Submitted:
28 October 2024
Posted:
28 October 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Insect Rearing and Treatment Methods
2.2. Total RNA Isolation and cDNA Synthesis
2.3. Selection of Candidate References and Primer Design
2.4. Quantitative Real-Time PCR (qRT-PCR)
2.5. Assessment of Reference Gene Stability
2.6. Validation of Selected Reference Genes Under Diverse Adult Tissues
3. Results
3.1. Selection of Candidate Reference Genes
3.2. Expression Levels of Reference Genes
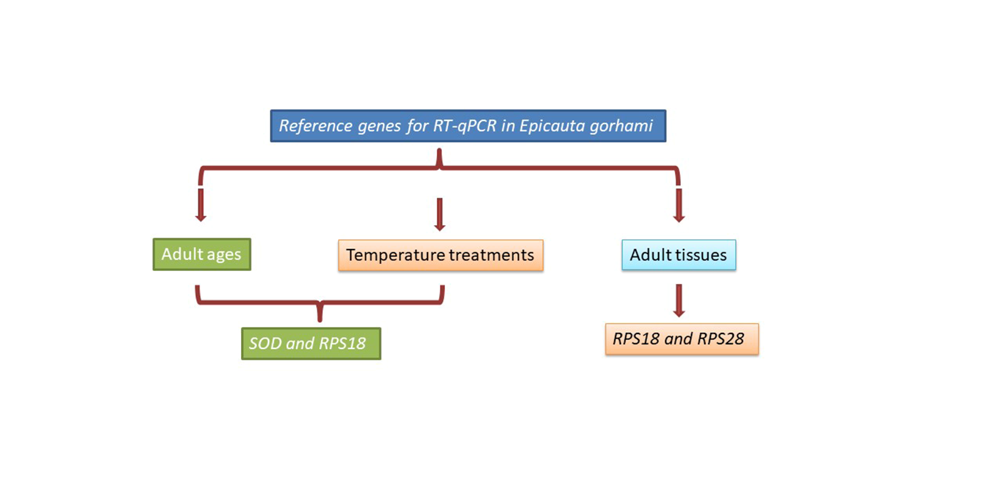
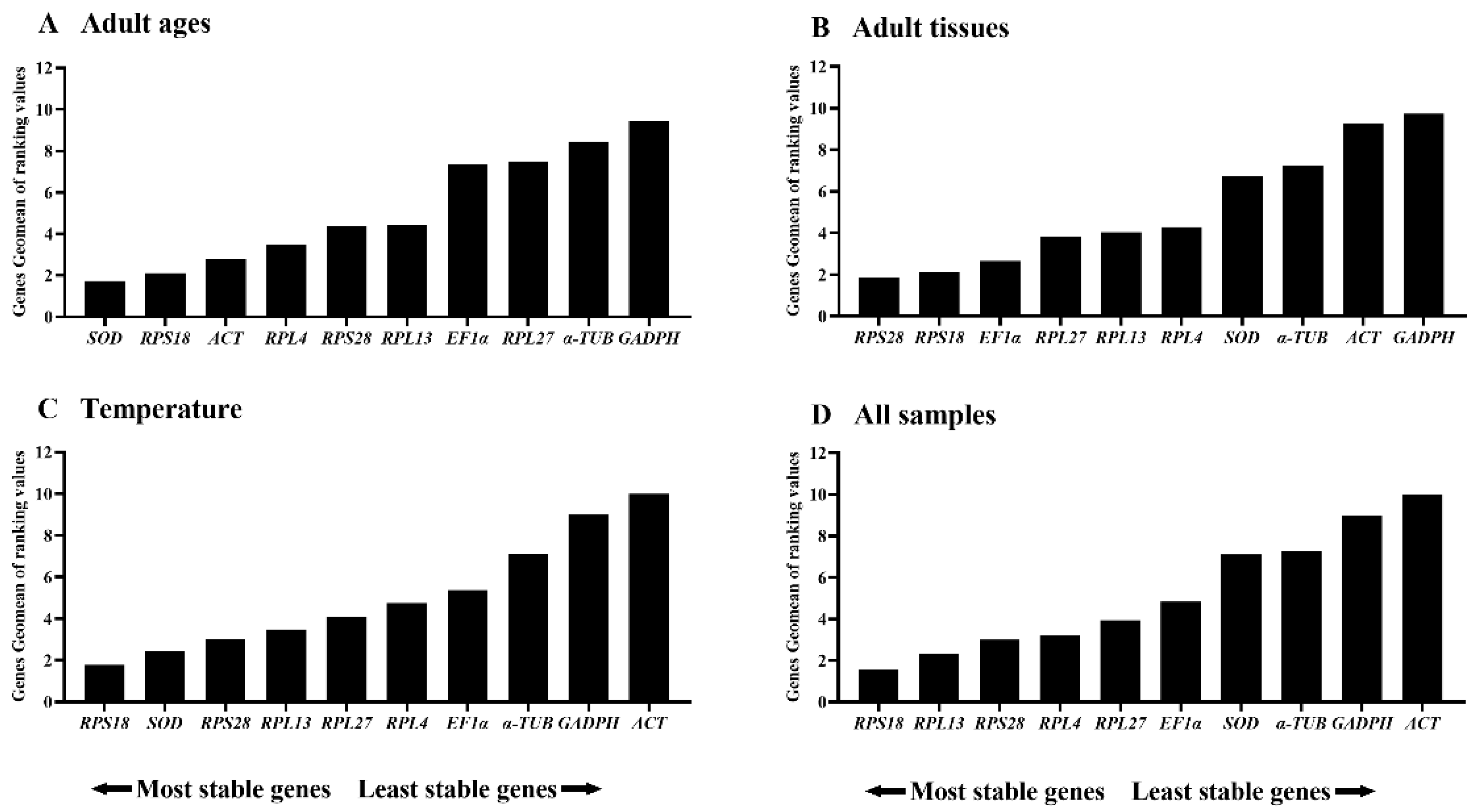
3.3. Stability of the Ten HKGs Among Adult Ages
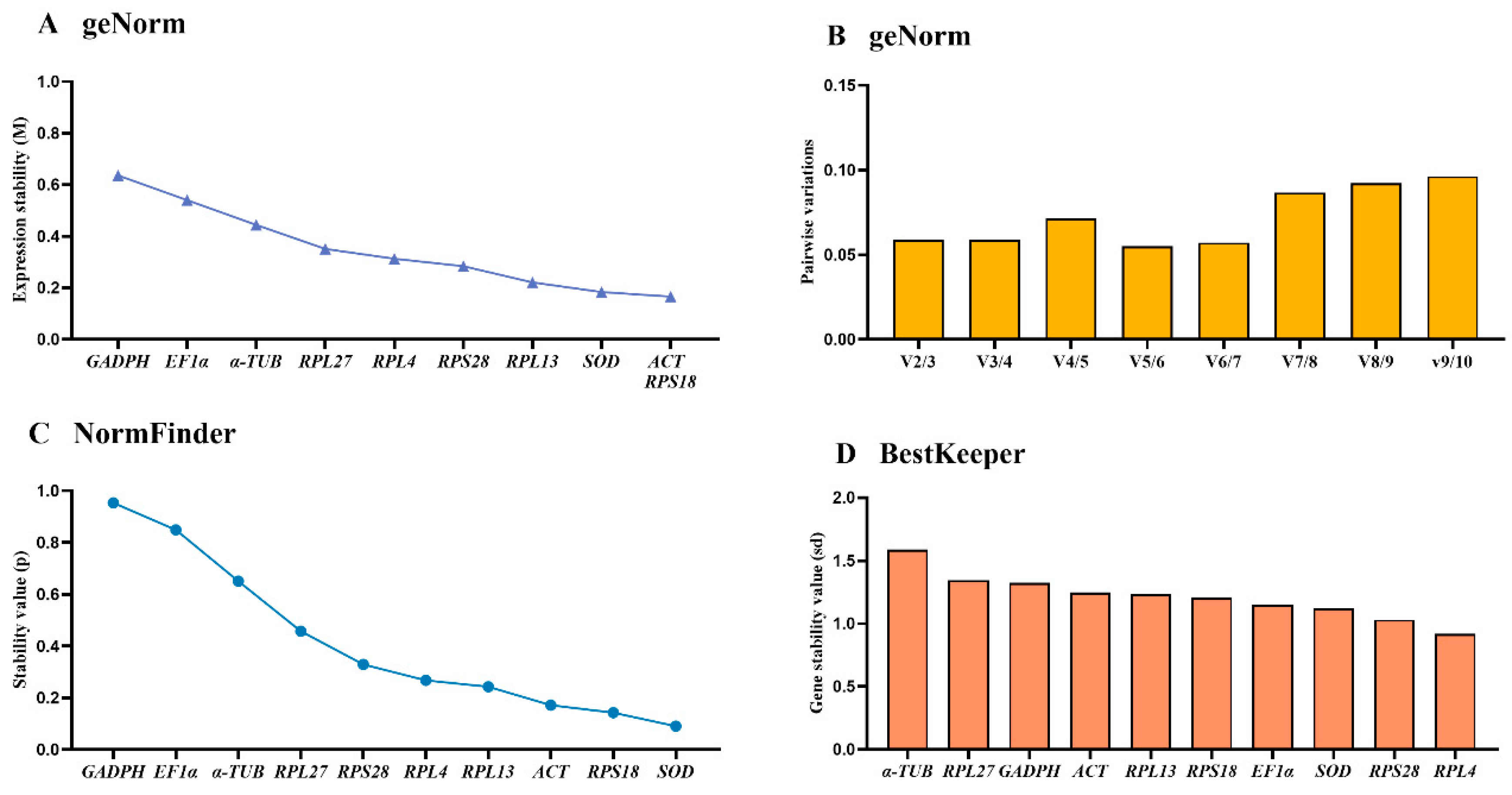
3.4. Stability of the Ten HKGs Across Various Adult Tissues
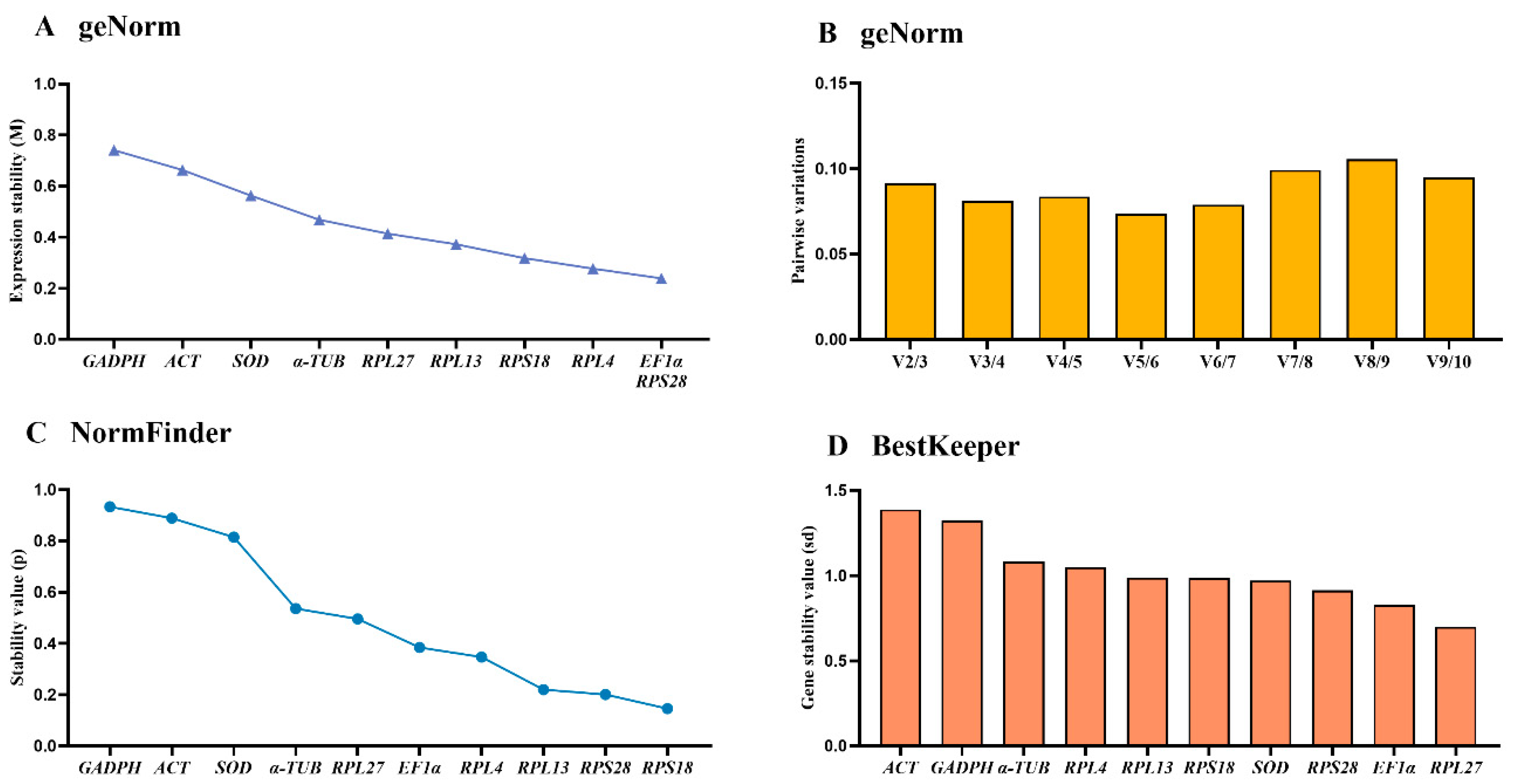
3.5. Stability of the Ten HKGs at Diverse Temperature Conditions
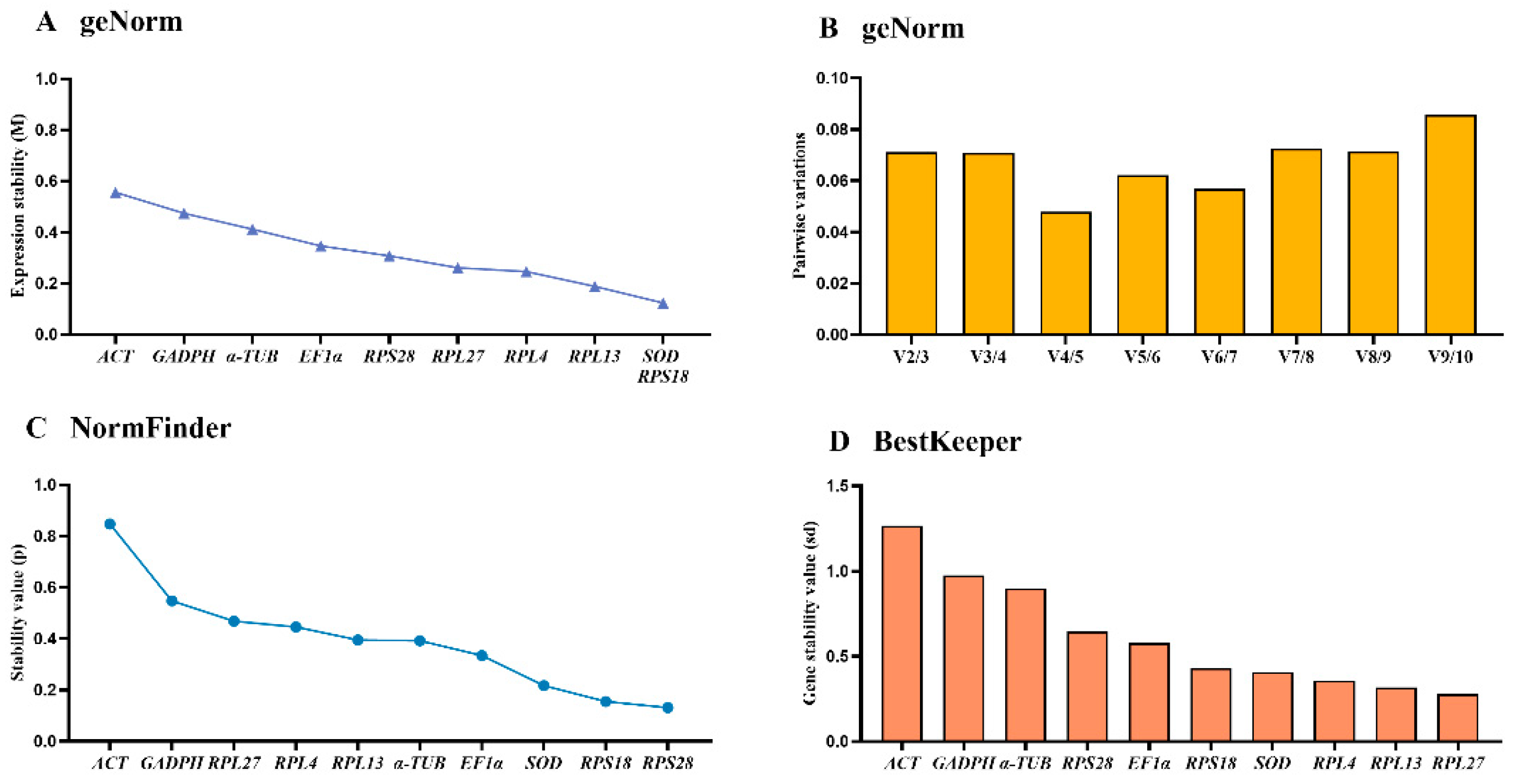
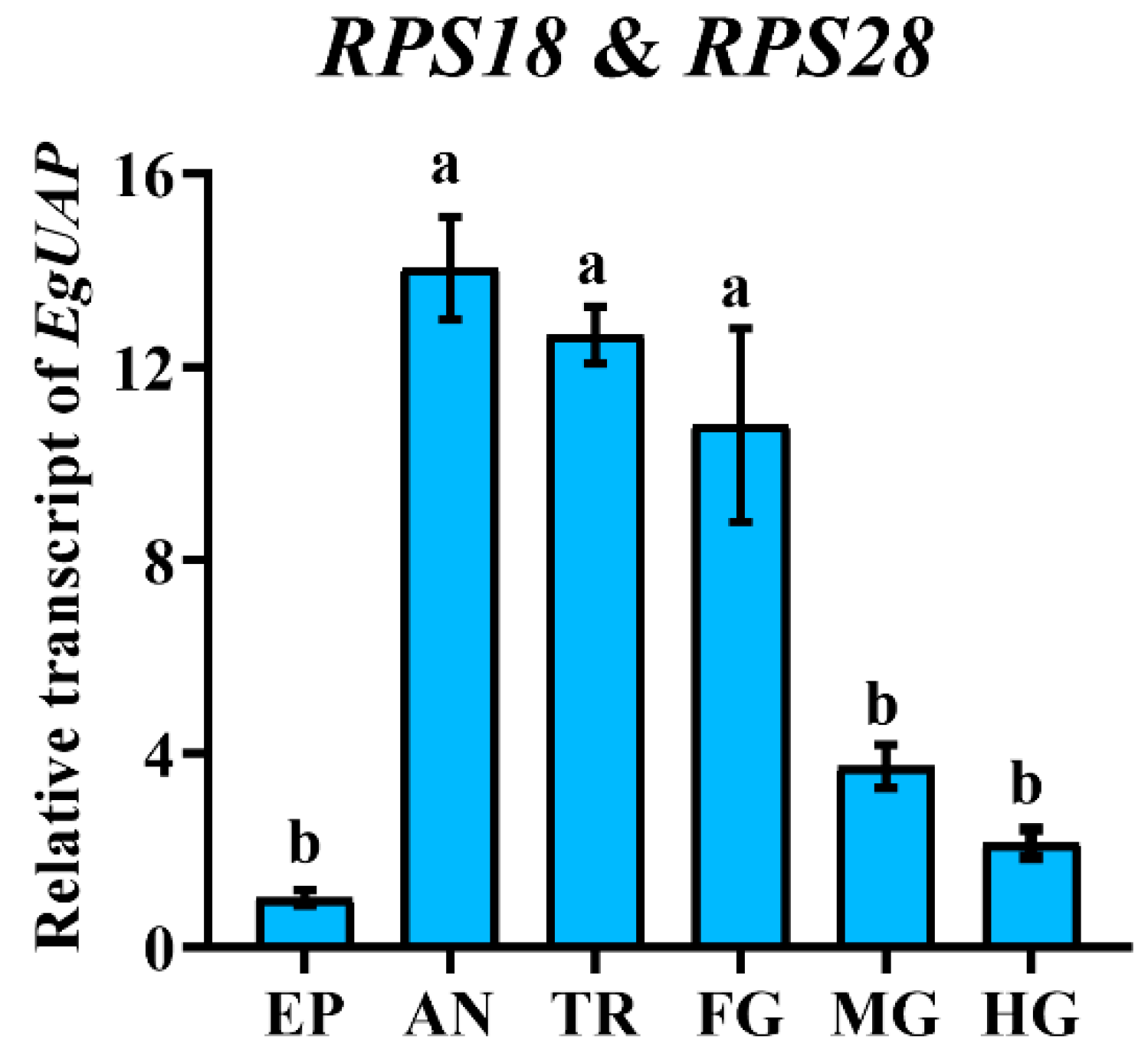
3.6. Validation of the Selected Reference Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Terao, M.; Hirose, Y.; Shintani, Y. Effects of temperature and photoperiod on termination of pseudopupal diapause in the bean blister beetle, Epicauta gorhami. J. Insect Physiol. 2012, 58, 737–742. [CrossRef]
- Shintani, Y.; Terao, M.; Tanaka, S. Adaptive significance of precocious pupation in the bean blister beetle, Epicauta gorhami (Coleoptera: Meloidae), a hypermetamorphic insect. J. Insect Physiol. 2017, 99, 107–112. [CrossRef]
- Terao, M.; Tokuda, M.; Shintani, Y. Geographic Variation in Photoperiodic Response for Induction of Pseudopupal Diapause in Epicauta gorhami (Coleoptera: Meloidae). Environ. Èntomol. 2021, 50, 1145–1150. [CrossRef]
- Zhou, Z.; Liu, Y.; Chen, X. Structural Features and Phylogenetic Implications of Three New Mitochondrial Genomes of Blister Beetles (Coleoptera: Meloidae). J. Insect Sci. 2021, 21. [CrossRef]
- Hina, A.; Razzaq, M.K.; Abbasi, A.; Shehzad, M.B.; Arshad, M.; Sanaullah, T.; Arshad, K.; Raza, G.; Ali, H.M.; Hayat, F.; et al. Genomic blueprints of soybean (Glycine max) pathogen resistance: revealing the key genes for sustainable agriculture. Funct. Plant Biol. 2024, 51. [CrossRef]
- Vianna, G.; Cunha, N.; Rech, E. Soybean seed protein storage vacuoles for expression of recombinant molecules. Curr. Opin. Plant Biol. 2023, 71, 102331. [CrossRef]
- ozefczuk, J.; Adjaye, J. Quantitative real-time PCR-based analysis of gene expression. Methods Enzymol. 2011, 500, 99-109.
- Derveaux, S.; Vandesompele, J.; Hellemans, J. How to do successful gene expression analysis using real-time PCR. Methods 2010, 50, 227–230. [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, Research0034.
- Valasek, M.A.; Repa, J.J. The power of real-time PCR. Adv. Physiol. Educ. 2005, 29, 151–159. [CrossRef]
- Shen, C.-H.; Tang, M.; Li, X.-F.; Zhu, L.; Li, W.; Deng, P.; Zhai, Q.; Wu, G.; Yan, X.-H. Evaluation of reference genes for quantitative expression analysis in Mylabris sibirica (Coleoptera, Meloidae). Front. Physiol. 2024, 15, 1345836. [CrossRef]
- Shen, C.H.; Peng, L.J.; Zhang, Y.X.; Zeng, H.R.; Yu, H.F.; Jin, L.; Li, G.Q. Reference genes for expression analyses by qRT-PCR in Phthorimaea operculella (Lepidoptera: Gelechiidae). Insects. 2022, 13, 140.
- Zhang, Y.-X.; Tan, Q.; Shen, C.-H.; Wu, J.-J.; Wu, Y.-K.; Li, W.-Z.; Jin, L.; Li, G.-Q. Reference gene selection for transcriptional profiling by RT-qPCR in the 28-spotted larger potato ladybird. J. Asia Pac. Entomol. 2022, 25, 101900.
- Wang, Z.; Shang, X.; Wei, J.; Tian, X.; Liu, Y.; Zhang, G. Evaluation and validation of reference genes for gene expression analysis using qRT-PCR in the sugarcane stem borer Chilo sacchariphagus (Lepidoptera: Pyralidae). Insects. 2024, 15, 594.
- Lü, J.; Yang, C.; Zhang, Y.; Pan, H. Selection of reference genes for the normalization of RT-qPCR data in gene expression studies in insects: A systematic review. Front. Physiol. 2018, 9, 1560.
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509-515.
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of Real-Time Quantitative Reverse Transcription-PCR Data: A Model-Based Variance Estimation Approach to Identify Genes Suited for Normalization, Applied to Bladder and Colon Cancer Data Sets. Cancer Res. 2004, 64, 5245–5250. [CrossRef]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. [CrossRef]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [CrossRef]
- Xie, F.; Wang, J.; Zhang, B. RefFinder: a web-based tool for comprehensively analyzing and identifying reference genes. Funct. Integr. Genom. 2023, 23, 1–5. [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [CrossRef]
- Landry-Voyer, A.-M.; Hassani, Z.M.; Avino, M.; Bachand, F. Ribosomal Protein uS5 and Friends: Protein–Protein Interactions Involved in Ribosome Assembly and Beyond. Biomolecules 2023, 13, 853. [CrossRef]
- Shi, X.-Q.; Guo, W.-C.; Wan, P.-J.; Zhou, L.-T.; Ren, X.-L.; Ahmat, T.; Fu, K.-Y.; Li, G.-Q. Validation of reference genes for expression analysis by quantitative real-time PCR in Leptinotarsa decemlineata (Say). BMC Res. Notes 2013, 6, 93–10. [CrossRef]
- Ma, L.; Jiang, T.; Liu, X.; Xiao, H.; Peng, Y.; Zhang, W. Evaluation of candidate reference genes for gene expression analysis in the brassica leaf beetle, Phaedon brassicae (Coleoptera: Chrysomelidae). PLOS ONE 2021, 16, e0251920. [CrossRef]
- Toutges, M.J.; Hartzer, K.; Lord, J.; Oppert, B. Evaluation of Reference Genes for Quantitative Polymerase Chain Reaction across Life Cycle Stages and Tissue Types of Tribolium castaneum. J. Agric. Food Chem. 2010, 58, 8948–8951. [CrossRef]
- Sellamuthu, G.; Amin, S.; Bílý, J.; Synek, J.; Modlinger, R.; Sen, M.K.; Chakraborty, A.; Roy, A. Reference Gene Selection for Normalizing Gene Expression in Ips Sexdentatus (Coleoptera: Curculionidae: Scolytinae) Under Different Experimental Conditions. Front. Physiol. 2021, 12. [CrossRef]
- Yuan, F.; Xie, Z.; Li, Z.; Lian, P.; Wei, C. Screening of reference genes for gene expression study in different tissues from the transcriptome data of the vector leafhopper Psammotettix striatus. Gene 2024, 927, 148696. [CrossRef]
- Li, M.; Li, X.; Wang, C.; Li, Q.; Zhu, S.; Zhang, Y.; Li, X.; Yang, F.; Zhu, X. Selection and validation of reference genes for qRT-PCR analysis of Rhopalosiphum padi (Hemiptera: Aphididae). Front. Physiol. 2021, 12, 663338.
- Bansal, R.; Mamidala, P.; Mian, M.A.R.; Mittapalli, O.; Michel, A.P. Validation of Reference Genes for Gene Expression Studies in Aphis glycines (Hemiptera: Aphididae). J. Econ. Èntomol. 2012, 105, 1432–1438. [CrossRef]
- Bansal, R.; Haviland, D.R.; Hunter, W.B. Selection and validation of reference genes for quantifying gene expression in the Gill's mealybug. J. Econ. Entomol. 2023, 116, 2166-2172.
- Bassan, M.M.; Angelotti-Mendonça, J.; Alves, G.R.; Yamamoto, P.T.; Filho, F.d.A.A.M. Selection of Reference Genes for Expression Studies in Diaphorina citri (Hemiptera: Liviidae). J. Econ. Èntomol. 2017, 110, 2623–2629. [CrossRef]
- Pinheiro, D.H.; Moreira, R.O.; Leite, N.A.; Redoan, A.C.; Xavier, A.D.S.; Barros, B.A.; Carneiro, N.P. Suitable reference genes for RT-qPCR analysis in Dichelops melacanthus (Hemiptera: Pentatomidae). Mol. Biol. Rep. 2020, 47, 4989-5000.
- Liu, Z.; Xiao, J.; Xia, Y.; Wu, Q.; Zhao, C.; Li, D. Selection and validation of reference genes for RT-qPCR-based analyses of Anastatus japonicus Ashmead (Hymenoptera: Helicopteridae). Front. Physiol. 2022, 13, 1046204.
- Wang, L.; Yang, C.; Liu, Q.; Zhang, X.; Mei, X.; Zhang, T.; Ning, J. Validation and Evaluation of Reference Genes for Quantitative Real-Time PCR Analysis in Mythimna loreyi (Lepidoptera: Noctuidae). Insects 2024, 15, 185. [CrossRef]
- Fu, W.; Xie, W.; Zhang, Z.; Wang, S.; Wu, Q.; Liu, Y.; Zhou, X.; Zhou, X.; Zhang, Y. Exploring Valid Reference Genes for Quantitative Real-time PCR Analysis in Plutella xylostella (Lepidoptera: Plutellidae). Int. J. Biol. Sci. 2013, 9, 792–802. [CrossRef]
- Wu, S.; Luo, Y.; Zeng, Z.; Yu, Y.; Zhang, S.; Hu, Y.; Chen, L. Determination of internal controls for quantitative gene expression of Spodoptera litura under microbial pesticide stress. Sci. Rep. 2024, 14, 1–13. [CrossRef]
- Dalai, M.; Jagota, A. Identification of specific reference gene for normalization of RT-qPCR data in rhythmic gene expression studies of the effect of developmental hormone antagonist in postembryonic development in Bombyx mori. Front. Insect Sci. 2024, 4, 1362473.
- Zhang, S.; An, S.; Li, Z.; Wu, F.; Yang, Q.; Liu, Y.; Cao, J.; Zhang, H.; Zhang, Q.; Liu, X. Identification and validation of reference genes for normalization of gene expression analysis using qRT-PCR in Helicoverpa armigera (Lepidoptera: Noctuidae). Gene. 2015, 555, 393-402.
- Shen, X.X.; Zhang, G.Q.; Zhao, Y.X.; Zhu, X.X.; Yu, X.F.; Yang, M.F.; Zhang, F. Selection and validation of optimal reference genes for RT-qPCR analyses in Aphidoletes aphidimyza Rondani (Diptera: Cecidomyiidae). Front. Physiol. 2023, 14, 1277942.
- Yang, J.; Jiang, Z.; Xu, Q.; Liu, X.; Dai, M.; Li, B.; Wei, J. Evaluation of suitable reference genes for expression analysis using quantitative real-time polymerase chain reaction in the parasitoid Exorista sorbillans (Diptera: Tachinidae). Arch. Insect Biochem. Physiol. 2023, 113, e22009. [CrossRef]
- Tian, P.; Qiu, L.; Zhou, A.; Chen, G.; He, H.; Ding, W.; Li, Y. Evaluation of Appropriate Reference Genes For Investigating Gene Expression in Chlorops oryzae (Diptera: Chloropidae). J. Econ. Èntomol. 2019, 112, 2207–2214. [CrossRef]
- Yang, Q.; Li, Z.; Cao, J.; Zhang, S.; Zhang, H.; Wu, X.; Zhang, Q.; Liu, X. Selection and Assessment of Reference Genes for Quantitative PCR Normalization in Migratory Locust Locusta migratoria (Orthoptera: Acrididae). PLOS ONE 2014, 9, e98164. [CrossRef]
- Hou, Q.; Yuan, L.; Jin, H.; Yan, H.; Li, F.; Wu, S. Identification and validation of reference genes for normalization of gene expression analysis using qRT-PCR in Megalurothrips usitatus (thysanoptera: thripidae). Front. Physiol. 2023, 14, 1161680.
- Zheng, Y.T.; Li, H.B.; Lu, M.X.; Du, Y.Z. Evaluation and validation of reference genes for qRT-PCR normalization in Frankliniella occidentalis (Thysanoptera: Thripidae). PLoS One. 2014, 9, e111369.
- Yamamoto, K.; Yamaguchi, M. Characterization of a novel superoxide dismutase in Nilaparvata lugens. Arch. Insect Biochem. Physiol. 2021, 109, e21862. [CrossRef]
- Han, S.; Qin, Q.; Wang, D.; Zhou, Y.; He, Y. Selection and evaluation of reference genes for qRT-PCR in Spodoptera frugiperda (Lepidoptera: Noctuidae). Insects. 2021, 12, 902.
- Singh, S.; Gupta, M.; Pandher, S.; Kaur, G.; Goel, N.; Rathore, P.; Palli, S.R. RNA sequencing, selection of reference genes and demonstration of feeding RNAi in Thrips tabaci (Lind.) (Thysanoptera: Thripidae). BMC Mol. Biol. 2019, 20, 1–21. [CrossRef]
- Wang, L.; Liu, Q.; Guo, P.; Gao, Z.; Chen, D.; Zhang, T.; Ning, J. Evaluation of Reference Genes for Quantitative Real-Time PCR Analysis in the Bean Bug, Riptortus pedestris (Hemiptera: Alydidae). Insects 2023, 14, 960. [CrossRef]
- Zhu, X.; Yuan, M.; Shakeel, M.; Zhang, Y.; Wang, S.; Wang, X.; Zhan, S.; Kang, T.; Li, J. Selection and evaluation of reference genes for expression analysis using qRT-PCR in the beet armyworm Spodoptera exigua (Hübner) (Lepidoptera: Noctuidae). PLoS One. 2014, 9, e84730.
- Zhang, Y.; Chen, J.; Chen, G.; Ma, C.; Chen, H.; Gao, X.; Tian, Z.; Cui, S.; Tian, Z.; Guo, J.; et al. Identification and Validation of Reference Genes for Quantitative Gene Expression Analysis in Ophraella communa. Front. Physiol. 2020, 11, 355. [CrossRef]
- Yang, C.; Preisser, E.L.; Zhang, H.; Liu, Y.; Dai, L.; Pan, H.; Zhou, X. Selection of reference genes for RT-qPCR analysis in Coccinella septempunctata to assess un-intended effects of RNAi transgenic plants. Front. Plant. Sci. 2016, 7, 1672.
- Tan, Q.-Q.; Zhu, L.; Li, Y.; Liu, W.; Ma, W.-H.; Lei, C.-L.; Wang, X.-P. A De Novo Transcriptome and Valid Reference Genes for Quantitative Real-Time PCR in Colaphellus bowringi. PLOS ONE 2015, 10, e0118693–e0118693. [CrossRef]

| Gene | Primer sequences (5'to 3´) | Length (bp) | Slope | R2 | Efficiency (%) |
|---|---|---|---|---|---|
| EF1α | F- ATCATTGACGCACCTGGACA | 99 | -3.435 | 0.999 | 95.50 |
| R- ACCAGTACCAGCAGCAACAA | |||||
| GAPDH | F-ACAGTACATGCCACCACAGC | 180 | -3.351 | 0.999 | 98.82 |
| R-TGGTACACGGAAGGCCATAC | |||||
| RPL4 | F- TCGATGAACCACCGTCAACC | 80 | -3.308 | 0.993 | 100.60 |
| R- CGACCGGTACCCCATGATTC | |||||
| RPL13 | F-AAGCCGCCGGTATTAACAGC | 172 | -3.318 | 0.998 | 100.16 |
| R-TCACCAGGACGCAACTTCTT | |||||
| RPL27 | F-TCGTATTGGTCTTGGCAGGC | 112 | -3.373 | 0.999 | 98.03 |
| R-TCAATGCCGGCAACAATAGC | |||||
| SOD | F-AGTTGTCCATGCTGATCCGG | 95 | -3.32 | 0.998 | 100.08 |
| R-TAACACCACAGGCCAAACGT | |||||
| ACT | F- TACGTGTGGCACCTGAAGAA | 169 | -3.284 | 0.997 | 101.60 |
| R- CCAGTTGTACGACCAGAAGCA | |||||
| α-TUB | F-ATGGGCACGTCTTGATCACA | 174 | -3.258 | 0.999 | 102.73 |
| R-TCATTTTCACCTTCGCCCGA | |||||
| RPS18 | F-AGGTGTTGGTCGTCGTTACTC | 210 | -3.323 | 0.999 | 99.96 |
| R-TCTAAGGTAGCCGATGTGAGC | |||||
| RPS28 | F-GGTGAGCAAAACCGTCAGATC | 90 | -3.347 | 0.997 | 98.97 |
| R-TGCTTCACGTTCAGACTCCA |
| Conditions | CRGs* | geNorm | Normfinder | Normfinder | ΔCt | ||||
|---|---|---|---|---|---|---|---|---|---|
| Stability | Rank | Stability | Rank | Stability | Rank | Stability | Rank | ||
| Developmental ages | EF1α | 0.540 | 8 | 0.849 | 9 | 1.149 | 4 | 0.931 | 9 |
| GADPH | 0.636 | 9 | 0.954 | 10 | 1.325 | 8 | 1.024 | 10 | |
| RPL4 | 0.313 | 5 | 0.268 | 5 | 0.920 | 1 | 0.543 | 5 | |
| RPL13 | 0.221 | 3 | 0.243 | 4 | 1.236 | 6 | 0.500 | 4 | |
| RPL27 | 0.351 | 6 | 0.458 | 7 | 1.344 | 9 | 0.619 | 7 | |
| SOD | 0.184 | 2 | 0.091 | 1 | 1.121 | 3 | 0.443 | 1 | |
| ACT | 0.166 | 1 | 0.172 | 3 | 1.249 | 7 | 0.469 | 3 | |
| α-TUB | 0.444 | 7 | 0.651 | 8 | 1.589 | 10 | 0.782 | 8 | |
| RPS18 | 0.166 | 1 | 0.143 | 2 | 1.209 | 5 | 0.476 | 2 | |
| RPS28 | 0.284 | 4 | 0.329 | 6 | 1.033 | 2 | 0.557 | 6 | |
| Adult tissues | EF1α | 0.239 | 1 | 0.384 | 5 | 0.828 | 2 | 0.621 | 5 |
| GADPH | 0.741 | 9 | 0.933 | 10 | 1.326 | 9 | 1.055 | 10 | |
| RPL4 | 0.277 | 2 | 0.347 | 4 | 1.051 | 7 | 0.614 | 4 | |
| RPL13 | 0.372 | 4 | 0.220 | 3 | 0.989 | 6 | 0.608 | 3 | |
| RPL27 | 0.414 | 5 | 0.496 | 6 | 0.700 | 1 | 0.710 | 6 | |
| SOD | 0.563 | 7 | 0.815 | 8 | 0.971 | 4 | 0.944 | 8 | |
| ACT | 0.663 | 8 | 0.889 | 9 | 1.389 | 10 | 1.012 | 9 | |
| α-TUB | 0.468 | 6 | 0.536 | 7 | 1.085 | 8 | 0.744 | 7 | |
| RPS18 | 0.318 | 3 | 0.146 | 1 | 0.987 | 5 | 0.549 | 1 | |
| RPS28 | 0.239 | 1 | 0.201 | 2 | 0.914 | 3 | 0.552 | 2 | |
| Temparature treatment | EF1α | 0.346 | 6 | 0.334 | 4 | 0.58 | 6 | 0.532 | 5 |
| GADPH | 0.474 | 8 | 0.547 | 9 | 0.976 | 9 | 0.675 | 9 | |
| RPL4 | 0.246 | 3 | 0.446 | 7 | 0.357 | 3 | 0.549 | 6 | |
| RPL13 | 0.188 | 2 | 0.395 | 6 | 0.317 | 2 | 0.522 | 4 | |
| RPL27 | 0.261 | 4 | 0.468 | 8 | 0.278 | 1 | 0.555 | 7 | |
| SOD | 0.123 | 1 | 0.217 | 3 | 0.407 | 4 | 0.431 | 3 | |
| ACT | 0.556 | 9 | 0.847 | 10 | 1.265 | 10 | 0.886 | 10 | |
| α-TUB | 0.412 | 7 | 0.392 | 5 | 0.899 | 8 | 0.570 | 8 | |
| RPS18 | 0.123 | 1 | 0.155 | 2 | 0.429 | 5 | 0.415 | 1 | |
| RPS28 | 0.307 | 5 | 0.131 | 1 | 0.646 | 7 | 0.430 | 2 | |
| Experimental conditions | The recommended reference genes | |
|---|---|---|
| Adult ages | SOD | RPS18 |
| Adult tissues | RPS28 | RPS18 |
| Temperature | SOD | RPS18 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





