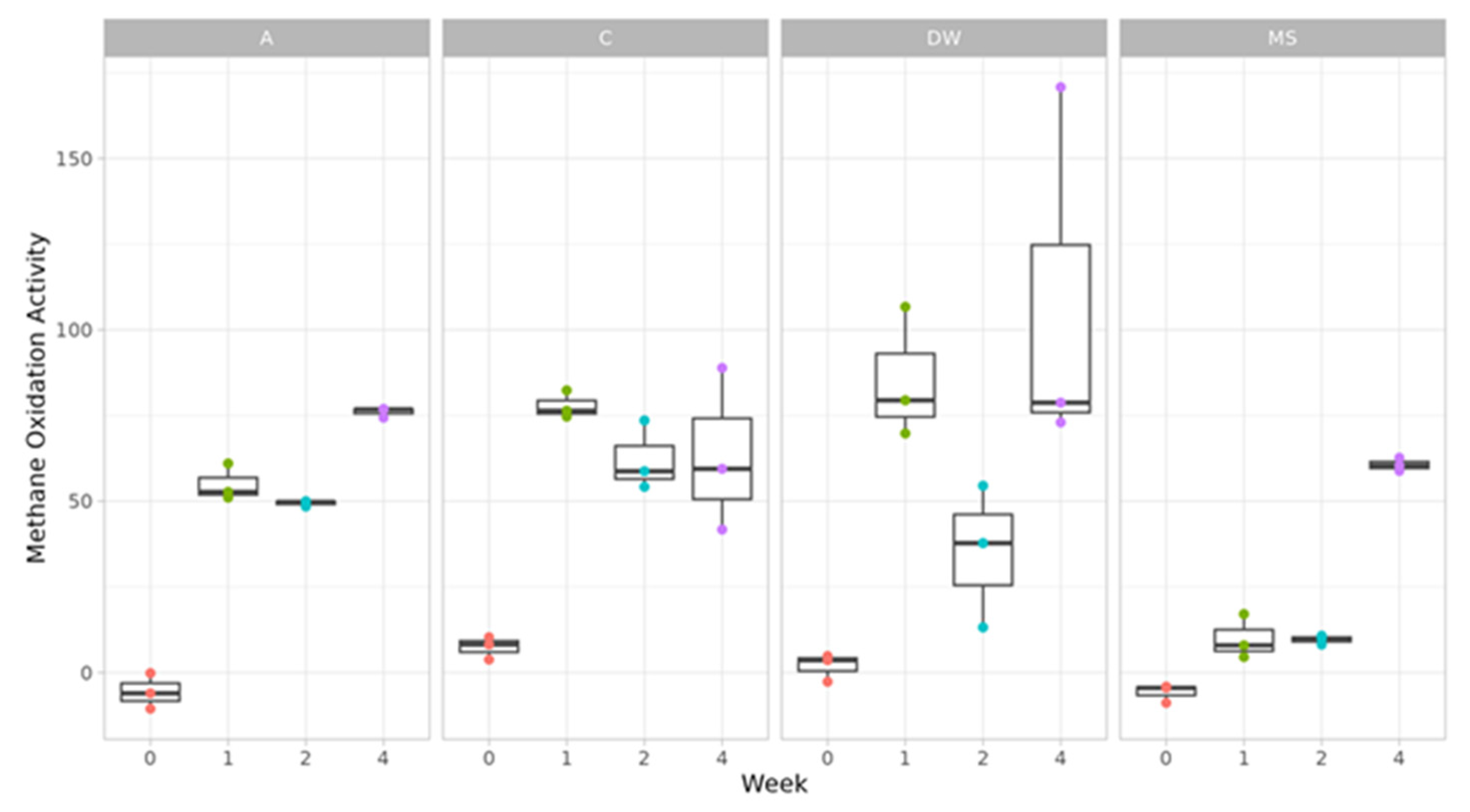
3.2. Methane Oxidation Response to Drying-Rewetting, Ammomium Addition and Their Combined Action
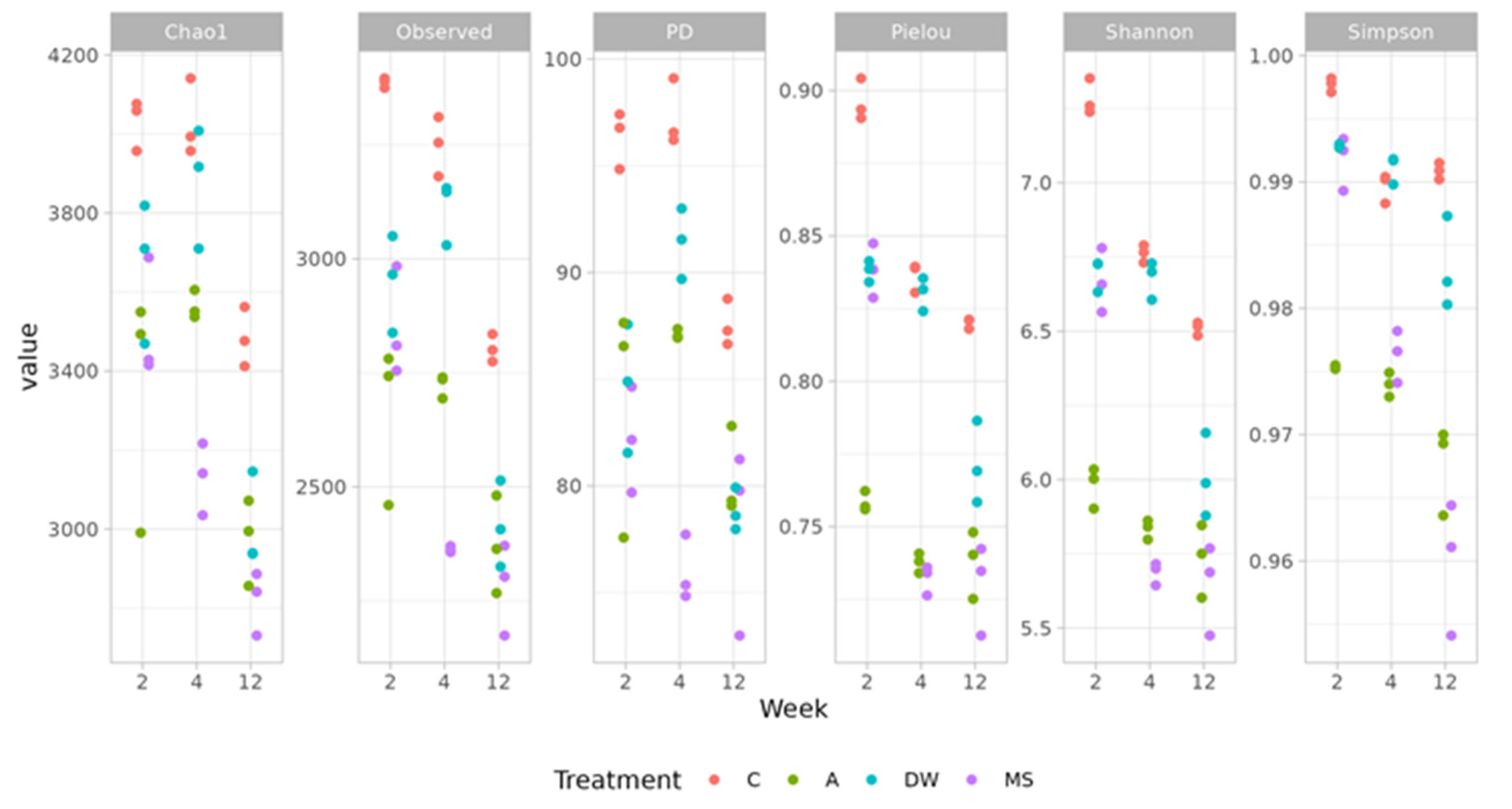
3.3. Sequencing Data and Bacterial α-Diversity of Microbial Communities
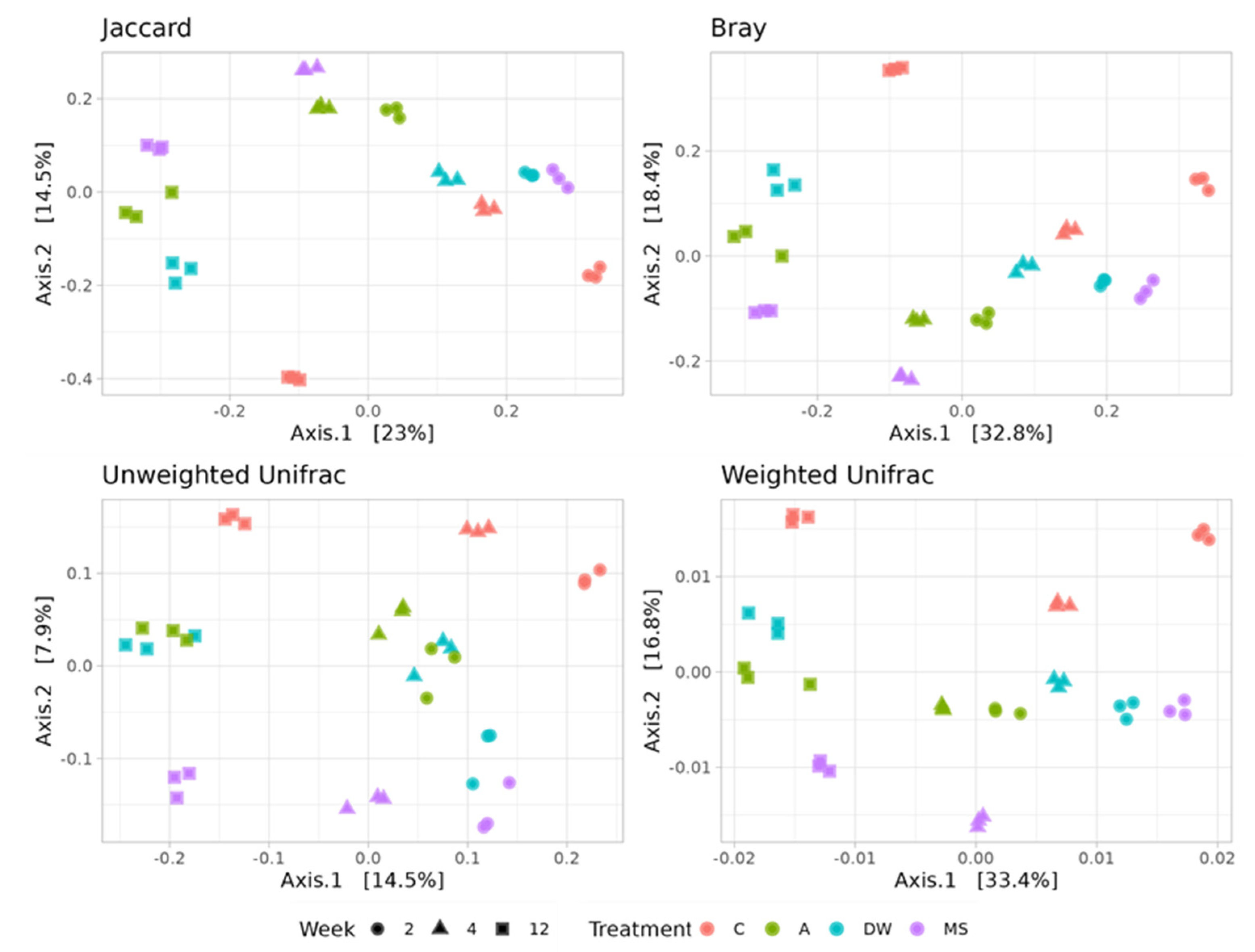
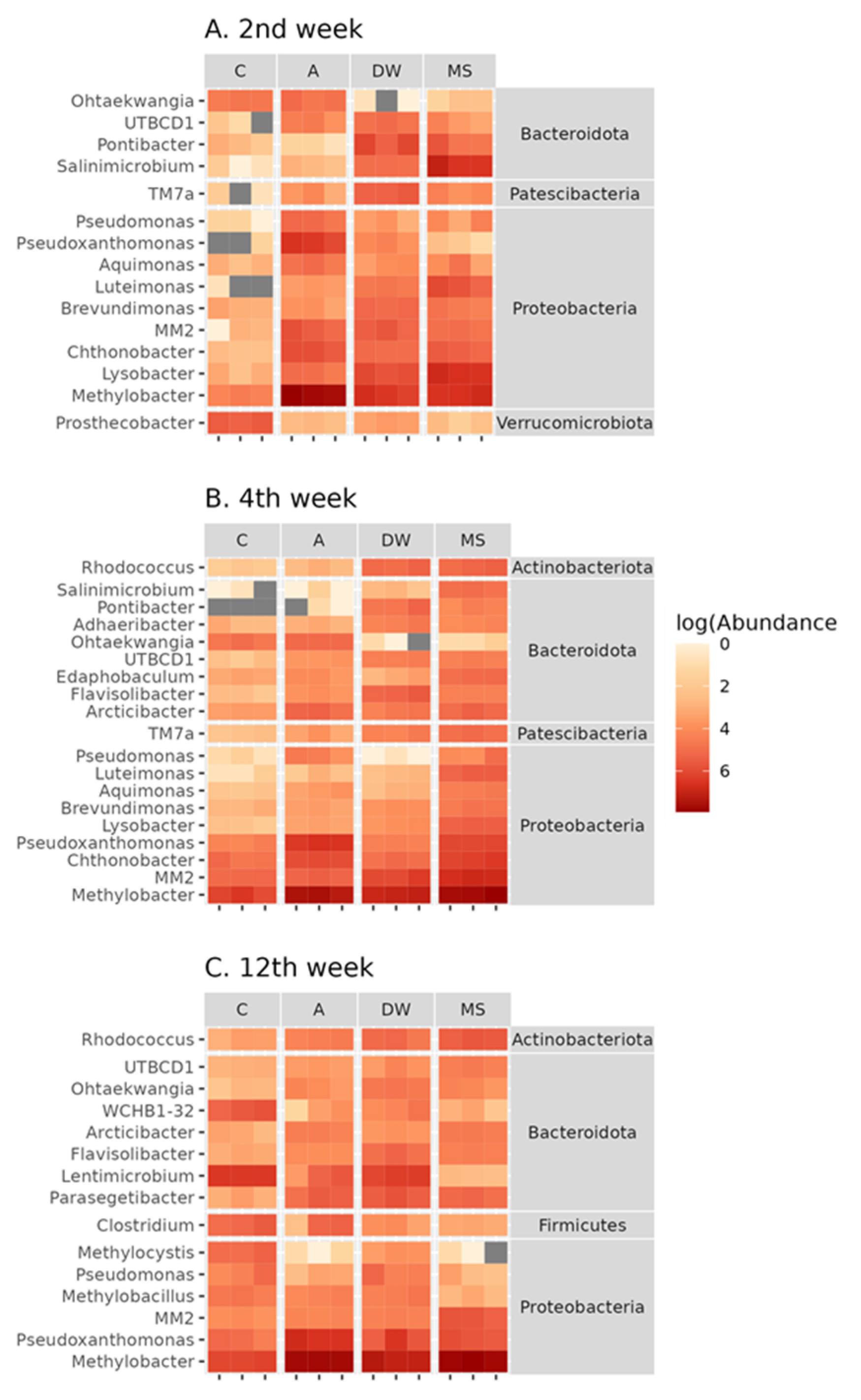
3.4. Bacterial Community Structure
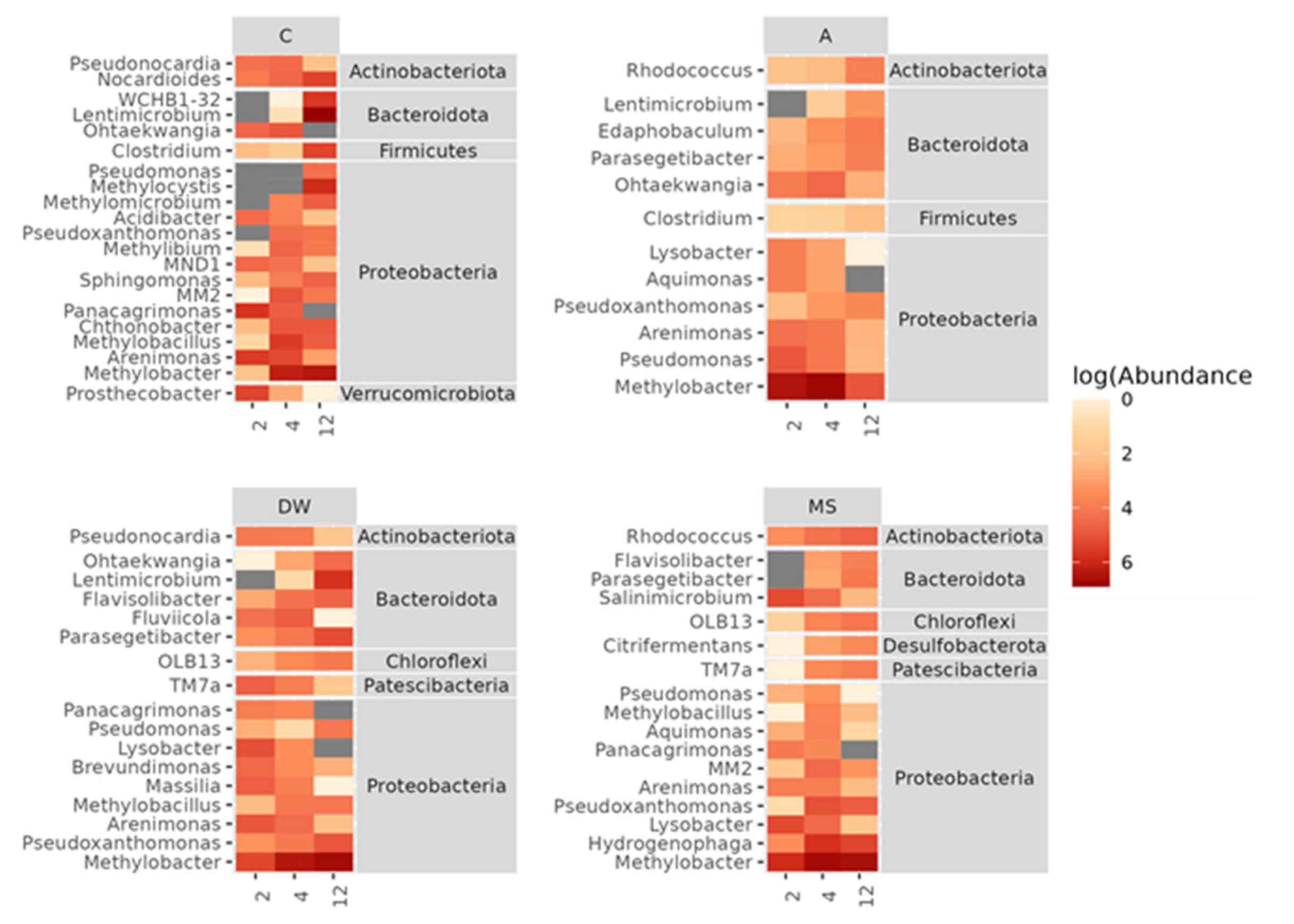
3.5. Effects of Stressors on the Composition of Soil Bacterial Community
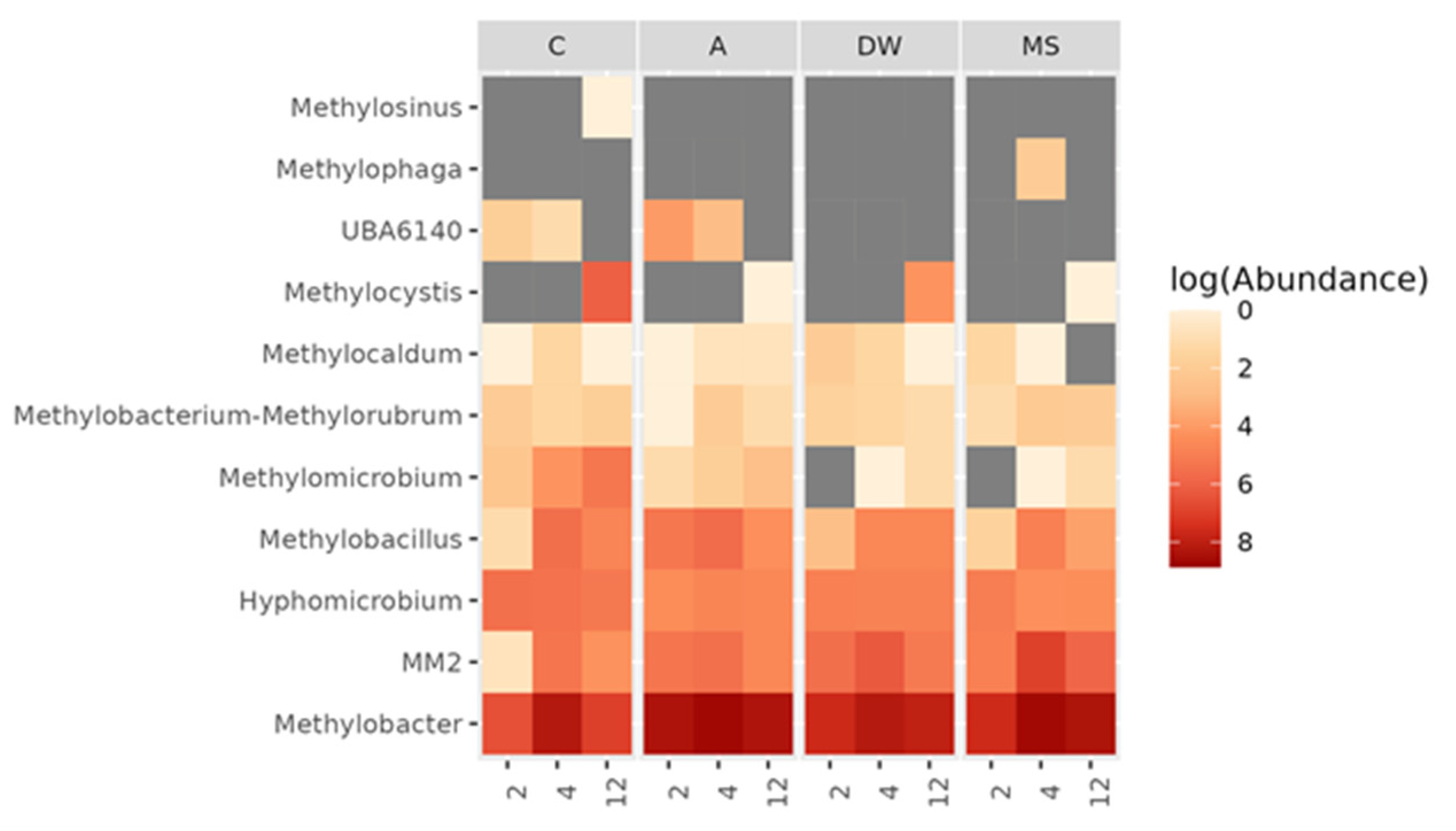
3.6. Structure and Composition of Methylotrophic Soil Bacteria Community
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- IPCC, 2013: Climate Change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. Stocker, T.F., Qin, D., Plattner, G.-K., Tignor, M., Allen, S.K., Boschung, J., Nauels, A., Xia, Y., Bex, V., Midgley, P.M., Eds.; Cambridge University Press: Cambridge, United Kingdom and New York, NY, USA, 1535 pp.
- Saunois, M.; Stavert, A.R.; Poulter, B.; Bousquet, P.; Canadell, J.G.; Jackson, R.B.; Raymond, P.A.; Dlugokencky, E.J.; Houweling, S.; Patra, P.K.P.; et al. The global methane budget 2000–2017. ESSD 2020, Earth 12, 1561–1623. [Google Scholar] [CrossRef]
- IPCC, 2021: Climate Change 2021: The Physical Science Basis. Contribution of Working Group I to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change. Masson-Delmotte, V., Zhai, P., Pirani, A., Connors, S.L., Péan, C., Berger, S., Caud, N., Chen, Y., Goldfarb, L., Gomis, M.I., Huang, M., Leitzell, K., Lonnoy, E., Matthews, J.B.R., Maycock, T.K., Waterfield, T., Yelekçi, O., Yu, R., Zhou, B., Eds.; Cambridge University Press: Cambridge, United Kingdom and New York, NY, USA, 2391 pp.
- Cowan, N.; Maire, J.; Krol, D.; Cloy, J.M.; Hargreaves, P.; Murphy, R.; Carswell, A.; Jones, S.K.; Hinton, N.; Anderson, M.; et al. Agricultural soils: a sink or source of methane across the British Isles? Eur. J. Soil Sci. 2021, 72, 1842–1862. [Google Scholar] [CrossRef]
- Kravchenko, I.K. Microbial oxidation of atmospheric methane in natural and agricultural upland soils. In Agro-Environmental Sustainability. Singh, J.S., Seneviratne,G., Eds.; Springer International Publishing AG: Cham, Switzerland, 2017; Volume 2, pp. 183–211. [Google Scholar]
- Hansen, L.V.; Brændholt, A.; Tariq, A.; Jensen, L.S.; Peixoto, L.E.K.; Petersen, S.O.; Bruun, S. Methane uptake rates across different soil types and agricultural management practices in Denmark. Agric. Ecosyst. Environ. 2024, 363, 108878. [Google Scholar] [CrossRef]
- Kravchenko, I.K.; Boeckx, P.; Galchenko, V.; Van Cleemput, O. Short- and medium term effect of NH4+ on CH4 and N2O fluxes in arable soils with a different texture. Soil Biol. Biochem. 2002, 34, 669–678. [Google Scholar] [CrossRef]
- Khan, N.; Ray, R.L.; Sargani, G.R.; Ihtisham, M.; Khayyam, M.; Ismail, S. Current progress and future prospects of agriculture technology: gateway to sustainable agriculture. Sustainability 2021, 13, 4883. [Google Scholar] [CrossRef]
- Thakur, M.P.; Reich, P.B.; Hobbie, S.E.; Stefanski, A.; Rich, R.; Rice, K.E.; Eddy, W.C.; Eisenhauer, N. Reduced feeding activity of soil detritivores under warmer and drier conditions. Nat. Clim. Change 2017, 8, 75–78. [Google Scholar] [CrossRef]
- Reichstein, M.; Bahn, M.; Ciais, P.; Frank, D.; Mahecha, M.; Seneviratne, S.I.; Zscheischler, J.; Beer, C.; Buchmann, N.; Frank, D.C.; et al. Climate extremes and the carbon cycle. Nature 2013, 500, 287–295. [Google Scholar] [CrossRef]
- Beierkuhnlein, C.; Thiel, D.; Jentsch, A.; Willner, E.; Kreyling, J. Ecotypes of European grass species respond differently to warming and extreme drought. J. Ecol. 2011, 99, 703–713. [Google Scholar] [CrossRef]
- Wu, H.; Yan, L.; Li, Y.; Zhang, K.; Hao, Y.; Wang, J.; Zhang, X.; Yan, Z.; Zhang, Y.; Kang, X. Drought-induced reduction in methane fluxes and its hydrothermal sensitivity in alpine peatland. Peer, J. 2020, 8, e8874. [Google Scholar] [CrossRef]
- Lavallee, J.M.; Chomel, M.N.; Segura, A.; deCastro, F.; Magilton, M.; Goodall, T.; Griffiths, R.I.; Baggs, E.M.; Rhymes, J.M.; Delgado-Baquerizo, M.; et al. Land management shapes drought responses of dominant soil microbial taxa across grasslands. Nat. Commun. 2024, 15, 29. [Google Scholar] [CrossRef]
- Reinsch, S.; Robinson, D.A.; van Soest, M.A.J.; Keith, A.M.; Parry, S.; Tye, A.M. Temperate soils exposed to drought—key processes, impacts, indicators, and unknowns. Land 2024, 13, 1759. [Google Scholar] [CrossRef]
- Wang, Y.F.; Chen, H.; Zhu, Q.A.; Peng, C.H.; Wu, N.; Yang, G.; Zhu, D.; Tian, J.; Kang, X.; He, Y.; et al. Soil methane uptake by grasslands and forests in China. Soil Biol. Biochem. 2014, 74, 70–81. [Google Scholar] [CrossRef]
- Ni, X.; Groffman, P.M. Declines in methane uptake in forest soils. Proc. Natl. Acad. Sci. USA. 2018, 115, 8587–8590. [Google Scholar] [CrossRef]
- Gu, X.; Zhang, Q.; Li, J.; Singh, V.P.; Liu, J.; Sun, P.; Cheng, C. Attribution of global soil moisture drying to human activities: a quantitative viewpoint. Geophys. Res. Lett. 2019, 46, 2573–2582. [Google Scholar] [CrossRef]
- Chan, A.S.K.; Parkin, T.B. Methane oxidation and production activity in soils from natural and agricultural ecosystems. J Environ. Qual. 2001, 30, 1896–1903. [Google Scholar] [CrossRef] [PubMed]
- Nyerges, G.; Stein, L.Y. Ammonia cometabolism and product inhibition vary considerably among species of methanotrophic bacteria. FEMS Microbiol. Lett. 2009, 297, 131–136. [Google Scholar] [CrossRef]
- Versantvoort, W.; Pol, A.; Jetten, M.S.M.; van Niftrik, L.; Reimann, J.; Kartal, B.; Op den Camp, H.J.M. Multiheme hydroxylamine oxidoreductases produce NO during ammonia oxidation in methanotrophs. Proc. Natl. Acad. Sci. USA 2020, 117, 24459–24463. [Google Scholar] [CrossRef]
- King, G.M.; Schnell, S. Effects of ammonium and non-ammonium salt additions on methane oxidation by Methylosinus trichosporium OB3b and main forest soils. Appl. Environ. Microbiol. 1998, 64, 253–257. [Google Scholar] [CrossRef]
- Bodelier, P.L.E. , Laanbroek, H.J. Nitrogen as a regulatory factor of methane oxidation in soils and sediments. FEMS Microbiol. Ecol. 2004, 47, 265–277. [Google Scholar] [CrossRef]
- Kaupper,T. ; Luehrs, J.; Lee, H.J.; Mo, Y.; Jia, Z.; Horn, M.A.; Ho, A. Disentangling abiotic and biotic controls of aerobic methane oxidation during re-colonization. Soil Biol. Biochem. 2020, 142, 107729. [Google Scholar]
- Krause, S.; Lüke, C.; Frenzel, P. Methane source strength and energy flow shape methanotrophic communities in oxygen-methane counter-gradients. Environ. Microbiol. Rep. 2012, 4, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Mohanty, S.R.; Bodelier, P.L.E.; Floris, V.; Conrad, R. Differential effects of nitrogenous fertilizers on methane-consuming microbes in rice fled and forest soils. Appl. Environ. Microbiol. 2006, 72, 1346–1354. [Google Scholar] [CrossRef]
- Smith, P.; House, J.I.; Bustamante, M.; Sobocká, J.; Harper. R.; Pan, G.; West, P.C.; Clark, J.M.; Adhya, T.; Rumpel, C. et al. Global change pressures on soils from land use and management. Glob. Chang. Biol. 2016, 22, 1008–1028. [Google Scholar] [CrossRef] [PubMed]
- Corning, A.P. Nature’s magic: synergy in evolution and the fate of humankind. Cambridge University Press: New York, USA, 2003; 431 p.
- Azarbad, H.; Van Gestel, C.A.; Niklinska, M.; Laskowski, R.; Röling, W.F.; Van Straalen, N.M. Resilience of soil microbial communities to metals and additional stressors: DNA-based approaches for assessing “stress-on-stress” responses. Int. J. Mol. Sci. 2016, 17, 933. [Google Scholar] [CrossRef] [PubMed]
- Van Kruistum, H.; Bodelier, P.L.E.; Ho, A.; Meima-Franke, M.; Veraart, A.J. Resistance and recovery of methane-oxidizing communities depends on stress regime and history; a microcosms study. Front. Microbiol. 2018, 9, 1714. [Google Scholar] [CrossRef]
- Tobor-Kapłon, M.A.; Bloem, J.; De Ruiter, P.C. Functional stability of microbial communities from long-term stressed soils to additional disturbance. Environ. Toxicol. Chem. 2006, 25, 1993–1999. [Google Scholar] [CrossRef]
- Shishov, L.L.; Tonkonogov, V.D.; Gerasimova, M.I.; Lebedeva, I.I. New classification system of Russian soils. Eurasian Soil Sci. 2005, 38, S35–S43. [Google Scholar]
- Kravchenko, I.; Sukhacheva, M. Methane oxidation and diversity of aerobic methanotrophs in forest and agricultural soddy-podzolic soils. Appl Soil Ecol. 2017, 119, 267–274. [Google Scholar] [CrossRef]
- Pinaev, A.G.; Kichko, A.A.; Aksenova, T.S.; Safronova, V.I.; Kozhenkova, E.V.; Andronov, E.E. RIAM: a universal accessible protocol for the isolation of high purity DNA from various soils and other humic substances. Methods Protoc. 2022, 5, 99. [Google Scholar] [CrossRef]
- Bates, S.T.; Berg-Lyons, D.; Caporaso, J.G.; Walters, W.A.; Knight, R.; Fierer, N. Examining the global distribution of dominant archaeal populations in soil. ISME J. 2010, 5, 908–917. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Wright, E.S. Using DECIPHER v2. 0 to analyze big biological sequence data in R. The R Journal 2016, 8, 352–359. [Google Scholar]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. NAR 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello,E. K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of highthroughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Wickham, H.; Averick, M.; Bryan,J. ; Chang, W.; D’Agostino McGowan, L.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J. et al. Welcome to the Tidyverse. Journal of Open Source Software 2019, 4, 1686. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet,F. G.; Friendly, M.; Kindt, R.; Legendre,P.; McGlinn, D.; Minchin,P.R.; O’Hara,R. B.; Simpson, G.L.; Solymos, P.; et al. vegan: Community Ecology Package. R package version 2.5-6. 2019. Community ecol. 2020, 8, 732–740. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014, 15, 550. [Google Scholar] [CrossRef]
- Cotrufo, M.F.; Lavallee, J.M.; Zhang, Y.; Hansen, P.M.; Paustian, K.H.; Schipanski, M.; Wallenstein, M.D. In-N-Out: A hierarchical framework to understand and predict soil carbon storage and nitrogen recycling. Glob. Chang. Biol. 2021, 27, 4465–4468. [Google Scholar] [CrossRef]
- Kolb, S.; Knief, C.; Stubner, S.; Conrad, R. Quantitative detection of methanotrophs in soil by novel pmoA-targeted real-time PCR assays. Appl Environ Microbiol. 2003, 69, 2423–2429. [Google Scholar] [CrossRef]
- Seghers,D. ; Top, E.M.; Reheul, D.; Bulcke, R.; Boeckx, P.; Verstraete, W.; Siciliano, S.D. Long-term effects of mineral versus organic fertilizers on activity and structure of the methanotrophic community in agricultural soils. Environ Microbiol. 2003, 5, 867–877. [Google Scholar] [CrossRef] [PubMed]
- Kravchenko, I.K.; Semenov, V.M.; Kuznetsova, T.V.; Bykova, S.A.; Dulov, L.E.; Pardini, G.; Gispert, M.; Boeckx, P.; Van Cleemput, O.; Gal’chenko, V.F. Physicochemical and biological factors affecting atmospheric methane oxidation in gray forest soils. Microbiology (Russian Federation) 2005, 74, 216–220. [Google Scholar] [CrossRef]
- Dorr, N.; Glaser, B.; Kolb, S. Methanotrophic communities in Brazilian ferralsols from naturally forested, afforested, and agricultural sites. Appl Environ Microbiol 2010, 76, 1307–1310. [Google Scholar] [CrossRef] [PubMed]
- Dunfield, P.F.; Knowles, R. Kinetics of inhibition of methane oxidation by nitrate, nitrite, and ammonium in a humisol. Appl. Environ. Microbiol. 1995, 61, 3129–3135. [Google Scholar] [CrossRef]
- Ho, A.; Kerckhof, F.-M.; Luke, C.; Reim, A.; Krause, S.; Boon, N.; Bodelier, P.L.E. Conceptualizing functional traits and ecological characteristics of methane-oxidizing bacteria as life strategies. Environ. Microbiol. Rep. 2013, 5, 335–345. [Google Scholar] [CrossRef]
- van Kruistum, H.; Bodelier, P.L.E.; Ho, A.; Meima-Franke, M.; Veraart, A.J. Resistance and recovery of methane-oxidizing communities depends on stress regime and history; a microcosm study. Front. Microbiol. 2018, 9, 1714. [Google Scholar] [CrossRef]
- Collet, S.; Reim, A.; Ho, A.; Frenzel, P. Recovery of paddy soil methanotrophs from long term drought. Soil Biol. Biochem. 2015, 88, 69–72. [Google Scholar] [CrossRef]
- Bodelier, P.L.E. Interactions between nitrogenous fertilizers and methane cycling in wetland and upland soils. Curr. Opinion Environ. Sustain. 2011, 3, 379–388. [Google Scholar] [CrossRef]
- Bykova, S.; Boeckx, P.; Kravchenko, I.; Galchenko, V.; Van Cleemput, O. Impact of fertilization type and CH4 concentration on methane oxidation and methanotrophic diversity in arable soils. Biol. Fertil. Soils 2007, 43, 341–348. [Google Scholar] [CrossRef]
- Acton, S.D.; Baggs, E.M. Interactions between N application rate, CH4 oxidation and N2O production in soil. Biogeochemistry 2011, 103, 15–26. [Google Scholar] [CrossRef]
- Tate, K.R.; Ross, D.J.; Scott, N.A.; Rodda, N.J.; Townsend, J.A.; Arnold, G.C. Post-harvest patterns of carbon dioxide production, methane uptake and nitrous oxide production in a Pinus radiata D. Don plantation. For. Ecol. Manag. 2006, 228, 40–50. [Google Scholar] [CrossRef]
- Jacinthe, P.A.; Lal, R. Labile carbon and methane uptake as affected by tillage intensity in a Mollisol. Soil Tillage Res. 2006, 80, 35–45. [Google Scholar] [CrossRef]
- Aronson, E.L.; Dubinsky, E.A.; Helliker, B.R. Effects of nitrogen addition on soil microbial diversity and methane cycling capacity depend on drainage conditions in a pine forest soil. Soil Biol. Biochem. 2013, 62, 119–128. [Google Scholar] [CrossRef]
- Pátek, M.; Grulich, M.; Nešvera, J. Stress response in Rhodococcus strains. Biotechnol. Adv. 2021, 53, 107698. [Google Scholar] [CrossRef] [PubMed]
- Naylor, D.; Coleman-Derr, D. Drought stress and root-associated bacterial communities. Front. Plant Sci. 2018, 8, 785–852. [Google Scholar] [CrossRef]
- Bowman, J.P. Methylobacter. In: Bergey’s Manual of Systematics of Archaea and Bacteria.; Trujillo, M.E., Dedysh, S., DeVos, P., Hedlund, B., Kämpfer, P., Rainey, F.A., Whitman, W.B., Eds. [CrossRef]
- Wang, H.; Zhang, S.; Zhang, J. The copper resistance mechanism in a newly isolated Pseudoxanthomonas spadix ZSY-33. Environ Microbiol. Rep. 2023, 15, 484–496. [Google Scholar] [CrossRef]
- Maxfield, P.J.; Hornibrook, E.R.C.; Evershed, R.P. Acute impact of agriculture on high-affinity methanotrophic bacterial populations. Environ. Microbiol 2008, 10, 1917–1924. [Google Scholar] [CrossRef]
- Seghers, D.; Verthe, K.; Reheul, D.; Bulcke, R.; Siciliano, S.D.; Verstraete, W.; Top, E.M. Effect of longterm herbicide applications on the bacterial community structure and function in an agricultural soil. FEMS Microb. Ecol 2006, 46, 139–146. [Google Scholar] [CrossRef]
- Ho, A.; Mendes, L.W.; Lee, H.J.; Kaupper, T.; Mo,Y. ; Poehlein, A.; Bodelier, P.L.E.; Jia, Z.; Horn, M.A. Response of a methane-driven interaction network to stressor intensification. FEMS Microbiol. Ecol. 2020, 96, fiaa180. [Google Scholar] [CrossRef]
- Kizilova, A.; Yurkov, A.; Kravchenko, I. Aerobic methanotrophs in natural and agricultural soils of European Russia. Diversity 2013, 5, 541–556. [Google Scholar] [CrossRef]
- Kravchenko, I.K.; Sizov, L.R.; Tikhonova, E.N.; Lysak, L.V. The effect of lanthanum on the composition of methanotrophic community of sod-podzolic soil. Microbiology (Moscow) 2022, 91, 604–608. [Google Scholar] [CrossRef]
- Danilova, O.V.; Ivanova, A.A.; Terent’eva, I.E.; Glagolev, M.V.; Sabrekov, A.F. Microbial community composition of floodplains shallow-water seeps in the Bolshaya Rechka floodplain, Western Siberia. Microbiology (Russian Federation) 2021, 90, 632–642. [Google Scholar] [CrossRef]
- Oshkin, I. , Beck, D., Lamb, A., Tchesnokova,V.; Benuska, G.; McTaggart,T.L.; Kalyuzhnaya,M.G.; Dedysh,S.N.; Lidstrom, M.E.; Chistoserdova, L. Methane-fed microbial microcosms show differential community dynamics and pinpoint taxa involved in communal response. ISME J 2015, 9, 1119–1129. [Google Scholar] [CrossRef]
- Karthikeyan, O.P.; Chidambarampadmavathy, K.; Nadarajan, S.; Lee, P.K.; Heimann, K. Effect of CH4/O2 ratio on fatty acid profile and polyhydroxybutyrate content in a heterotrophic–methanotrophic consortium. Chemosphere 2015, 141, 235–242. [Google Scholar] [CrossRef] [PubMed]
| Soil’s parameter, dimension | Parameter`s value |
|---|---|
| Texture | Loamy sanda, coarseb |
| Clay, % | 4 |
| Sand, % | 87 |
| Loam, % | 9 |
| pHH2O | 7.6±0.1 |
| SOC, % TN,% |
5.97±0.9 0.37± 0.02 |
| C:N | 16.14 |
| Soil density, g/ cm3 | 2.55 |
| Na+, mmol/100g | 0.283±0.01 |
| К+, mmol/100g | 0.550±0.01 |
| Ca2+, mmol/100g | 18.15±0.21 |
| Mg2+, mmol/100g | 5.852±0.073 |
| Basal microbial respiration, µg С-СО2 g-1 h-1 | 0.82 ±0.02 |
| Microbial biomass, µg С g-1 | 1252±65 |
| Metabolic quotient (QCO2) | 1.35±0.03 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





