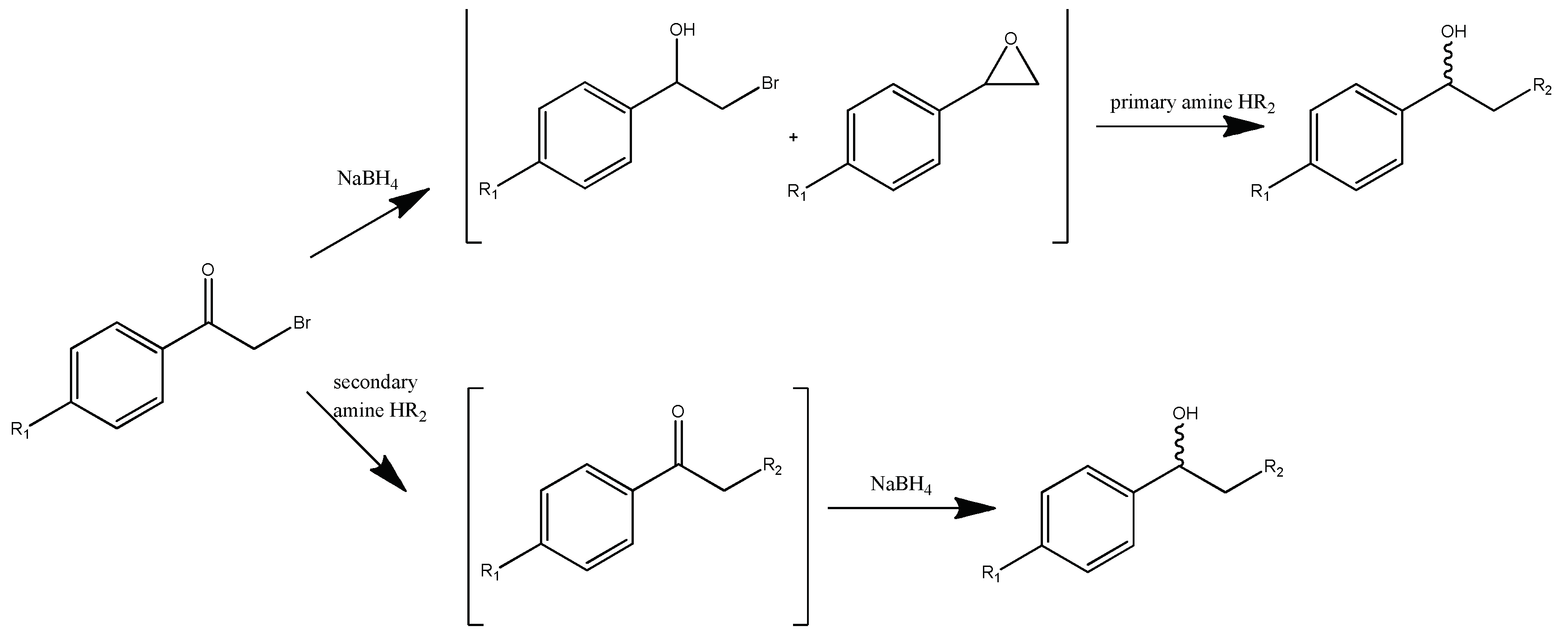
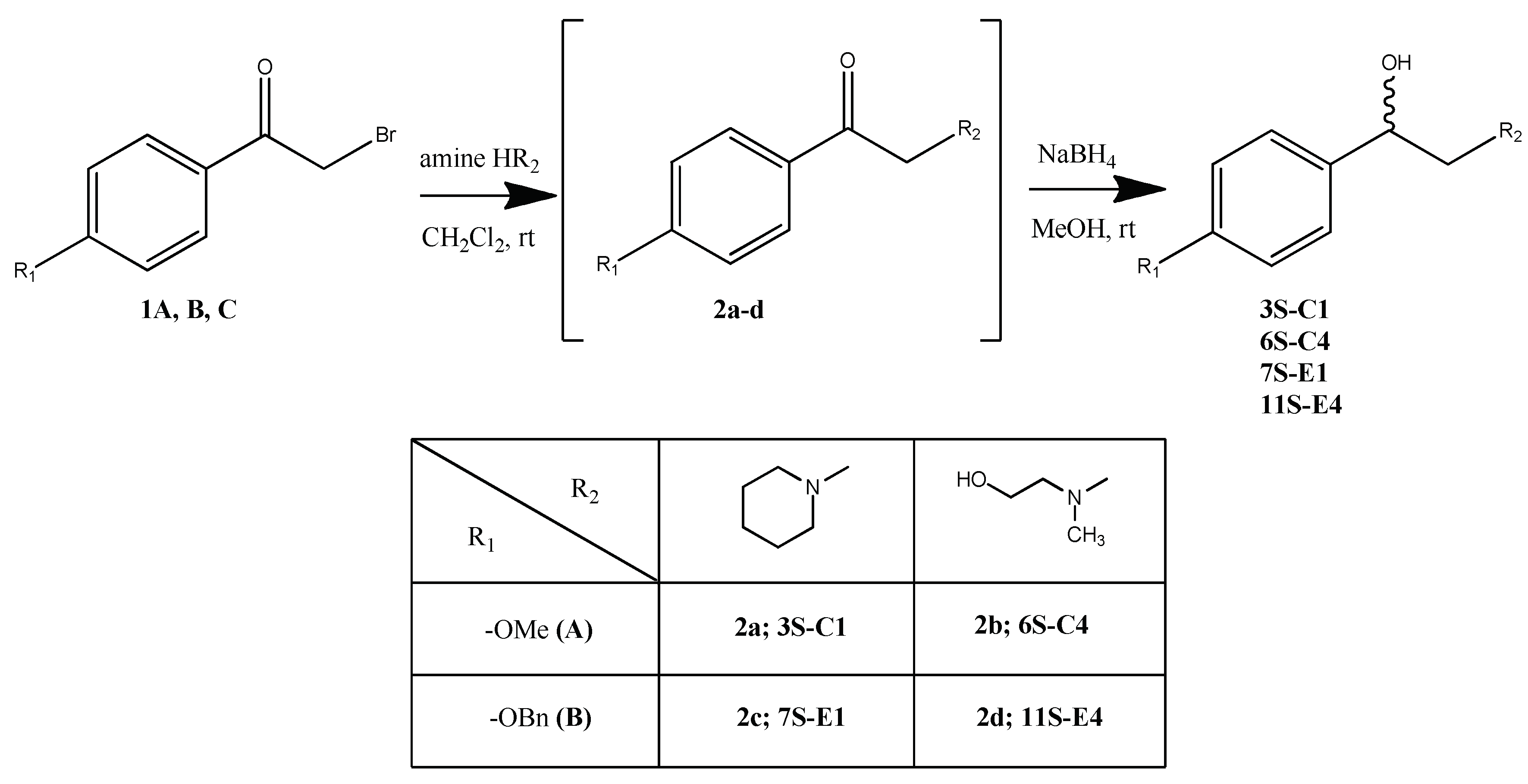
2.1.2. General Procedure for Compounds 3S-C1, 6S-C4, 7S-E1, 11S-E4 Synthesis
A solution of bromoacetophenone 1 in methylene chloride (1 ml of solvent per 100 mg 1) were added drop by drop to a solution of amine in methylene chloride (10 ml of solvent per 1 ml of amine). The reaction was controlled by the initial bromoacetophenone 1 conversion by TLC. After bromoacetophenone 1 conversion was complete, volatile components were evaporated. The residue was dissolved in 5 ml of water and extracted with methylene chloride (3 x 10 ml). The organic layers are combined and dried with anhydrous calcium chloride, filtered off and evaporated. The resulting oil was dissolved in 10 ml of methanol, after which NaBH4 was added in portions during cooling. After intermediate conversion (control by TLC), the reaction mixture was acidified with 10% aqueous HCl solution to pH 3, the volatile components was evaporated. The extraction and drying process was repeated. The products 3S-C1, 6S-C4, 7S-E1, 11S-E4 were isolated by column chromatography (methanol gradient from 0 to 15%).
1-(4-methoxyphenyl)-2-piperedine-1-yl ethanol (3S-C1)
From 0.63 ml (6.30 mmol) piperedine, 0.42 g (1.80 mmol) of 4-methoxybromoacetophenone 1А, 0.07 g (1.80 mmol) NaBH4 the product 3S-C1 0.31 g (74%) was obtained. Rf = 0.45 (10% methanol in chloroform). The content of the target substance according to HPLC data is 97.5%. 1H NMR spectrum (CDCl3) δ: 7.31-7.28 (m, 2H, Ph); 6.83-6.80 (m, 2H, Ph); 5.33-5.30 (m, 1H, -CH); 4.60 (br. s, 1H, -OH); 3.76-3.69 (m, 2H,-CH2-OH); 3.74 (s, 1H, -O-CH3); 3.19-2.99 (m, 2H, piperedine -CH2-); 2.83-2.76 (m, 2H, piperedine -CH2-); 2.29-2.16 (m, 2H, piperedine -CH2-); 1.83-1.77 (m, 3H, piperedine 2×-CH2-); 1.47-1.37 (m, 1H, piperedine -CH2-). 13C NMR spectrum (CDCl3) δ: 159.42; 132.18; 127.15; 114.03; 67.43; 65.52; 55.51; 55.26; 54.04; 22.72; 22.67; 21.80. For C14H21NO2 [M+H]+ calculated: 236.1650, found: 236.1652.
2-(2-hydroxyethyl)(methyl) amino)-1-(4-methoxyphenyl)ethanol (6S-C4)
From 0.55 ml (7.00 mmol) N-methyl ethanolamine, 0.45 g (2.00 mmol) of 4-methoxybromoacetophenone 1А, 0.08 g (2.00 mmol) NaBH4 the product 6S-C4 0.20 g (45%) was obtained. Rf = 0.75 (5% methanol in chloroform). The content of the target substance according to HPLC data is 99%. 1H NMR spectrum (CDCl3) δ: 7.28-7.25 (m, 2H, Ph); 6.87-6.84 (m, 2H, Ph); 4.53-4.49 (m, 1H, -CH-); 3.99-3.84 (m, 2H, -CH2-OH); 3.77 (s, 3H,-O-CH3); 2.88-2.72 (m, 2H, -N(CH3)-CH2-); 2.32 (s, 3H, -N-CH3); 2.28-2.06 (m, 2H, -CH(OH)-CH2-). 13C NMR-spectrum (CDCl3) δ: 159.11; 132.16; 127.34; 113.61; 77.48; 66.74; 61.86; 55.15; 54.46; 45.92. For C12H19NO3 [M+H]+ calculated: 226.1443, found: 226.1445.
1-(4-(benzyloxy) phenyl)-2-piperidine-1-yl-ethanol (7S-E1)
From 0.63 ml (6.35 mmol) piperidine, 0.39 g (1.27 mmol) 4-benzyloxybromoacetophenone 1В and 0.05 g (1.27 mmol) NaBH4 the product 7S-E1 0.30 g (76%) was obtained. Rf = 0.45 (5% methanol in chloroform). The content of the target substance according to HPLC data is 98%. 1H NMR spectrum (CDCl3) δ: 7.43-7.27 (m, 7H, Ph); 6.96-6.93 (m, 2H, Ph); 5.04 (s, 2H, -CH2-O-); 4.91-4.86 (m, 1H, -CH-OH); 4.62 (br.s., 1H, -OH); 2.86 (br.s., 2H, -CH2-); 2.71-2.59 (m, 4H, piperidine 2×CH2); 1.78-1.74 (m, 4H, piperidine 2×CH2); 1.56-1.50 (m, 2H, piperidine CH2). 13C NMR spectrum (CDCl3) δ: 158.31; 136.89; 133.80; 128.54; 127.42; 127.13; 114.77; 69.96; 67.96; 66.57; 54.62; 24.94; 23.42. For C20H25NO2 [M+H]+ calculated: 312.1963, found: 312.1965.
1-(4-(benzyloxy)phenyl)-2-((2-hydroxyethyl)(methyl)amino)ethanol (11S-E4)
From 0.39 ml (4.9 mmol) N-methyl ethanolamine, 0.30 g (0.98 mmol) of 4-benzyloxybromoacetophenone 1В and 0.04 g (0.98 mmol) NaBH4 the product 11S-E4 0.18 g (61%) was obtained. Rf = 0.55 (7% methanol in chloroform). The content of the target substance according to HPLC data is 98%.1H NMR spectrum (CDCl3) δ: 7.43-7.27 (m, 7H, Ph); 6.96-6.93 (m, 2H, Ph); 5.04 (s, 2H, -O-CH2-); 4.91-4.86 (m, 1H, -CH(OH)-); 4.63 (br.s., 1H, -OH); 2.87 (br.s., 2H, -CH(OH)-CH2-) 2.71-2.59 (m, 2H, -N(СН3)-CH2-); 1.76 (s, 3H, -N-CH3); 1.54-1.52 (m, 2H, -CH2-OH). 13C NMR spectrum (CDCl3) δ: 158.93; 136.88; 130.18; 128.60; 128.02; 127.44; 123.29; 114.94; 70.15; 68.97; 61.42; 58.63; 55.24; 37.62. For C18H23NO3 [M+H]+ calculated: 302.1756, found: 302.1760.
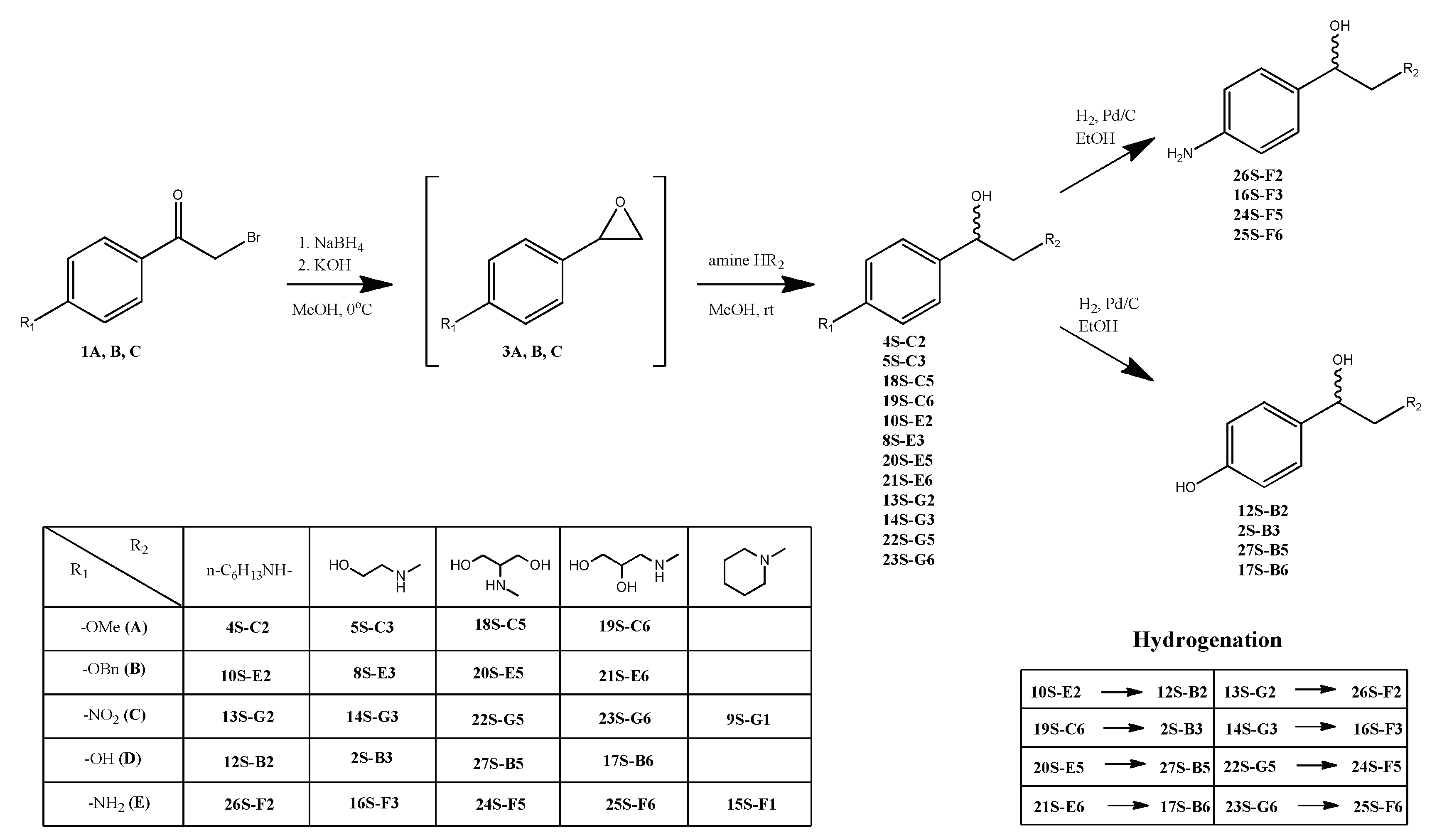
2.1.3. General procedure for compounds 4S-C2, 5S-C3, 18S-C5, 19S-C6, 10S-E2, 8S-E3, 20S-E5, 21S-E6, 13S-G2, 14S-G3, 22S-G5, 23S-G6, 9S-G1 synthesis
To a solution of bromoacetophenone 1 in methanol (for 4S-C2, 5S-C3, 18S-C5, 19S-C6, 10S-E2, 8S-E3, 20S-E5, 21S-E6) or 1,4-dioxane (for 13S-G2, 14S-G3, 22S-G5, 23S-G6, 9S-G1) (1 ml per 100 mg) while cooling in an ice bath (in the case of compounds 9S-G1 obtaining, the reaction was carried out at room temperature), 1 eq NaBH4 was added in portions. After the bromoacetophenone 1 was converted (control by TLC), a solution of 5 eq of primary amine and 1.2 eq КОН in methanol (1 ml per 1 ml amine) was added. After 12 h stirring, the reaction mixture was acidified with 10% aqueous solution of HCl to pH 3, the volatile components was removed on a vacuum rotary evaporator, the residue was suspended in 5 ml of water and extracted with methylene chloride (3 x 10 ml). The organic layers were combined and dried with anhydrous calcium chloride, filtered off and evaporated. The product was isolated by column chromatography on silica gel in a chloroform-methanol solvent system (methanol gradient from 0 to 20%).
2-(hexylamino)-1-(4-methoxyphenyl)ethanol (4S-C2f)
From 0.5 g (2.70 mmol) 4-methoxybromoacetophenone 1A, 0.1 g (2.70 mmol) NaBH4, 1.75 ml (13.5 mmol) amine and 0.19 g (3.30 mmol) KOH obtained 0.37 g (54%) of product 4S-C2. Rf (10% methanol in chloroform) = 0.45. The content of the target substance according to HPLC data is 98%. 1H NMR spectrum (CDCl3) δ: 7.30-7.27 (m, 2H, Ph); 6.87-6.84 (m, 2H, Ph); 4.86-4.82 (m, 1H, -CH); 3.82 (br.s, 1H, -OH); 3.78 (s, 3H, O-CH3); 2.94-2.67 (m, 4H, 2×CH2); 1.59-1.52 (m, 2H, -CH2-); 1.29-1.23 (m, 6H, 3×CH2); 0.88-0.84 (m, 3H, -CH3). 13C NMR spectrum (CDCl3) δ: 159.13; 133.70; 127.06; 113.80; 70.24; 56.30; 55.24; 49.11; 31.47; 28.39; 26.67; 22.51; 13.99. For C15H25NO2 [M+H]+ calculated: 252.1963, found: 252.1966.
2-((2-hydroxyethyl)amino)-1-(4-methoxyphenyl)ethanol (5S-C3)
From 0.5 g (2.70 mmol) 4-methoxybromoacetophenone 1A, 0.1 g (2.70 mmol) NaBH4, 0.82 ml (13.5 mmol) ethanolamine and 0.19 g (3.30 mmol) KOH obtained 0.33 g (58%) of product 5S-C3. Product Rf = 0.40 (20% methanol in chloroform). The content of the target substance according to HPLC data is 99%. 1H NMR spectrum (DMSO-d6) δ: 7.44-7.41 (m, 2H, Ph); 6.96-6.93 (m, 2H, Ph); 5.40 (br.s., 1H, -CH2-OH-); 5.02 (br.s., 1H, -CH-OH-); 4.11-4.07 (m, 1H, -CH-); 3.74 (s, 3H, -O-CH3); 3.71-3.32 (m, 4H, -CH2-CH2-OH); 2.75-2.56 (m, 2H, -CH2-NH-). 13C NMR spectrum (DMSO-d6) δ: 159.30; 129.63; 127.28; 113.96; 62.92; 57.21; 55.11; 47.79. For C11H17NO3 [M+H]+ calculated: 212.1286, found: 212.1290.
2-((2-hydroxy-2-(4-methoxyphenyl)ethyl)amino)propane-1,3-diol (18S-C5)
From 0.5 g (2.20 mmol) 4-methoxybromoacetophenone 1A, 0.08 g (2.20 mmol) NaBH4, 1.0 g (11.0 mmol) 2-aminopropane-1,3-diol and 0.15 g (2.60 mmol) KOH obtained 0.07 g (13%) of product 18S-C5. Product Rf = 0.55 (15% methanol in chloroform). The content of the target substance according to HPLC data is 96%. 1H NMR spectrum (DMSO-d6) δ: 7.26-7.21 (m, 2H, Ph); 6.87-6.82 (m, 2H, Ph); 4.72 (br.s., 1H, -CH-OH); 4.31 (br.s., 1H, -CH2-OH); 4.21 (br.s., 1H, -CH-OH); 3.78-2.74 (m, 1H, -CH-NH-); 3.71 (s, 3H, -O-CH3); 3.42-3.23 (m, 5H, -CH2-NH- and -CH-(CH2-OH)2); 2.36-2.31 (m, 1H, -CH2-NH-). 13C NMR spectrum (DMSO-d6) δ: 158.17; 134.47; 128.42; 113.43; 66.90; 61.89; 61.35; 57.69; 54.93. For C12H20NO4 [M+H]+ calculated: 242.1392, found: 242.1395.
3-((2-hydroxy-2-(4-methoxyphenyl)ethyl)amino)propane-1,2-diol (19S-C6)
From 0.5 g (2.20 mmol) 4-methoxybromoacetophenone 1A, 0.08 g (2.20 mmol) NaBH4, 0.85 ml (11.0 mmol) 3-aminopropane-1,2-diol and 0.15 g (2.60 mmol) KOH obtained 0.05 g (9%) of product 19S-C6. Product Rf = 0.46 (13% methanol in chloroform). The content of the target substance according to HPLC data is 98%. 1H NMR spectrum (CD3OD) δ: 7.27-7.22 (m, 2H, Ph); 6.90-6.86 (m, 2H, Ph); 3.76 (s, 3H, -O-CH3); 3.61-3.43 (m, 4H, -CH2-CH(OH)-CH2-OH); 3.30-3.28 (m, 1H, -CH(OH)-CH2-OH); 2.65-2.40 (m, 2H, -CH2-NH-). 13C NMR spectrum (CD3OD) δ: 160.78; 132.86; 132.76; 129.92; 129.89; 115.06; 115.04; 67.41; 67.27; 66.26; 66.13; 66.03; 65.07; 55.73; 51.53; 50.75. For C12H20NO4 [M+H]+ calculated: 242.1392, found: 242.1394.
1-(4-(benzyloxy)phenyl)-2(hexylamino)ethanol (10S-E2)
From 0.5 g (1.92 mmol) of 4-benzyloxybromoacetophenone 1В, 0.07 g (1.92 mmol) NaBH4, 1.25 ml (9.6 mmol) hexylamine and 0.17 g (3.0 mmol) KOH obtained 0.17 g (27%) of product 10S-E2. Rf = 0.55 (20% methanol in chloroform). The content of the target substance according to HPLC data is 98%. 1H NMR spectrum (CDCl3) δ: 7.43-7.33 (m, 7H, Ph); 6.94-6.92 (m, 2H, Ph); 5.38 (br. s, 1H, -CH-OH-); 5.03 (s, 2H, -CH2-O-); 4.23 (br.s., 1H, -OH); 3.16-3.03 (m, 4H, -CH2-NH-CH2); 1.90 (br.s., 2H, -CH2-(CH2)3-CH3); 1.35-1.28 (m, 6H, 3×CH2); 0.89-0.85 (m, 3H, -CH3). 13C NMR spectrum (CDCl3) δ: 158.66; 136.76; 132.37; 128.56; 127.96; 127.41; 127.18; 69.97; 68.69; 55.11; 48.65; 31.13; 26.36; 25.82; 22.38; 13.92. For C21H29NO2 [M+H]+ calculated: 328.2277, found: 328.2281.
1-(4-(benzyloxy)phenyl)-2-((2-hydroxyethyl)amino)ethanol (8S-E3)
From 0.65 g (2.50 mmol) of 4-benzyloxybromoacetophenone 1В, 0.09 g (2.50 mmol) NaBH4, 0.75 ml (12.5 mmol) ethanolamine, and 0.17 g (3.0 mmol) KOH obtained 0.16 g (22%) of product 8S-E3. Rf = 0.35 (20% methanol in chloroform). The content of the target substance according to HPLC data is 98%. 1H NMR spectrum (DMSO-d6) δ: 7.45-7.29 (m, 7H, Ph); 7.03-7.00 (m, 2H, Ph); 5.27 (br.s, 1H, -CH-OH); 5.09 (s, 2H, -O-CH2-); 4.89 (br.s, 1H, -CH2-OH); 4.04-4.00 (m, 1H, -CH-); 3.64-3.55 (m, 4H, -CH2-CH2-OH); 2.75-2.54 (m, 2H, -CH2-CH(OH)). 13C NMR spectrum (DMSO-d6) δ: 158.28; 135.95; 129.42; 128.37; 127.78; 127.59; 114.73; 88.84; 69.18; 69.46; 63.00; 57.65; 48.01. For C17H21NO3 [M+H]+ calculated: 288.1599, found: 288.1602.
2-((2-(4-(benzyloxy)phenyl)-2-hydroxyethyl)amino)propane-1,3-diol (20S-E5)
From 1.0 g (3.30 mmol) of 4-benzyloxybromoacetophenone 1В, 0.12 g (3.30 mmol) NaBH4, 1.5 g (16.5 mmol) 2-aminopropane-1,3-diol, and 0.22 g (4.0 mmol) KOH obtained 0.27 g (27%) of product 20S-E5. Rf = 0.67 (13% methanol in chloroform). The content of the target substance according to HPLC data is 98%. 1H NMR spectrum (DMSO-d6) δ: 7.45-7.35 (m, 5H, Ph); 7.25-7.22 (m, 2H, Ph); 6.96-6.91 (m, 2H, Ph); 5.18 (br.s., 1H, -CH-OH); 5.07 (s., 2H, -O-CH2-); 4.53-4.49 (m., 1H, - CH-OH); 4.44-4.26 (m, 2H, 2×-CH2-OH-); 3.44-3.24 (m, 5H, -CH-(-CH2-OH)2); 2.67-2.51 (m, 2H, -CH2-NH-). 13C NMR spectrum (DMSO-d6) δ: 157.25; 137.24; 136.87; 128.43; 127.77; 127.63; 127.07; 114.25; 71.61; 69.12; 61.36; 61.24; 61.08; 55.64. For C18H24NO4 [M+H]+ calculated: 318.1705, found: 318.1707.
3-((2-(4-(benzyloxy)phenyl)-2-hydroxyethyl)amino)propane-1,2-diol (21S-E6)
From 1.0 g (3.30 mmol) of 4-benzyloxybromoacetophenone 1В, 0.12 g (3.30 mmol) NaBH4, 1.27 ml (16.5 mmol) 3-aminopropane-1,2-diol, and 0.22 g (4.0 mmol) KOH obtained 0.12 g (12%) of product 21S-E6. Rf = 0.75 (15% methanol in chloroform). The content of the target substance according to HPLC data is 98%. 1H NMR spectrum (DMSO-d6) δ: 7.45-7.29 (m, 5H, Ph); 7.25-7.20 (m, 2H, Ph); 6.95-6.92 (m, 2H, Ph); 5.05 (s, 2H, -O-CH2-); 4.58-4.56 (m, 1H, -CH-OH); 4.34 (br.s, 4H, -CH(OH)-CH2-OH and HO-CH-CH2-NH-); 3.58-3.18 (m, 5H, -CH2-CH(OH)-CH2-OH). 13C NMR spectrum (DMSO-d6) δ: 157.38; 157.33; 137.24; 137.21; 134.11; 134.06; 128.40; 128.36; 127.71; 127.58; 127.54; 127.01; 126.98; 114.39; 71.01; 70.15; 69.14; 66.75; 66.60; 64.76; 64.48; 63.89; 51.15; 50.35. For C18H24NO4 [M+H]+ calculated: 318.1705, found: 318.1708.
2-(hexylamino)-1-(4-nitrophenyl)ethanol (13S-G2)
From 0.85 g (3.50 mmol) of 4-nitrobromoacetophenone 1С, 0.13 g (3.50 mmol) NaBH4, 2.30 ml (17.5 mmol) hexylamine, and 0.23 g (4.2 mmol) KOH obtained 0.32 g (34%) of product 13S-G2. Rf = 0.61 (15% methanol in chloroform). The content of the target substance according to HPLC data is 98%. 1H NMR spectrum (DMSO-d6) δ: 8.18-8.16 (m, 2H, Ph); 7.62-7.59 (m, 2H, Ph); 5.58 (br.s, 1H,-OH); 4.77-4.73 (m, 1H, -CH); 2.69-2.53 (m, 4H, -CH2-NH-CH2-); 1.40-1.35 (m, 2H, -NH-CH2-CH2-); 1.26-1.22 (m, 6H, -(CH2)3-CH3); 0.86-0.81 (m, 3H, -CH3).13C NMR spectrum (DMSO-d6) δ: 152.80; 146.37; 127.11; 123.08; 70.78; 57.20; 49.01; 31.25; 29.53; 26.48; 22.12; 13.94. For C14H23N2O3 [M+H]+ calculated: 267.1709, found: 267.1712.
2-((2-hydroxyethyl)amino)-1-(4-nitrophenyl)ethanol (14S-G3)
From 1.25 g (5.14 mmol) of 4-nitrobromoacetophenone 1С, 0.19 g (5.14 mmol) NaBH4, 1.55 ml (25.7 mmol) ethanolamine, and 0.34 g (6.17 mmol) KOH obtained 0.41 g (35%) of product 14S-G3. Rf = 0.55 (13% methanol in chloroform). The content of the target substance according to HPLC data is 95%. 1H NMR spectrum (DMSO-d6) δ: 8.19-8.16 (m, 2H, Ph); 7.63-7.60 (m, 2H, Ph); 5.61 (br.s, 1H, -CH-OH); 4.78-4.74 (m, 1H, -CH-OH); 4.49 (br.s, 1H, -CH2-OH); 3.45-3.37 (m, 2H, -CH2- OH); 2.72-2.53 (m, 4H, -CH2-NH-CH2-). 13C NMR spectrum (DMSO-d6) δ: 152.76; 146.39; 127.12; 123.11; 70.96; 60.42; 57.15; 51.45. For C10H15N2O4 [M+H]+ calculated: 227.1032, found: 227.1034.
2-((2-hydroxy-2-(4-nitrophenyl)ethyl)amino)propane-1,3-diol (22S-G5)
From 1.7 g (7.00 mmol) of 4-nitrobromoacetophenone 1С, 0.265 g (7.00 mmol) NaBH4, 3.18 g (35.0 mmol) 2-aminopropane-1,3-diol, and 0.47 g (8.4 mmol) KOH obtained 0.72 g (40%) of product 22S-G5. Rf = 0.58 (10% methanol in chloroform). The content of the target substance according to HPLC data is 98%. 1H NMR spectrum (DMSO-d6) δ: 8.18-8.15 (m, 2H, Ph); 7.64-7.61 (m, 2H, Ph); 5.51 (br.s., 1H, -CH-OH); 4.74-4.72 (m, 1H, -CH-OH); 4.31-4.27 (m, 2H, 2×-CH2-OH); 3.43-3.24 (m, 4H, 2×-CH2-OH-); 2.84-2.64 (m, 2H, -CH2-NH); 2.55-2.51 (m, 1H, -CH-NH-). 13C NMR spectrum (DMSO-d6) δ: 152.69; 146.39; 127.13; 123.13; 71.39; 61.39; 61.23; 60.98; 55.10. For C11H17N2O5 [M+H]+ calculated: 257.1137, found: 257.1140.
3-((2-hydroxy-2-(4-nitrophenyl)ethyl)amino)propane-1,2-diol (23S-G6)
From 1.7 g (7.00 mmol) of 4-nitrobromoacetophenone 1С, 0.265 g (7.00 mmol) NaBH4, 2.7 ml (35.0 mmol) 3-aminopropane-1,2-diol, and 0.47 g (8.4 mmol) KOH obtained 0.52 g (29%) of product 23S-G6. Rf = 0.49 (10% methanol in chloroform). The content of the target substance according to HPLC data is 98%. 1H NMR spectrum (DMSO-d6) δ: 8.19-8.16 (m, 2H, Ph); 7.63-7.60 (m, 2H, Ph); 5.62 (br.s, 1H, -CH-OH); 4.77-4.57 (m, 1H, -CH-OH); 4.57 (br.s, 1H, -CH2-OH); 3.49 (br.s, 1H, -CH-OH); 3.34-3.23 (m, 3H, -CH(OH)-CH2-OH); 2.72-2.52 (m, 3H, -CH2-NH-CH2-); 2.46-2.38 (m, 1H, -CH2-NH). 13C NMR spectrum (DMSO-d6) δ: 152.73; 146.41; 127.11; 123.13; 71.08; 70.68; 64.54; 57.48; 52.73. For C11H17N2O5 [M+H]+ calculated: 257.1137, found: 257.1141.
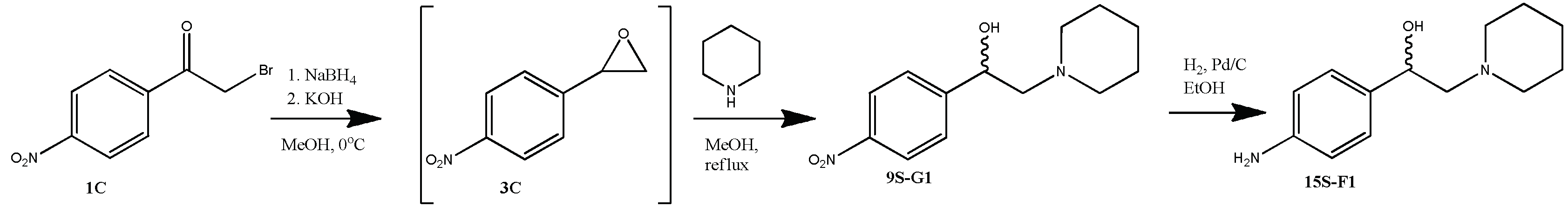
1-(4-nitrophenyl)-2-piperidin-1-yl ethanol (9S-G1)
From 0.5 g (2.05 mmol) of 4-nitrobromoacetophenone 1С, 0.08 g (2.05 mmol) NaBH4, 1.0 ml (10.2 mmol) piperedine and 0.14 g (2.46 mmol) KOH obtained 0.20 g (40%) of product 9S-G1. Rf = 0.67 (5% methanol in chloroform). The content of the target substance according to HPLC data is 97%. 1H NMR spectrum (CDCl3) δ: 8.19-8.16 (m, 2H, Ph); 7.57-7.55 (m, 2H, Ph); 5.05-5.00 (m, 1H, -CH-); 4.40 (br.s., 1H, -OH); 2.91-2.84 (m, 2H, -CH2-CH-); 2.70-2.49 (m, 4H, piperidine 2×CH2); 1.78-1.70 (m, 4H, piperidine 4×CH2); 1.54-1.50 (m, 2H, piperidine CH2). 13C NMR spectrum (CDCl3) δ: 149.41; 147.30; 126.53; 123.65; 67.71; 65.99; 54.63; 25.11; 23.45. For C13H18N2O3 [M+H]+ calculated: 251.1396, found: 251.1398.
2.1.3. General Procedure for Compounds 12S-B2, 2S-B3, 27S-B5, 17S-B6 26S-F2, 16S-F3, 24S-F5, 25S-F6, 15S-F1 Synthesis
10% Pd on carbon (0.05 eq of Pd) was added to a solution 12S-B2, 2S-B3, 27S-B5, 17S-B6 26S-F2, 16S-F3, 24S-F5, 25S-F6, 15S-F1 in ethanol and mixed in an atmosphere of H2 at room temperature overnight. The reaction mass was filtered through a layer of celite, which was washed with 10-15 ml ethanol, and combined organic solutions were evaporated. The product was isolated by column chromatography on silica gel in a chloroform-methanol solvent system (methanol gradient from 0 to 35%).
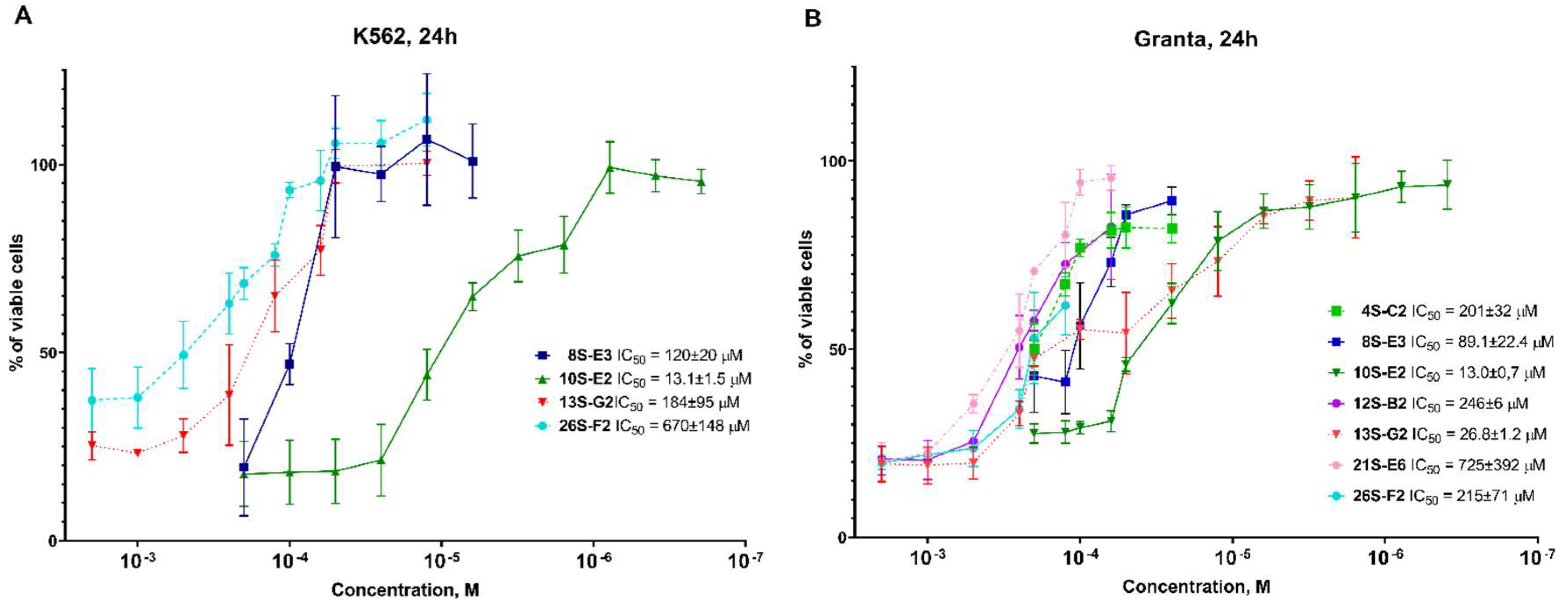
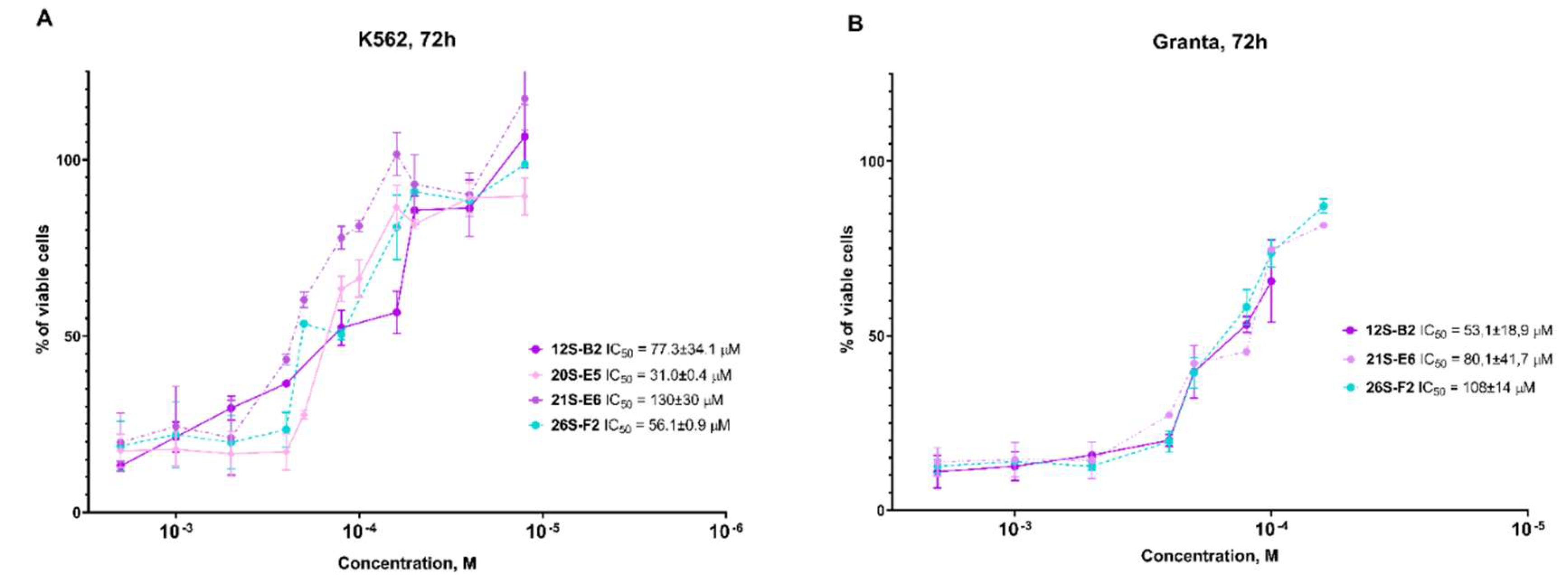
4-(2-(hexylamino)-1-hydroxyethyl)phenol (12S-B2)
From 0.10 g (0.3 mmol) 10S-E2 0.03 g (41%) of the product 12S-B2 was obtained. Rf = 0.34 (20% methanol in chloroform). The content of the target substance according to HPLC data is 97%. 1H NMR spectrum (DMSO-d6) δ: 7.16-7.12 (m, 2H, Ph); 6.76-6.71 (m, 2H, Ph); 4.73-4.68 (m, 1H, -CH); 3.86-2.71 (m, 4H, -CH2-NH-CH2-); 1.58-1.48 (m, 2H, -NH-CH2-CH2-); 1.30-1.25 (m, 6H, -(CH2)3-CH3); 1.29-0.87-0.84 (m, 3H, -CH3). 13C NMR spectrum (DMSO-d6) δ: 156.75; 133.09; 127.06; 114.91; 69.18; 55.24; 47.78; 30.93; 26.88; 26.02; 21.98; 13.90. For C14H24NO2 [M+H]+ calculated: 238.1807, found: 238.1810.
4-(1-hydroxy-2-((2-hydroxyethyl)amino)ethyl)phenol (2S-B3)
From 0.1 g (0.35 mmol) 19S-C6 0.05 g (73%) of the product 2S-B3 was obtained. Rf = 0.35 (30% methanol in chloroform). The content of the target substance according to HPLC data is 97%.1H NMR spectrum (CD3OD) δ: 7.36-7.33 (m, 2H, Ph); 6.87-6.85 (m, 2H, Ph); 5.01 (br.s, 2H, 2 × OH); 4.33-4.28 (m, 1H, -CH-OH); 3.99-3.86 (m, 2H, -CH2-OH); 3.83-3.69 (m, 2H, -CH2-NH-); 3.05-2.91 (m, 2H, -CH2-CH-); 2.15 (s, 1H, -OH). 13C NMR spectrum (CD3OD) δ: 159.90; 130.97; 124.11; 117.02; 64.74; 63.39; 57.78; 30.76. For C10H15NO3 [M+H]+ calculated: 198.1130, found: 198.1134.
2-((2-hydroxy-2-(4-hydroxyphenyl)ethyl)amino)propane-1,3-diol (27S-B5)
From 0.17 g (0.5 mmol) 20S-E5 0.03 g (25%) of the product 27S-B5 was obtained. Rf = 0.23 (15% methanol in chloroform). The content of the target substance according to HPLC data is 96%. 1H NMR spectrum (DMSO-d6) δ: 7.13-7.09 (m, 2H, Ph); 6.70-6.65 (m, 2H, Ph); 4.47-4.43 (m, 1H, -CH-OH); 3.44-3.20 (m, 5H, -CH-(-CH2-OH)2); 2.69-2.52 (m, 2H, -CH2-NH). 13C NMR spectrum (DMSO-d6) δ: 156.14; 134.72; 126.96; 114.62; 71.67; 61.29; 61.17; 60.98; 55.51. For C11H18NO4 [M+H]+ calculated: 228.1235, found: 228.1239.
3-((2-hydroxy-2-(4-hydroxyphenyl)ethyl)amino)propane-1,2-diol (17S-B6)
From 0.1 g (0.3 mmol) 21S-E6 0.03 g (42%) of the product 17S-B6 was obtained. Rf = 0.37 (20% methanol in chloroform). The content of the target substance according to HPLC data is 98%. 1H NMR spectrum (DMSO-d6) δ: 7.12-7.08 (m, 2H, Ph); 6.70-6.66 (m, 2H, Ph); 4.82 (br.s, 1H, -CH-OH); 4.82 (br.s, 2H, -CH(OH)-CH2-OH); 4.06 (br.s, 1H, -CH-OH); 3.60-3.17 (m, 7H, -CH2-CH(OH)-CH2-OH and -CH-OH and Ph-OH); 2.47-2.19 (m, 2H, -CH2-NH). 13C NMR spectrum (DMSO-d6) δ: 156.38; 156.34; 131.28; 128.44; 128,33; 114,88; 70.70; 69.73; 64,69; 64,40; 63,84; 50,92; 50.10. For C11H18NO4 [M+H]+ calculated: 228.1235, found: 228.1238.
1-(4-aminophenyl)-2-(hexylamino)ethanol (26S-F2)
From 0.1 g (0.4 mmol) 13S-G2 0.035 g (37%) of the product 26S-F2 was obtained. Rf = 0.41 (10% methanol in chloroform). The content of the target substance according to HPLC data is 95%. 1H NMR spectrum (DMSO-d6) δ: 6.96-6.94 (m, 2H, Ph); 6.51-6.47 (m, 2H, Ph); 4.86 (br.s, 2H,-NH2); 4.42-4.37 (m, 1H, -CH); 2.59-2.46 (m, 4H, -CH2-NH-CH2-); 1.40-1.32 (m, 2H, -NH-CH2-CH2-); 1.27-1.22 (m, 6H, -(CH2)3-CH3); 0.87-0.83 (m, 3H, -CH3). 13C NMR spectrum (DMSO-d6) δ: 147.39; 131.63; 126.53; 113.40; 71.27; 57.73; 49.02; 31.18; 29.58; 26.42; 22.02; 13.84. For C14H25N2O [M+H]+ calculated: 237.1967, found: 237.1970.
1-(4-aminophenyl)-2-((2-hydroxyethyl)amino)ethanol (16S-F3)
From 0.09 g (0.4 mmol) 14S-G3 0.062 g (79%) of the product 16S-F3 was obtained. Rf = 0.31 (13% methanol in chloroform). The content of the target substance according to HPLC data is 98%. 1H NMR spectrum (DMSO-d6) δ: 6.97-6.94 (m, 2H, Ph); 6.49-6.47 (m, 2H, Ph); 4.90 (br.s, 2H, 2×-OH); 4.42-4.38 (m, 1H, -CH-OH); 3.43-3.40 (m, 2H, -CH2-OH); 2.61-2.54 (m, 4H, -CH2-NH-CH2-). 13C NMR spectrum (DMSO-d6) δ: 147.52; 131.66; 126.64; 113.46; 71.51; 60.43; 57.75; 51.53. For C10H17N2O2 [M+H]+ calculated: 197.1290, found: 197.1294.
2-((2-(4-aminophenyl)-2-hydroxyethyl)amino)propane-1,3-diol (24S-F5)
From 0.14 g (0.5 mmol) 22S-G5 0.1 g (88%) of the product 24S-F5 was obtained. Rf = 0.26 (10% methanol in chloroform). The content of the target substance according to HPLC data is 98%. 1H NMR spectrum (DMSO-d6) δ: 6.97-6.94 (m, 2H, Ph); 6.49-6.46 (m, 2H, Ph); 4.92-4.90 (m, 3H, -NH2 and -CH-OH); 4.41-4.35 (m, 3H, -CH-OH and 2×-CH2-OH); 3.30-3.23 (m, 5H, -CH-(CH2-OH)2); 2.64-2.56 (m, 2H, -CH2-NH). 13C NMR spectrum (DMSO-d6) δ: 147.49; 131.67; 126.65; 113.44; 71.99; 61.37; 61.28; 61.11; 55.68. For C11H19N2O3 [M+H]+ calculated: 227.1396, found: 227.1400.
3-((2-(4-aminophenyl)-2-hydroxyethyl)amino)propane-1,2-diol (25S-F6)
From 0.2 g (0.8 mmol) 23S-G6 0.095 g (53%) of the product 25S-F6 was obtained. Rf = 0.23 (10% methanol in chloroform). The content of the target substance according to HPLC data is 95%. 1H NMR spectrum (DMSO-d6) δ: 6.97-6.94 (m, 2H, Ph); 6.50-6.47 (m, 2H, Ph); 4.90 (br.s, 3H, -NH2 and -CH-OH); 4.43-4.38 (m, 1H, -CH-OH); 3.53-3.23 (m, 5H, -CH(OH)-CH2-OH and -CH2-NH); 2.63-2.52 (m, 3H, -CH2-NH-CH2-); 2.44-2.37 (m, 1H, -CH2-NH). 13C NMR spectrum (DMSO-d6) δ: 147.53; 131.66; 126.63; 113.48; 71.59; 70.65; 64.63; 58.09; 52.83. For C11H19N2O3 [M+H]+ calculated: 227.1396, found: 227.1399.
1-(4-aminophenyl)-2-piperidin-1-ylethanol (15S-F1)
From 0.1 g (0.4 mmol) 9S-G1 0.03 g (34%) of the product 15S-F1 was obtained. Rf = 0.35 (10% methanol in chloroform). The content of the target substance according to HPLC data is 95%. 1H NMR spectrum (DMSO-d6) δ: 7.04-7.01 (m, 2H, Ph); 6.54-6.51 (m, 2H, Ph); 5.86 (br.s., 1H, -OH); 5.09 (br.s., 2H, -NH2); 4.95-4.91 (m, 1H, -CH-); 3.36-2.98 (m, 6H, piperidine 2×CH2 and -CH-CH2-); 1.77-1.76 (m, 4H, piperidine 4×CH2); 1.51 (br.s., 2H, piperidine CH2). 13C NMR spectrum (DMSO-d6) δ: 148.24; 128.69; 126.70; 113.47; 66.72; 62.98; 52.49; 22.24; 21.46. For C13H21N2O [M+H]+ calculated: 221.1654, found: 221.1656.