Submitted:
07 November 2024
Posted:
08 November 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Sampling and Enrichment
2.2. Establishment of Clonal Cultures
2.3. Light Microscopical Analyses
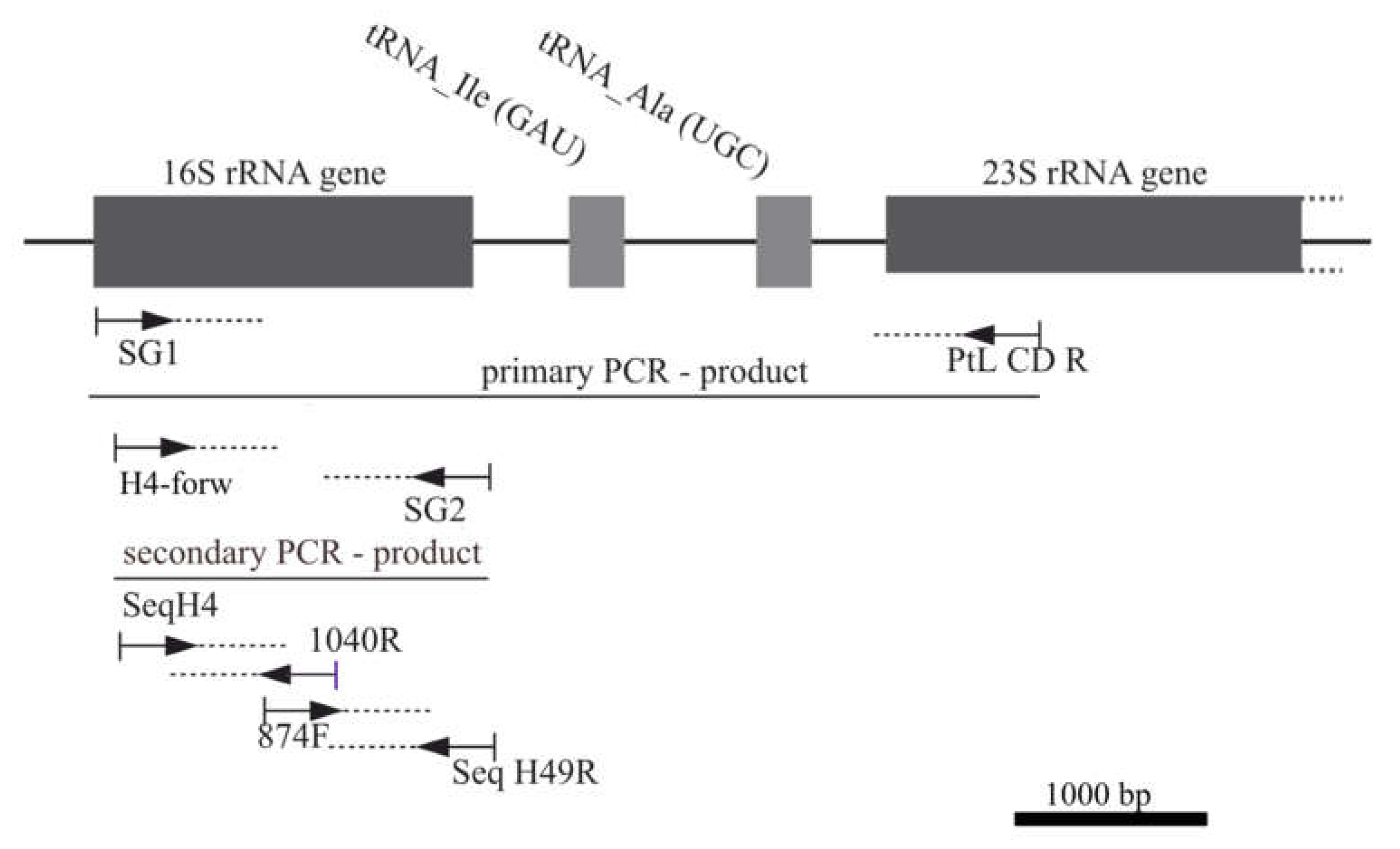
2.4. DNA Extraction and Sequencing
2.5. Phylogenetic Analyses
3. Results
3.1. Isolation of Strains
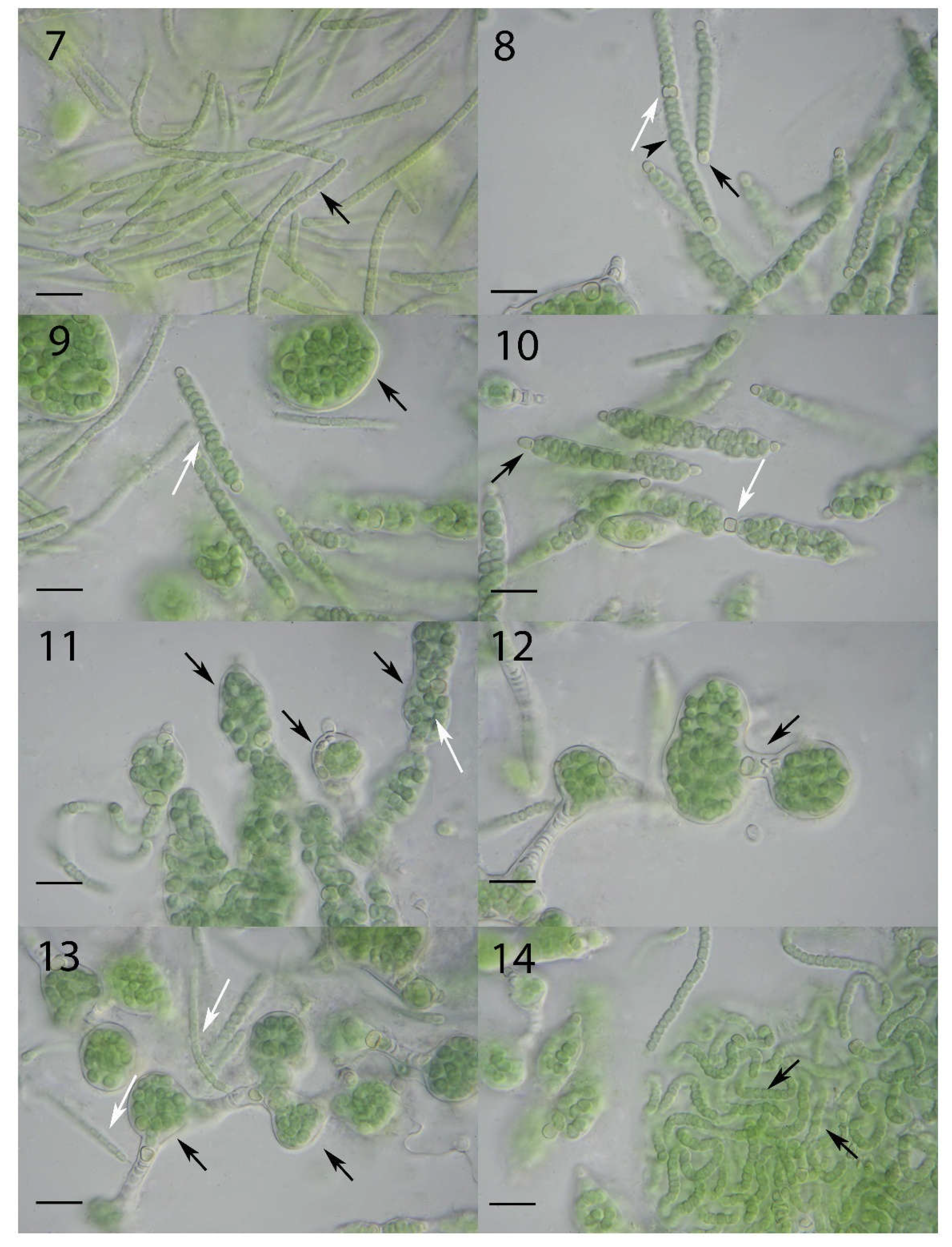
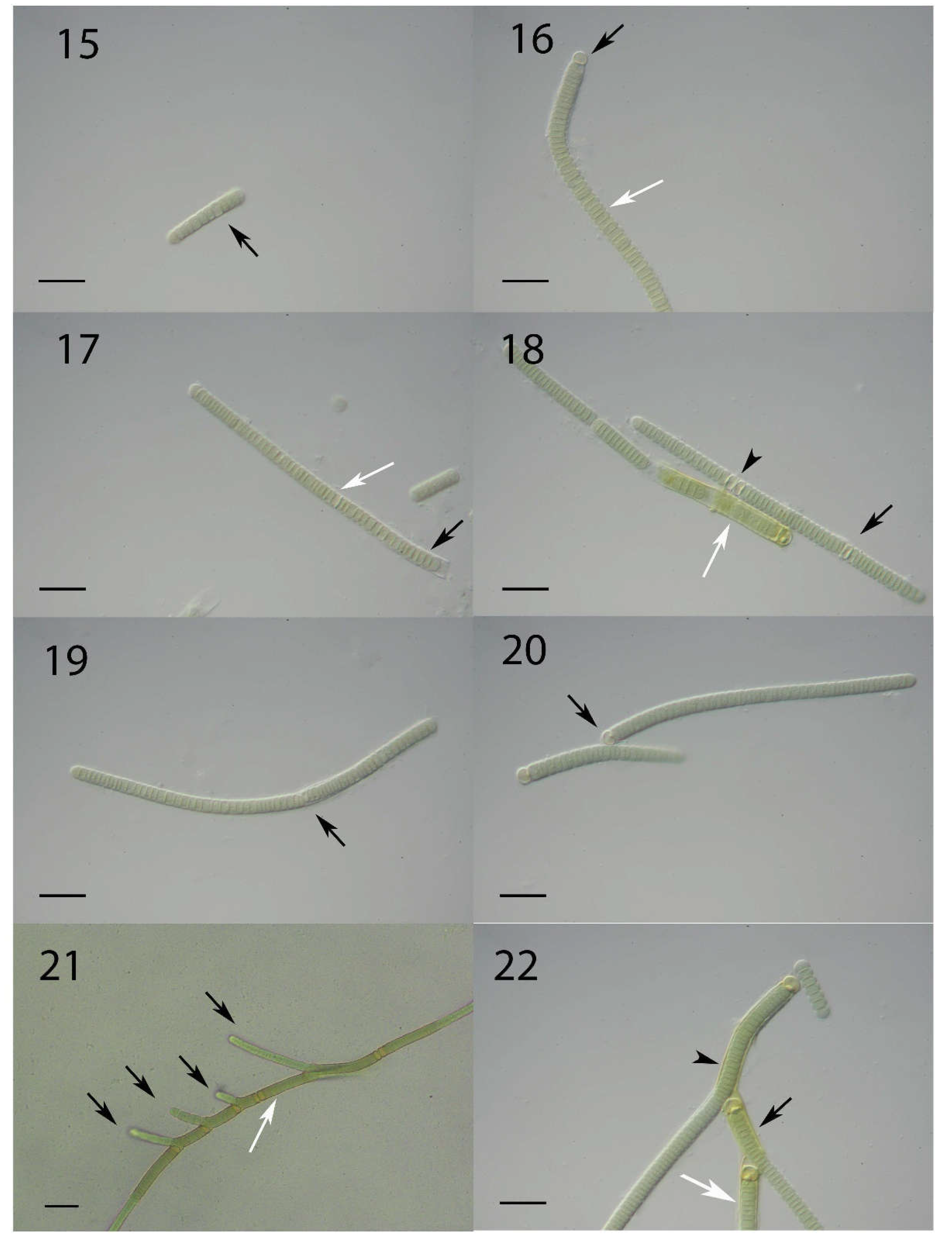
3.2. Light Microscopical Observations
3.3. Molecular Phylogeny using 16S DNA Gene Sequence Comparisons
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Morales, D.; Jimenez, M.S.; Gonzalez-Rodríguez, A.M.; Cermák, J. Laurel forest in Tenerife, Canary Islands. I. The site, stand structure and stand leaf area distribution. Trees 1996, 11, 34–40. [Google Scholar] [CrossRef]
- Nogué, S.; De Nascimento, L.; Fernández-Palacios, J.M.; Whittaker, R.J.; Willis, K.J. The ancient forests of La Gomera, Canary Islands, and their sensitivity to environmental change. J Ecol 2013, 101, 368–377. [Google Scholar] [CrossRef]
- Fernández-Palacios, J.M. Climatic responses of plant species on Tenerife, The Canary Islands. J Veg Sci 1992, 3, 595–603. [Google Scholar] [CrossRef]
- Del Arco Aguilar, M.J.; Rodriguez Delgado, O. Vegetation of the Canary Islands. In Vegetation of the Canary Islands. Plant and Vegetation, Volume 16, del Arco Aguilar M.J., Rodriguez Delgado, O., Eds.; Springer: Cham, Switzerland, 2018; pp. 83–319. [Google Scholar] [CrossRef]
- Fernández–Palacios, J.M.; De Nicolás, J.P. Altitudinal pattern of vegetation variation on Tenerife. J Veg Sci 1995, 6, 183–190. [Google Scholar] [CrossRef]
- Lindow, S.E.; Brandl, M.T. Microbiology of the phyllosphere. Appl Environ Microbiol 2003, 69, 1875–1883. [Google Scholar] [CrossRef]
- Koskella, B. The phyllosphere. Curr Biol 2020, 30, R1143–R1146. [Google Scholar] [CrossRef]
- Lambais, M.R.; Crowley, D.E.; Cury, J.C.; Büll, R.C.; Rodrigues, R.R. Bacterial diversity in tree canopies of the Atlantic forest. Science 2006, 312, 1917–1917. [Google Scholar] [CrossRef]
- Peñuelas, J.; Terradas, J. The foliar microbiome. Trends Plant Sci 2014, 19, 278–280. [Google Scholar] [CrossRef]
- Liu, B.W.; Li, S.Y.; Zhu, H.; Liu, G.X. Phyllosphere eukaryotic microalgal communities in rainforests: drivers and diversity. Plant Divers, 2023, 45, 45–53. [Google Scholar] [CrossRef]
- Coxson, D.S.; Nadkarni, N.M. Ecological roles of epiphytes in nutrient cycles of forest ecosystems. In Forest Canopies, Lowman, M.D.; Nadkarni, N.M., Ed.; Academic Press: San Diego, USA, 1995; pp. 495–543. [Google Scholar]
- Zhu, Y.G.; Peng, J.; Chen, C.; Xiong, C.; Li, S.; Ge, A.; Wang, E.; Liesack, W. Harnessing biological nitrogen fixation in plant leaves. Trends Plant Sci 2023, 28, 1391–1405. [Google Scholar] [CrossRef]
- González-González, R.; Léon, M.C.; Del Arco, M.J. Los helechos de la Reserva Natural Integral de El Pijaral. Consejería de Política Territorial y Medio Ambiente del Gobierno de Canarias. Sta Cruz de Tenerife, Spain, 2002; 184 pp.
- Beltrán-Tejera, E. (ed.) Hongos, líquenes y briófitos del Parque Nacional de Garajonay (La Gomera, Islas Canarias). O.A. Parques Nacionales, Serie Técnica. In Ministerio de Medio Ambiente. Madrid, Spain, 2006; 844 pp.
- Patiño, J.; González-Mancebo, J.M. Exploring the effect of host tree identity on epiphyte bryophyte communities in different Canarian subtropical cloud forests. Plant Ecol 2010, 212, 433–449. [Google Scholar] [CrossRef]
- Hernández-Hernández, R.; Castro, J.; Arco-Aguilar, D.; Fernández-López, Á.; González-Mancebo, J.M. Post-fire salvage logging imposes a new disturbance that retards succession: the case of bryophyte communities in a Macaronesian laurel forest. Forests 2017, 8, 252. [Google Scholar] [CrossRef]
- González-Montelongo, C.; Pérez-Vargas, I. Looking for a home: Exploring the potential of epiphytic lichens to colonize tree plantations in a Macaronesian laurel forest. For Ecol Manage 2019, 453, 117541. [Google Scholar] [CrossRef]
- González-Montelongo, C. , Pérez-Vargas, I. Is an invasive alien tree able to sustain a similar lichen diversity as the native forest? The case of the sweet chestnut (Castanea sativa Mill.) and the laurel forest in Macaronesia. For Ecol Manage 2021, 488, 119009. [Google Scholar] [CrossRef]
- Büdel, B.; Weber, H.M.; Porembski, S.; Barthlott, W. Cyanobacteria of inselbergs in the Atlantic rainforest zone of eastern Brazil. Phycologia 2002, 41, 498–506. [Google Scholar] [CrossRef]
- Gorbushina, A.A. Life on the rocks. Environ Microbiol 2007, 9, 1613–1631. [Google Scholar] [CrossRef]
- Sant’Anna, C.L.; Kaštovský, J.; Hentschke, G.S.; Komárek, J. Phenotypic studies on terrestrial stigonematacean cyanobacteria from the Atlantic Rainforest, Sao Paulo State, Brazil. Phytotaxa 2013, 89, 1–23. [Google Scholar] [CrossRef]
- Glime, J.M.; Pócs, T. Tropics: Epiphylls. Chapt. 8-6. In: Bryophyte Ecology. Glime, J.M., Ed.,Volume 4. Habitat and Role. Ebook sponsored by Michigan Technological University and the International Association of Bryologists. 2018. Last updated 23 July 2020 and available at .
- Freiberg, E. Microclimatic parameters influencing nitrogen fixation in the phyllosphere in a Costa Rican premontane rain forest. Oecologia 1998, 117, 9–18. [Google Scholar] [CrossRef]
- Vitousek, P.M.; Cassman, K.E.N.; Cleveland, C.; Crews, T.; Field, C.B.; Grimm, N.B.; Howarth, R.W.; Marino, R.; Martinelli, L.; Rastetter, E.B.; Sprent, J.I. Towards an ecological understanding of biological nitrogen fixation. Biogeochemistry 2002, 57/58, 1–45. [Google Scholar] [CrossRef]
- Bernap, J.; Prasse, R.; Harper, K. Influence of biological soil crusts on soil environments and vascular plants. In: Biological Soil Crusts: Structure, Function, and Management. Belnap J, Lange O.L., Eds., Springer, Berlin, Germany, 2003, pp 281–300.
- Fürnkranz, M.; Wanek, W.; Richter, A.; Abell, G.; Rasche, F.; Sessitsch, A. Nitrogen fixation by phyllosphere bacteria associated with higher plants and their colonizing epiphytes of a tropical lowland rainforest of Costa Rica. ISME J 2008, 2, 561–570. [Google Scholar] [CrossRef]
- Moreira, J.C.F.; Brum, M.; de Almeida, L.C.; Barrera-Berdugo, S.; de Souza, A.A.; de Camargo, P B.; Oliveira, R.S.; Alves, L.F.; Pimentel Rosado, B.H.; Lambais, M.R. Asymbiotic nitrogen fixation in the phyllosphere of the Amazon forest: Changing nitrogen cycle paradigms. Sci Total Environ 2021, 773, 145066. [Google Scholar] [CrossRef] [PubMed]
- Harrelson, M.A. Tropical Epiphyllae and Nitrogen Fixation. PhD thesis, University of Georgia, Athens, Georgia, USA, 1969.
- Bentley, B.L. . Nitrogen fixation by epiphylls in a tropical rainforest. Ann Missouri Bot Gard 1987, 74, 234–241. [Google Scholar] [CrossRef]
- Toomey, M.; Roberts, D.; Nelson, B. The influence of epiphylls on remote sensing of humid forests. Remote Sens Environ 2009, 113, 1787–1798. [Google Scholar] [CrossRef]
- Lemes-Da-Silva, N.M.; Branco, L.H.Z.; Necchi-Júnior, O. New aerophytic morphospecies of Cyanobacteria from tropical forest fragments in northwestern São Paulo state, Brazil. Acta Bot Brasil 2010, 24, 916–923. [Google Scholar] [CrossRef]
- Alvarenga, D.O.; Rigonato, J.; Branco, L.H.Z.; Fiore, M.F. Cyanobacteria in mangrove ecosystems. Biodivers Conserv 2015, 24, 799–817. [Google Scholar] [CrossRef]
- Branco, L.H.Z.; Hoffmann, L.; Teixeira, J.P.; Ferreira, V.; De Morais Filho, J.C. Aerophytic cyanoprokaryotes from the Atlantic rainforest region of São Paulo State, Brazil: Chroococcales and Oscillatoriales. Cryptogamie Algol 2009, 30, 135. [Google Scholar]
- Gama, W.A.JR.; Laughinghouse, H.D.IV.; Sant’Anna, CL. How diverse are coccoid cyanobacteria? A case study of terrestrial habitats from the Atlantic Rainforest (São Paulo, Brazil). Phytotaxa 2014, 178, 061–097. [Google Scholar] [CrossRef]
- Fiore, M.F.; Sant’Anna, C.L.; de Palva Azevedo, M.T.; Komárek, J.; Kaštovský, J.; Sulek, J.; Lorenzi, A.S. The cyanobacterial genus Brasilonema, gen. nov., a molecular and phenotypic evaluation. J Phycol 2007, 43, 789–798. [Google Scholar] [CrossRef]
- Sant’Anna, C. L.; de Palva Azevedo, T. M.; Kaštovský, J.; Komárek, J. Two form-genera of aerophytic heterocytous cyanobacteria from Brasilian rainy forest “Mata Atlântica”. Fottea 2010, 10, 217–228. [Google Scholar] [CrossRef]
- Rigonato, J.; Alvarenga, D.O.; Andreote, F.D.; Dias, A.C.F.; Melo, I.S.; Kent, A.; Fiore, M.F. Cyanobacterial diversity in the phyllosphere of a mangrove forest. FEMS Microbiol Ecol 2012, 80, 312–322. [Google Scholar] [CrossRef]
- Hentschke, G.S.; Johansen, J.R.; Pietrasiak, N.; Fiore, M.F.; Rigonato, J.; Sant’Anna, C.L.; Komárek, J. Phylogenetic placement of Dapisostemon gen. nov. and Streptostemon, two tropical heterocytous genera (Cyanobacteria). Phytotaxa 2016, 245, 129–143. [Google Scholar] [CrossRef]
- Johansen, J.R.; Mareš, J.; Pietrasiak, N.; Bohunická, M.; Zima, J.JR.; Štenclová, L.; Hauer, T. Highly divergent 16S rRNA sequences in ribosomal operons of Scytonema hyalinum (Cyanobacteria). PloS One 2017, 12, e0186393. [Google Scholar] [CrossRef] [PubMed]
- Alvarenga, D.O.; Rigonato, J.; Branco, L.H.Z.; Melo, I.S.; Fiore, M.F. Phyllonema aviceniicola gen. nov., sp. nov. and Foliisarcina bertiogensis gen. nov., sp. nov., epiphyllic cyanobacteria associated with Avicennia schaueriana leaves. Int J Syst Evol Microbiol 2016, 66, 689–700. [Google Scholar] [CrossRef]
- Alvarenga, D.O.; Andreote, A.P.D.; Branco, L.H.Z.; Fiore, M.F. Kryptousia macronema gen. nov., sp. nov. and Kryptousia microlepis sp. nov., nostocalean cyanobacteria isolated from phyllospheres. Int J Syst Evol Microbiol 2017, 67, 3301–3309. [Google Scholar] [CrossRef] [PubMed]
- Bohunická, M.; Johansen, J.R.; Villaneuva, C.D.; Mareš, M.; Štenclová, L.; Becerra-Absalòn, I.; Hauer, T.; Kaštovský, J. Revision of the pantropical genus Brasilonema (Nostocales, Cyanobacteria), with the description of 24 species new to science. Fottea 2024, 24, 137–184. [Google Scholar] [CrossRef]
- Hentschke, G.S.; De Souza Santos, K.R.; De Mattos, L.; Oliveira, F.; Vasconcelos, V.M. A journey through Cyanobacteria in Brazil: a review of novel genera and 16S rRNA sequences. Cryptogamie Algol 2024, 45, 63–75. [Google Scholar] [CrossRef]
- Rigonato, J.; Gama, W.A.; Alvarenga, D.O.; Branco, L.H.Z.; Brandini, F.P.; Genuario, D.B.; FIORE, M.F. Aliterella atlantica gen. nov., sp. nov., and Aliterella antarctica sp. nov., novel members of coccoid Cyanobacteria. Int J Syst Evol Microbiol 2016, 66, 2853–2861. [Google Scholar] [CrossRef]
- Saiki, R.K.; Gelfand, D.H.; Stoffel, S.; Scharf, S.J.; Higuchi, R.; Horn, G.T.; Mullis, K.B.; Erlich, H.A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 1988, 239, 487–491. [Google Scholar] [CrossRef]
- Marin, B.; Nowack, E.C.; Melkonian, M. A plastid in the making: evidence for a second primary endosymbiosis. Protist 2005, 156, 425–432. [Google Scholar] [CrossRef]
- Marin, B.; Melkonian, M. Molecular phylogeny and classification of the Mamiellophyceae class. nov. (Chlorophyta) based on sequence comparisons of the nuclear-and plastid-encoded rRNA operons. Protist 2010, 161, 304–336. [Google Scholar] [CrossRef]
- Maddison, WP. Mesquite: a modular system for evolutionary analysis version 2.75. 2011; http://mesquiteproject. org.
- Gouy, M.; Guindon, S.; Gascuel, O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 2010, 27, 221–224. [Google Scholar] [CrossRef] [PubMed]
- Sayers, E.W.; Cavanaugh, M.; Clark, K.; Pruitt, K.D.; Schoch, C.L.; Sherry, S.T.; Karsch-Mizrachi, I. GenBank. Nucleic Acids Res 2020, 49, D92–D96. [Google Scholar] [CrossRef] [PubMed]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J Mol Biol 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Rancel Rodríguez, N.M. Biodiversity of Epiphyllous Heterocyst-forming Cyanobacteria in the Laurel Forests of the Canary Islands. PhD Thesis, University of Cologne, Cologne, Germany.
- Struncecký, O.; Ivanova, A.P.; Mareš, J. An updated classification of cyanobacterial orders and families based on phylogenomic and polyphasic analysis. J Phycol 2023, 59, 12–51. [Google Scholar] [CrossRef]
- Rancel-Rodríguez, N.M.; Vieira, C.; Sansón, M. Terrestrial aerophytic cyanobacteria in the Canary Island laurel-forest (Laurisilva): Discovery of Brasilonema novocanariensis sp. nov. and Rhizonema melkonianarum sp. nov. from the Laurus phyllosphere. Diversity 2024, 16, 625. [Google Scholar] [CrossRef]
- Pham, H.T.; Nguyen, L.T.; Duong, T.A.; Bui, D.T.; Doan, Q.T.; Nguyen, H.T.; Mundt, S. Diversity and bioactivities of nostocacean cyanobacteria isolated from paddy soil in Vietnam. Syst Appl Microbiol 2017, 40, 470–481. [Google Scholar] [CrossRef]
- Nguyen, X.H.; Sumimoto, S.; Suda, S. Unexpected high diversity of terrestrial cyanobacteria from the campus of the University of the Ryukyus, Okinawa, Japan. Microorganisms 2017, 5, 69. [Google Scholar] [CrossRef]
- Sherwood, AR.; Carlile, A.L.; Vaccarino, M.A.; Johansen, J.R. Characterization of Hawaiian freshwater and terrestrial cyanobacteria reveals high diversity and numerous putative endemics. Phycol Res 2015, 63, 85–92. [Google Scholar] [CrossRef]
- Rigonato, J.; Kent, A.D.; Alvarenga, D.O.; Andreote, F.D.; Beirigo, R.M.; Vidal-Torrado, P.; Fiore, M.F. Drivers of cyanobacterial diversity and community composition in mangrove soils in south-east Brazil. Environ Microbiol 2013, 15, 1103–1114. [Google Scholar] [CrossRef] [PubMed]
- Sant'Anna, C.L.; Azevedo, M.P.; Fiore, M.F.; Lorenzi, A.S.; Kaštovský, J.; Komárek, J. Diversidade subgenérica de Brasilonema (Cyanobacteria, Scytonemataceae). Brazil J Bot 2011, 34, 51–62. [Google Scholar] [CrossRef]
- Hauer, T.; Bohunická, M.; Muehlsteinova, R. Calochaete gen. nov. (Cyanobacteria, Nostocales) a new cyanobacterial type from the “páramo” zone in Costa Rica. Phytotaxa 2013, 109, 36–44. [Google Scholar] [CrossRef]
- Hentschke, G.S.; Rigonato, J.; Genuário, D.B.; Laughinghouse, H.D.; Sant'Anna, C.L. Morphological and molecular characterization of Stigonema jureiensis sp. nov.(Nostocales, Cyanobacteria) from the Atlantic Rainforest, São Paulo, Brazil. Fottea 2019, 19, 185–191. [Google Scholar] [CrossRef]
- Villanueva, C.D.; Garvey, A.D.; Hašler, P.; Dvořák, P.; Poulíčková, A.; Norwich, A.R.; Casamatta, D.A. Descriptions of Brasilonema geniculatum and Calothrix dumus (Nostocales, Cyanobacteria): two new taxa isolated from cemetery tombstones. Phytotaxa 2019, 387, 1–20. [Google Scholar] [CrossRef]
- Cai, F.; Li, R. Purpureonostoc, a new name for a recently described genus of Nostoc-like cyanobacteria. Fottea 2020, 20, 111. [Google Scholar] [CrossRef]
- Mishra, D.; Saraf, A.; Kumar, N.; Pal, S.; Singh, P. Issues in cyanobacterial taxonomy: comprehensive case study of unbranched, false branched and true branched heterocytous cyanobacteria. FEMS Microbiol Lett 2021, 368, fnab005. [Google Scholar] [CrossRef]
- Kaštovský, J. Welcome to the jungle!: An overview of modern taxonomy of cyanobacteria. Hydrobiologia 2024, 851, 1063–1077. [Google Scholar] [CrossRef]
- Aguiar, R.; Fiore, M.F.; Franco, M.W.; Ventrella, M.C.; Lorenzi, A.S.; Vanetti, C.A.; Alfenas, A.C. A novel epiphytic cyanobacterial species from the genus Brasilonema causing damage to Eucalyptus leaves. J Phycol 2008, 44, 1322–1334. [Google Scholar] [CrossRef]
- Alvarenga, D.O.; Franco, M.W.; Sivonen, K.; Fiore, M.F.; Varani, A.M. Evaluating Eucalyptus leaf colonization by Brasilonema octagenarum (Cyanobacteria, Scytonemataceae) using in planta experiments and genomics. PeerJ 2020, 8, e9158. [Google Scholar] [CrossRef] [PubMed]
- Bentley, E.L.; Carpenter, E.J. Effects of desiccation and rehydration on nitrogen fixation by epiphylls in a tropical rainforest. Microb Ecol 1980, 6, 109–113. [Google Scholar] [CrossRef] [PubMed]
- Freiberg, E. Influence of microclimate on the occurrence of cyanobacteria in the phyllosphere in a premontane rain forest of Costa Rica. Plant Biol 1999, 1, 244–252. [Google Scholar] [CrossRef]
- Cleveland, C.C.; Townsend, A.R.; Schimel, D.S. Fisher, H., Howarth, R.W.; Hedin, L.O.; Perakis, S.S.; Latty, E.F.; von Fischer, J.C.; Elseroad, A.; Wasson, M.F. Global patterns of terrestrial biological nitrogen (N2) fixation in natural ecosystems. Global Biogeochem Cycl 1999, 13, 623–645. [Google Scholar] [CrossRef]
- Pons, J.; Barraclough, T.G.; Gomez-Zurita, J.; Cardoso, A.; Duran, D.P.; Hazell, S.; Kamoun, S.; Sumlin, W.D.; Vogler, A.P. Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Syst Biol 2006, 55, 595–609. [Google Scholar] [CrossRef] [PubMed]
- Alvarenga, D.O.; Clasen, L.A.; Thomsen, A.M.R.; Andersen, R.F.; Rousk, K. Light drives nitrogen fixation in tropical montane cloud forests in Costa Rica. Sci Total Environ 2024, 940, 173631. [Google Scholar] [CrossRef]
- Markham, J.; Otárola, M.F. Bryophyte and lichen biomass and nitrogen fixation in a high elevation cloud forest in Cerro de La Muerte, Costa Rica. Oecologia 2021, 195, 489–497. [Google Scholar] [CrossRef]
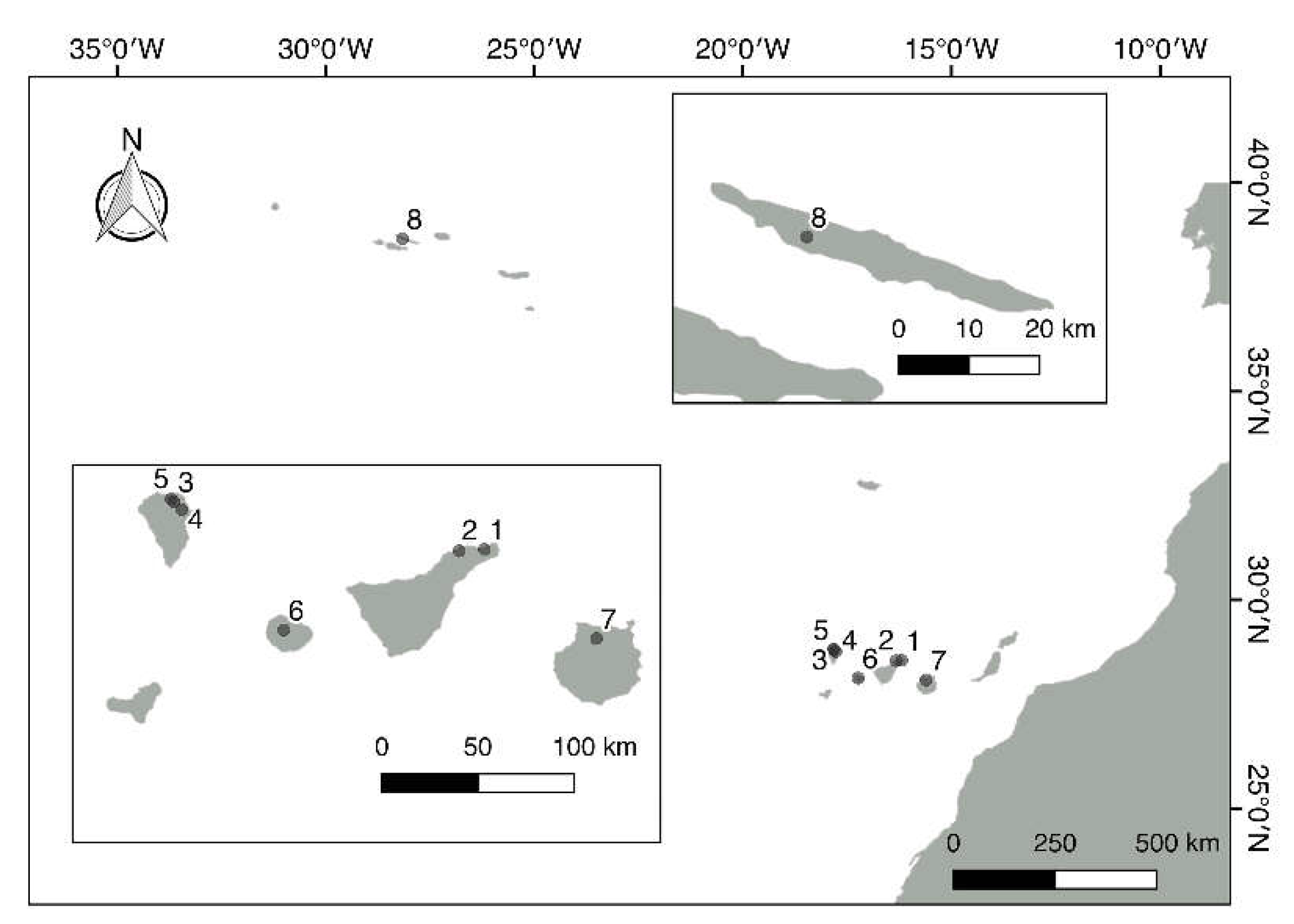

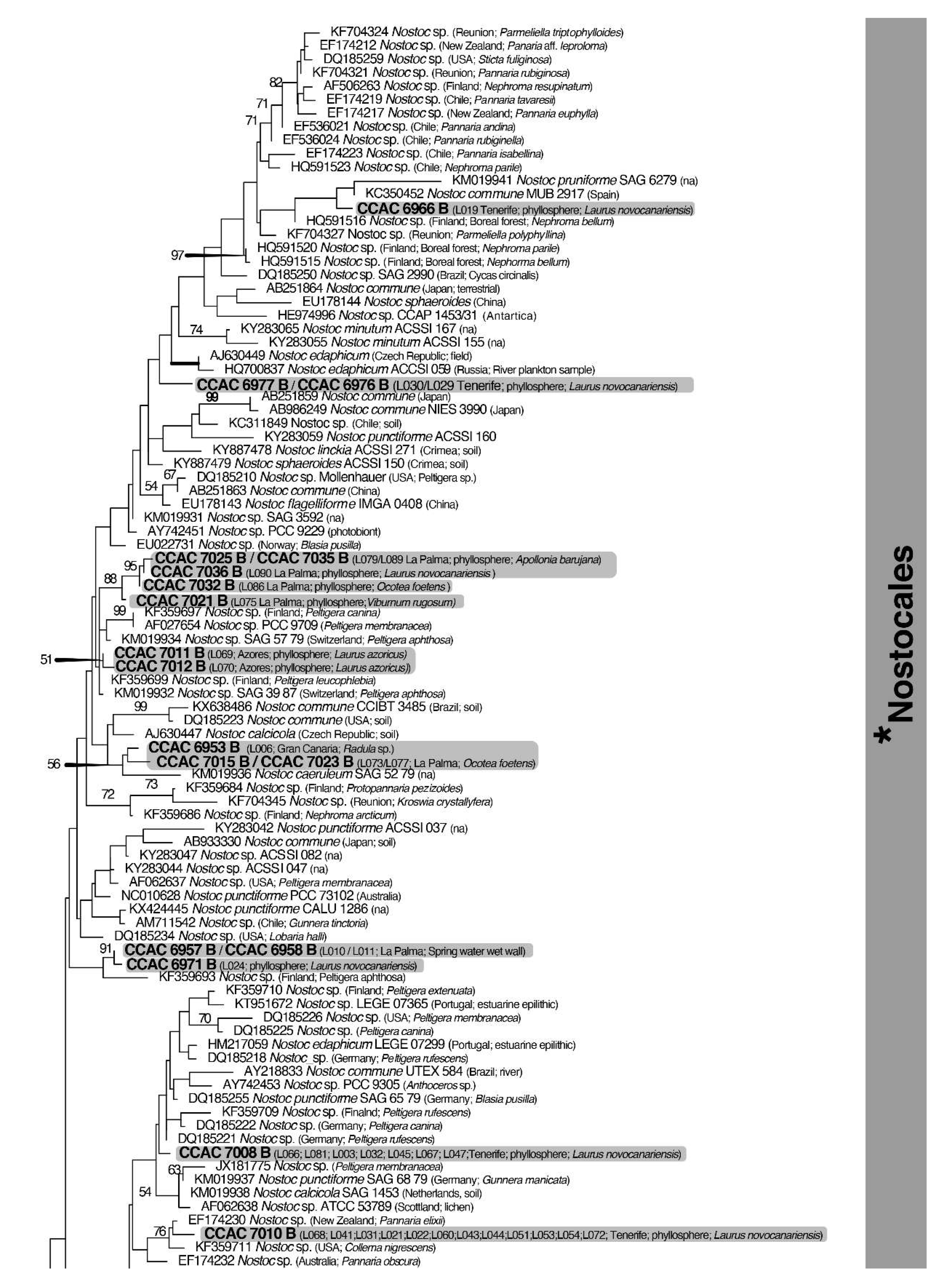
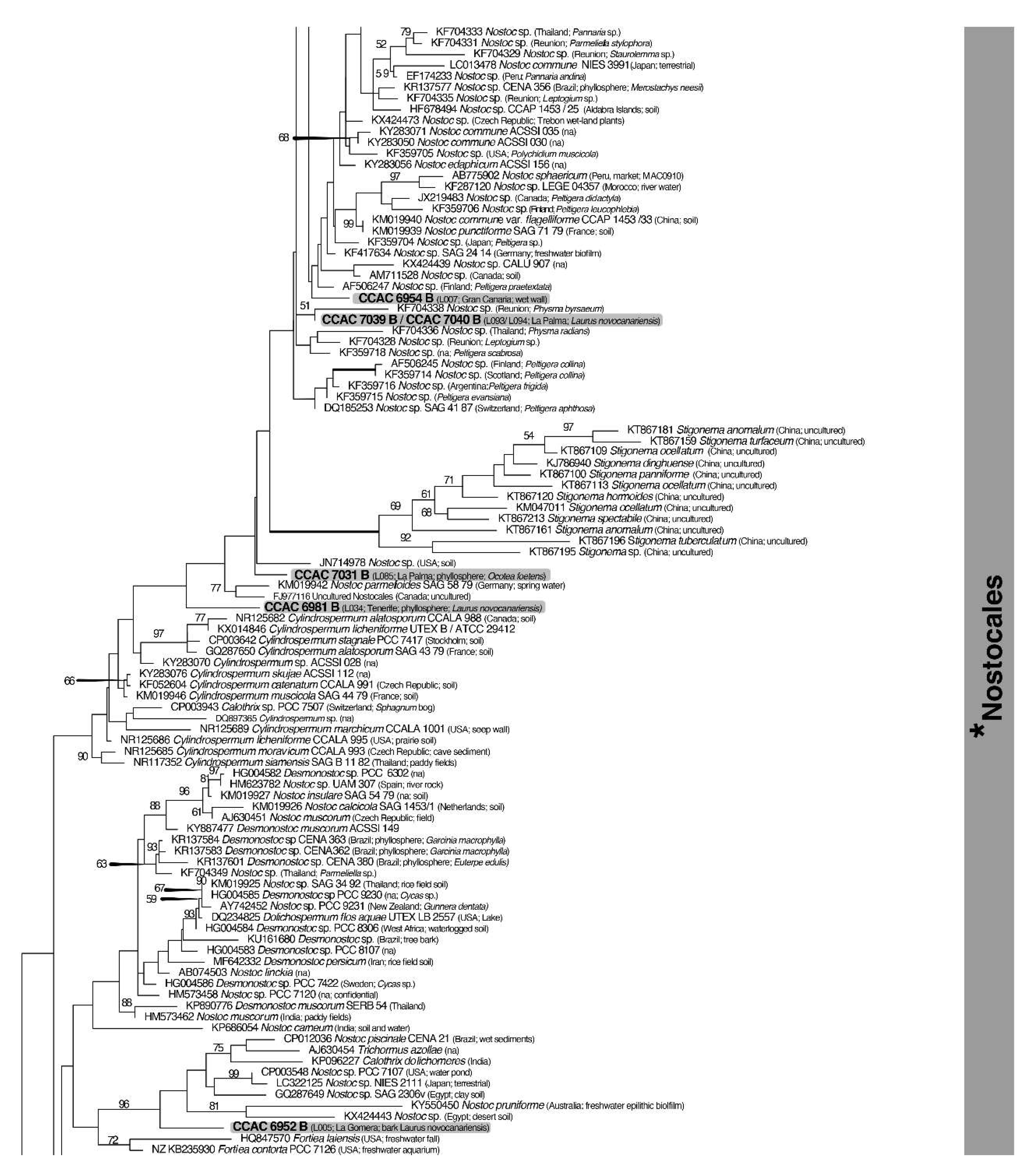

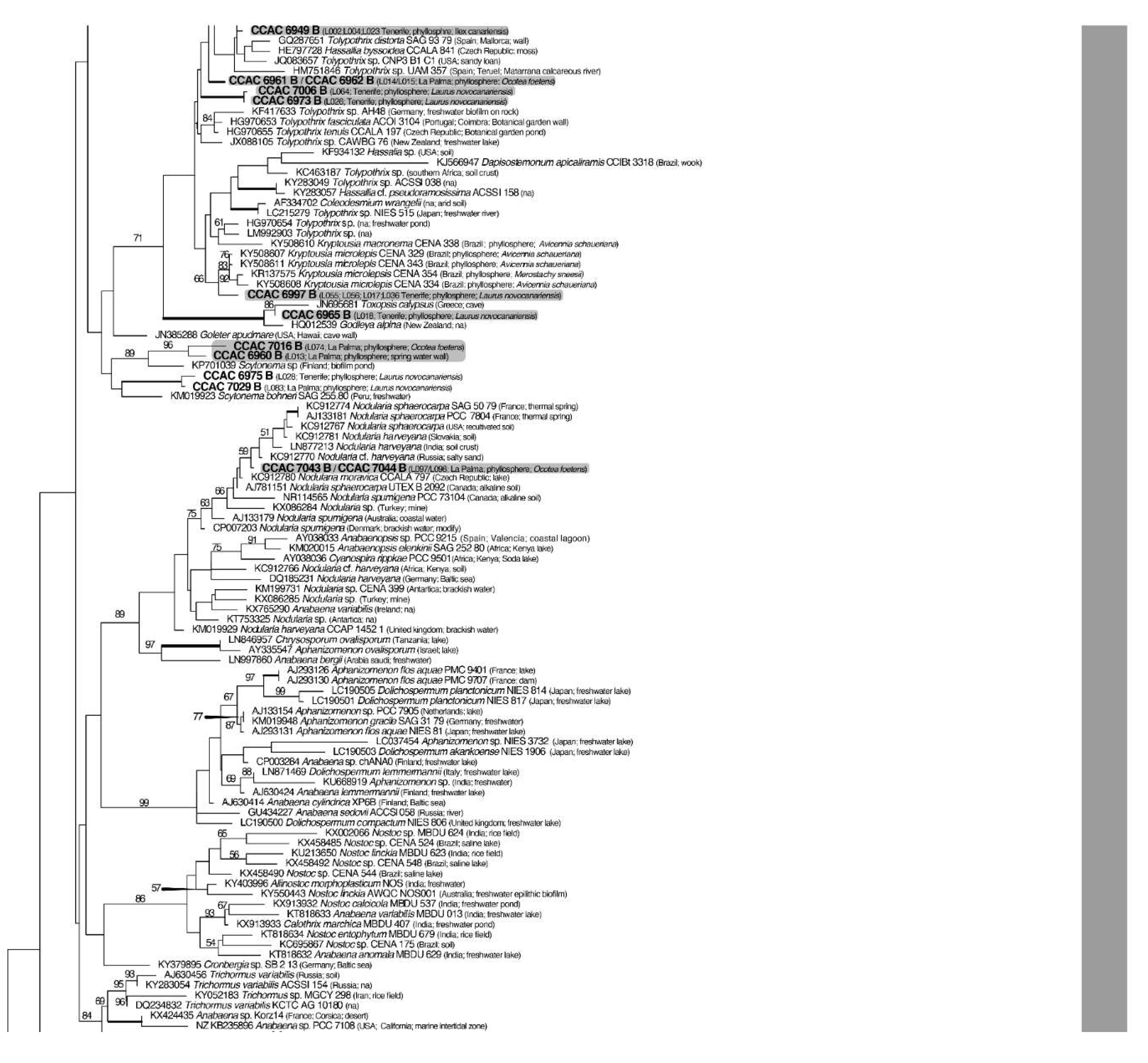
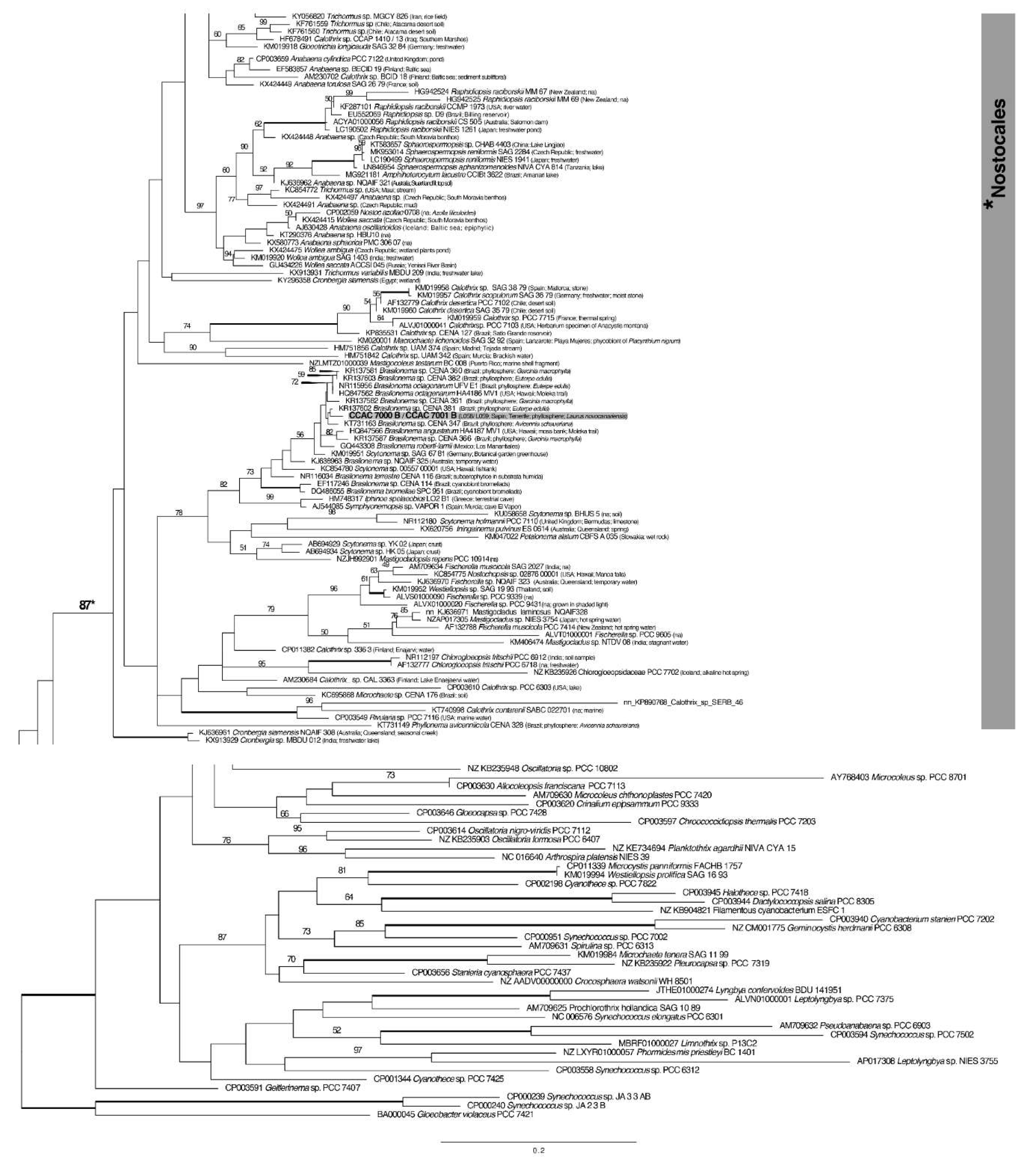
| Island | Locality | Geographic Coordinates and Altitude | Date of Sampling |
|---|---|---|---|
| Tenerife | Anaga National Park (1)* | 28º32´57´´N-16º11´09´´W 727 m |
17/10/2010 11/03/2012 |
| Pedro Álvarez forest (2)* | 28º32´28´´N-16º18´65´´W 780 m |
23/12/2012 | |
| La Palma | Los Tilos National Park (3)* | 28º48´00´´N-17º49´00´´W | 04/03/2011 |
| Cubo de la Galga (4)* | 28º45´28´´N-17º46´34´´W | 05/03/2011 | |
| Barranco del Agua (5)* | 28º48´46´´N-17º49´47´´W | 14/08/2012 | |
| La Gomera | Garajonay National Park (6)* | 28°8'3. 73" N-17°14'27. 53" O | 03/12/2010 07/12/2013 |
| Gran Canaria | Los Tilos de Moya (7)* | 28º04´53´´N-15º35´53´´W | 14/10/2010 17/11/2013 |
| Sao Jorge | Parque Natural de Sao Jorge (Azores)Reserva (8)* | 38º40´13´´ N- 28º81´54´´ W | 18/04/2012 |
| PCR-primer | Primer sequence (5´to 3´) |
|---|---|
| 16S_SG1_short_forw 16S H4_forw |
GCAGAGAGTTYGATCCTGGCTCAGG GATCCTKGCTCAGGATKAACGCTGGC |
| SG2_rev | CACGGATCCAAGGAGGTGATCCANCCNCACC |
| ptLSU C-D_rev | GCCGGCTCATTCTTCAAC |
| Sequencing-primer | Primer sequence (5´to 3´) |
| Seq_16S_H4_forw | TKGCTCAGGATKAACGCTGGC |
| Seq_16S_pos874_forw | ACTCAAAGGAATTGACG |
| Seq_16S_1040_rev | ACTTAACCCRACATCTCACGACACG |
| Seq_16S_49_rev | TACGGCTACCTTGTTACGACTTC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





