Submitted:
10 October 2024
Posted:
10 October 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
Animals and Nutrition
Fattening Performance Test
Carcass Traits
Genotyping
Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mohammadabadi M, Bordbar F, Jensen J, Du M, Guo W. Key Genes Regulating Skeletal Muscle Development and Growth in Farm Animals. Animals (Basel). 2021 Mar 16;11(3):835. [CrossRef] [PubMed]
- Yan E, Guo J, Yin J. Nutritional regulation of skeletal muscle energy metabolism, lipid accumulation and meat quality in pigs. Anim Nutr. 2023 Jun 7;14:185-192. [CrossRef]
- Rothschild MF, Hu ZL, Jiang Z. Advances in QTL mapping in pigs. Int J Biol Sci. 2007 Feb 10;3(3):192-7. [CrossRef]
- Zeng Q, Du ZQ. Advances in the discovery of genetic elements underlying longissimus dorsi muscle growth and development in the pig. Anim Genet. 2023 Dec;54(6):709-720. [CrossRef]
- Rohrer GA, Nonneman DJ, Miller RK, Zerby H, Moeller SJ. Association of single nucleotide polymorphism (SNP) markers in candidate genes and QTL regions with pork quality traits in commercial pigs. Meat Sci. 2012 Dec;92(4):511-8. [CrossRef]
- Fujii, J. , Otsu K., Zorzato F., De Leon S., Khanna V.K., Weiler J.E., O’Brien P.J., MacLennan D.H. (1991) – Identification of a mutation in porcine ryanodine receptor associated with malignant hyperthermia. Science 253, 448-451. [CrossRef]
- Nezer C, Moreau L, Brouwers B, Coppieters W, Detilleux J, Hanset R, Karim L, Kvasz A, Leroy P, Georges M. An imprinted QTL with major effect on muscle mass and fat deposition maps to the IGF2 locus in pigs. Nat Genet. 1999 Feb;21(2):155-6. [CrossRef]
- Van Laere AS, Nguyen M, Braunschweig M, Nezer C, Collette C, Moreau L, Archibald AL, Haley CS, Buys N, Tally M, Andersson G, Georges M, Andersson L. A regulatory mutation in IGF2 causes a major QTL effect on muscle growth in the pig. Nature. 2003 Oct 23;425(6960):832-6. [CrossRef] [PubMed]
- Zhang J, Chai J, Luo Z, He H, Chen L, Liu X, Zhou Q. Meat and nutritional quality comparison of purebred and crossbred pigs. Anim Sci J. 2018 Jan;89(1):202-210. [CrossRef]
- Xiang G, Ren J, Hai T, Fu R, Yu D, Wang J, Li W, Wang H, Zhou Q. Editing porcine IGF2 regulatory element improved meat production in Chinese Bama pigs. Cell Mol Life Sci. 2018 Dec;75(24):4619-4628. [CrossRef]
- López-Buesa P, Burgos C, Galve A, Varona L. Joint analysis of additive, dominant and first-order epistatic effects of four genes (IGF2, MC4R, PRKAG3 and LEPR) with known effects on fat content and fat distribution in pigs. Anim Genet. 2014 Feb;45(1):133-7. [CrossRef]
- Li S, Lei H, Li J, Sun A, Ahmed Z, Duan H, Chen L, Zhang B, Lei C, Yi K. Analysis of genetic diversity and selection signals in Chaling cattle of southern China using whole-genome scan. Anim Genet. 2023 Jun;54(3):284-294. [CrossRef]
- Ron M, Weller JI. From QTL to QTN identification in livestock--winning by points rather than knock-out: a review. Anim Genet. 2007 Oct;38(5):429-39. [CrossRef]
- Wang M, Wang Q, Pan Y. From QTL to QTN: candidate gene set approach and a case study in porcine IGF1-FoxO pathway. PLoS One. 2013;8(1):e53452. [CrossRef]
- Salas RC, Mingala CN. Genetic Factors Affecting Pork Quality: Halothane and Rendement Napole Genes. Anim Biotechnol. 2017 Apr 3;28(2):148-155. [CrossRef]
- Ropka-Molik K, Pawlina-Tyszko K, Żukowski K, Tyra M, Derebecka N, Wesoły J, Szmatoła T, Piórkowska K. Identification of Molecular Mechanisms Related to Pig Fatness at the Transcriptome and miRNAome Levels. Genes (Basel). 2020 May 29;11(6):600. [CrossRef]
- Pocrnic I, Lourenco D, Misztal I. SNP profile for quantitative trait nucleotide in populations with small effective size and its impact on mapping and genomic predictions. Genetics. 2024 Jun 24:iyae103. [CrossRef]
- Liu X, Zhang J, Xiong X, Chen C, Xing Y, Duan Y, Xiao S, Yang B, Ma J. An Integrative Analysis of Transcriptome and GWAS Data to Identify Potential Candidate Genes Influencing Meat Quality Traits in Pigs. Front Genet. 2021 Oct 21;12:748070. [CrossRef]
- Blicharski T, Polok P, Snopkiewicz M. Results of the pig assessment in 2017 [Wyniki oceny trzody chlewnej w 2017 roku]. Polish Association of Pig Breeders and Producers POLSUS [Polski Związek Hodowców i Producentów Trzody Chlewnej POLSUS], Warsaw 2018, pp. 5-35.
- Różycki, M. , Tyra M. Results of fattening and slaughter performance assessment of pigs at control stations [Wyniki oceny użytkowości tucznej i rzeźnej świń w stacjach kontroli]. Stan Hodowli i Wyniki Oceny Świń w roku 2012. Zespół Wydawnictw i Poligrafii IZ PIB, Kraków 2013, s. 49 -72. (in polish).
- Tyra M, Żak G. Analysis of relationships between fattening and slaughter performance of pigs and the level of intramuscular fat (IMF) in longissimus dorsi muscle. Ann Anim Sci. 2012 Apr 12;12(2):169-178. [CrossRef]
- Karpesiuk, K. Current problems and new challenges in pig nutrition [Aktualne problemy i nowe wyzwania w w zakresie żywienia świń] in Basic issues in the field of pig breeding and husbandry – current problems and new challenges [Podstawowe zagadnienia w zakresie chowu i hodowli trzody chlewnej – aktualne problemy i nowe wyzwania]; Pawłowski R., Ed.; Publisher Warmińsko-Mazurski Ośrodek Doradztwa Rolniczego in Olsztyn, Poland; 2020; pp. 41-59.
- Terman, A. , Woźniak-Męch K., Korpal A., Polasik D., Tyra M., Szyndler-Nędza, M., Żak G., Rybarczyk A., Dybus A. Association Between ATP Citrate Lyase (ACLY) Gene Polymorphism and Fattening, Slaughter and Pork Quality Traits in Polish Pigs. Ann Anim Sci. 2021 Oct 28;21(4):1301-1313. [CrossRef]
- Polasik D, Tyra M, Szyndler-Nędza M, Korpal A, Woźniak-Męch K, Terman A. Relationship between VRTN gene polymorphism and growth, slaughter and meat quality traits in three polish pig breeds. Cienc Agrotec. 2018; 42(5):540-549. [CrossRef]
- Ahmed Z, Xiang W, Wang F, Nawaz M, Kuthu ZH, Lei C, Xu D. Whole-genome resequencing deciphers patterns of genetic diversity, phylogeny, and evolutionary dynamics in Kashmir cattle. Anim Genet. 2024 May 10. [CrossRef]
- Piórkowska K, Żukowski K, Ropka-Molik K, Tyra M. Deep sequencing of a QTL-rich region spanning 128-136Mbp of pig chromosome 15. Gene. 2018 Mar 20;647:268-275. [CrossRef]
- Zhang J, Chen W, Chen G, Flannick J, Fikse E, Smerin G, Degner K, Yang Y, Xu C; Consortium AMP-T2D-GENES; Li Y, Hanover JA, Simonds WF. Ancestry-specific high-risk gene variant profiling unmasks diabetes-associated genes. Hum Mol Genet. 2024 Apr 8;33(8):655-666. [CrossRef]
- Cheng L, Tachibana K, Iwasaki H, Kameyama A, Zhang Y, Kubota T, Hiruma T, Tachibana K, Kudo T, Guo JM, Narimatsu H. Characterization of a novel human UDP-GalNAc transferase, pp-GalNAc-T15. FEBS Lett. 2004 May 21;566(1-3):17-24. [CrossRef] [PubMed]
- Lin B, Qing X, Liao J, Zhuo K. Role of Protein Glycosylation in Host-Pathogen Interaction. Cells. 2020 Apr 20;9(4):1022. [CrossRef]
- Bennett EP, Mandel U, Clausen H, Gerken TA, Fritz TA, Tabak LA. Control of mucin-type O-glycosylation: a classification of the polypeptide GalNAc-transferase gene family. Glycobiology. 2012 Jun;22(6):736-56. [CrossRef]
- Chia J, Tay F, Bard F. The GalNAc-T Activation (GALA) Pathway: Drivers and markers. PLoS One. 2019 Mar 19;14(3):e0214118. [CrossRef]
- Dang K, Jiang S, Gao Y, Qian A. The role of protein glycosylation in muscle diseases. Mol Biol Rep. 2022 Aug;49(8):8037-8049. [CrossRef]
- Cieniewski-Bernard C, Montel V, Berthoin S, Bastide B (2012) Increasing O-GlcNAcylation level on organ culture of soleus modulates the calcium activation parameters of muscle fibers. PLoS One 7:e48218. [CrossRef]
- Cieniewski-Bernard C, Lambert M, Dupont E, Montel V, Stevens L, Bastide B (2014) O-GlcNAcylation, contractile protein modifications and calcium affinity in skeletal muscle. Front Physiol 5:421. [CrossRef]
- Valdés-Hernández J, Folch JM, Crespo-Piazuelo D, Passols M, Sebastià C, Criado-Mesas L, Castelló A, Sánchez A, Ramayo-Caldas Y. Identification of candidate regulatory genes for intramuscular fatty acid composition in pigs by transcriptome analysis. Genet Sel Evol. 2024 Feb 12;56(1):12. Erratum in: Genet Sel Evol. 2024 Feb 26;56(1):14. doi: 10.1186/s12711-024-00885-8. [CrossRef]
- Liu X, Wang LG, Luo WZ, Li Y, Liang J, Yan H, Zhao KB, Wang LX, Zhang LC. Genome-wide SNP scan in a porcine Large White×Minzhu intercross population reveals a locus influencing muscle mass on chromosome 2. Anim Sci J. 2014 Dec;85(12):969-75. [CrossRef]
- Rodriguez A, Hilvo M, Kytömäki L, Fleming RE, Britton RS, Bacon BR, Parkkila S. Effects of iron loading on muscle: genome-wide mRNA expression profiling in the mouse. BMC Genomics. 2007 Oct 19;8:379. [CrossRef]
- Jozefczuk J, Kashofer K, Ummanni R, Henjes F, Rehman S, Geenen S, Wruck W, Regenbrecht C, Daskalaki A, Wierling C, Turano P, Bertini I, Korf U, Zatloukal K, Westerhoff HV, Lehrach H, Adjaye J. A Systems Biology Approach to Deciphering the Etiology of Steatosis Employing Patient-Derived Dermal Fibroblasts and iPS Cells. Front Physiol. 2012 Sep 3;3:339. [CrossRef]
- Funari VA, Day A, Krakow D, Cohn ZA, Chen Z, Nelson SF, Cohn DH. Cartilage-selective genes identified in genome-scale analysis of non-cartilage and cartilage gene expression. BMC Genomics. 2007 Jun 12;8:165. [CrossRef]
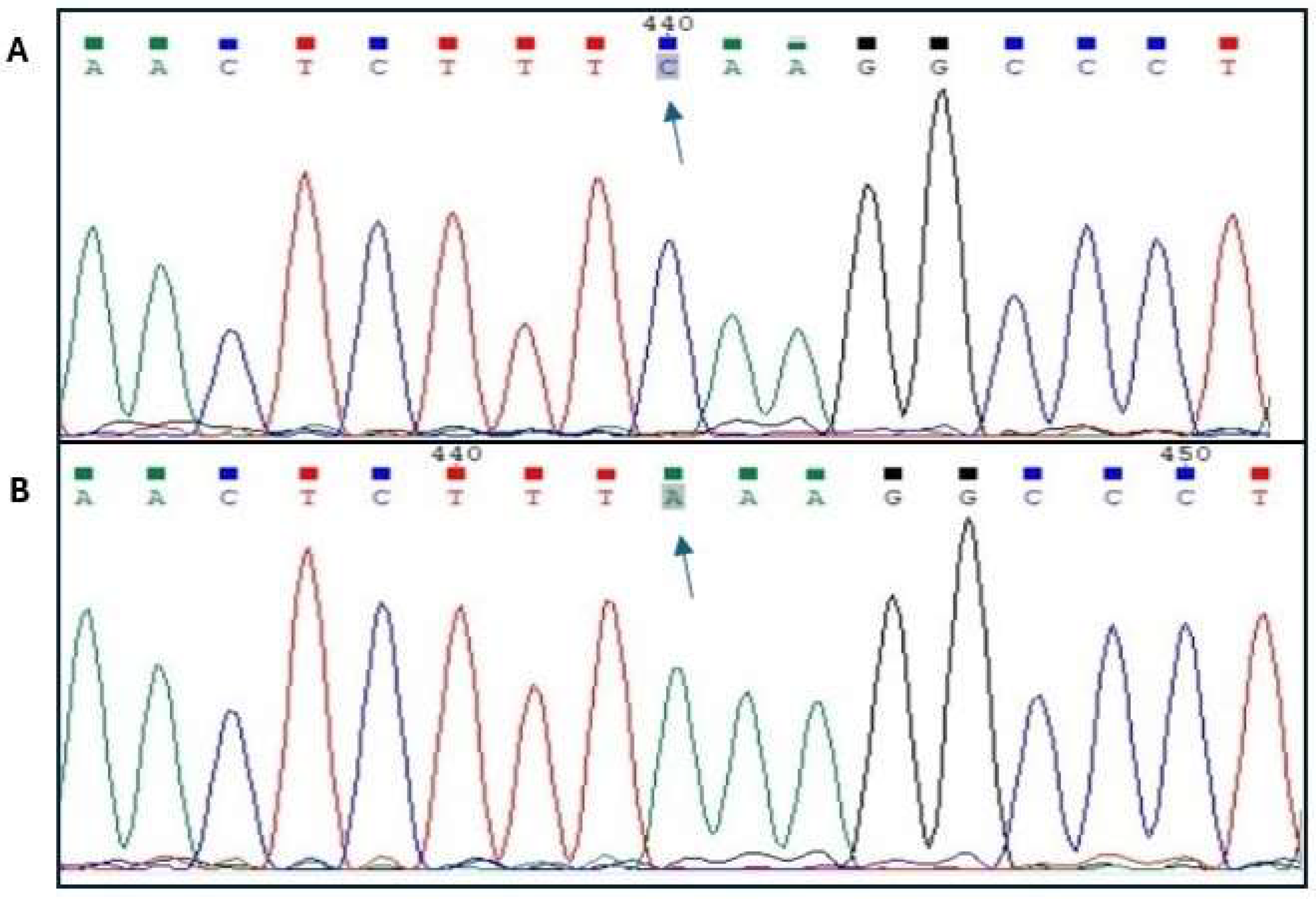
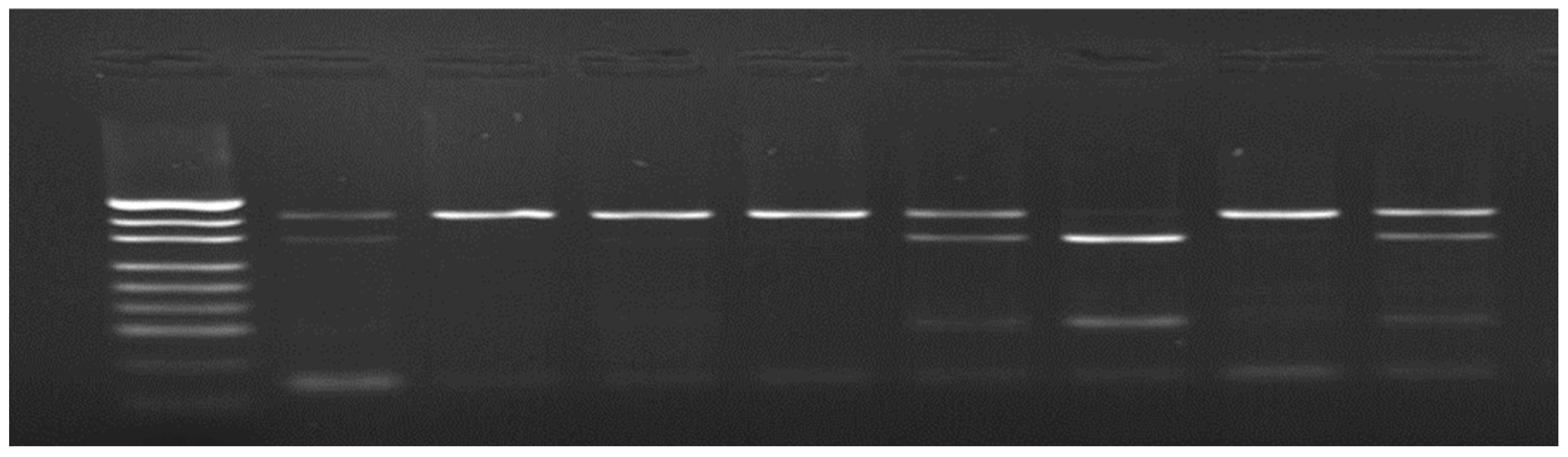
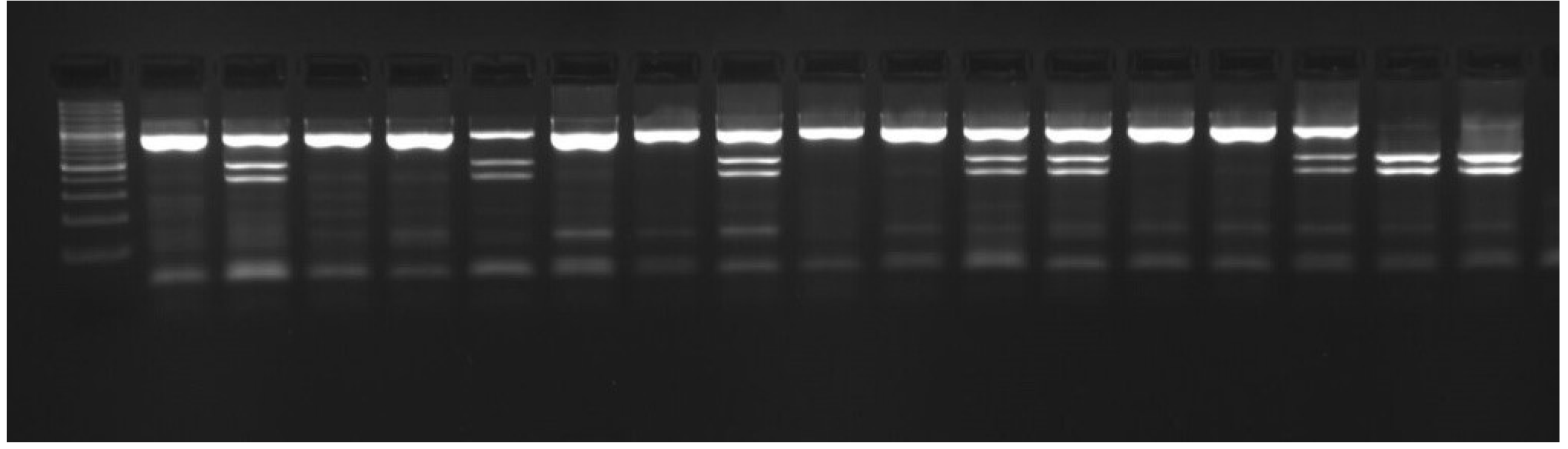
| Breed | Polish Large White Polish Landrace |
Pulawska |
| Forward Reverse |
5’-GTGCCTTCCTAGTGTCCCTT-3’ 5’-TCATGGACCACACACTCTAACA -3’ |
5’- CCACCCCAGACCTCTTGAAT-3’ 5’- GACTCTAGACTGAAGGCCCC-3’ |
| Amplicon length (bp) | 424 | 842 |
| Restriction fragments lengths (bp) |
AA – 316 and 108 AC – 424, 316 and 108 CC – 424 (no digestion) |
AA – 474 and 368 AC – 842, 474 and 368 CC – 842 (no digestion) |
| Breed | N | Genotype | Allele | HWE | ||||
|---|---|---|---|---|---|---|---|---|
| AA | AC | CC | A | C | ꭓ2 | p | ||
| Polish Landrace | 192 | 0.27 (n = 52) |
0.73 (n = 140) |
- | 0.36 | 0.64 | 63.2083 | ≤0.0001 |
| Polish Large White | 187 | 0.04 (n = 7) |
0.42 (n = 79) |
0.54 (n = 101) |
0.25 | 0.75 | 3.1896 | 0.0741 |
| Pulawska | 48 | 0.12 (n = 6) |
0.48 (n = 23) |
0.40 (n = 19) |
0.36 | 0.64 | 0.0561 | 0.8127 |
| Trait | Genotype | Polish Large White | Polish Landrace | Pulawska | Whole population |
|---|---|---|---|---|---|
| Number of days on test [days] |
AA AC CC |
79.354 ± 5.39 83.738 ± 3.65 82.628 ± 3.72 |
77.804 ± 1.54 78.660 ± 1.15 - |
100.333 ± 3.40A 85.217 ± 1.74B 88.105 ± 1.91B |
82.599 ± 1.68 81.960 ± 1.17 81.108 ± 1.39 |
| Daily feed intake [kg] |
AA AC CC |
2.396 ± 0.15 2.319 ± 0.10 2.365 ± 0.10 |
2.536 ± 0.04 2.477 ± 0.03 - |
2.138 ± 0.06 2.277 ± 0.03 2.244 ± 0.03 |
2.438 ± 0.04 2.398 ± 0.03 2.439 ± 0.04 |
| Lifetime daily gain [g/day] |
AA AC CC |
592.653 ± 36.73 566.227 ± 24.86 574.518 ± 25.33 |
603.930 ± 8.46 595.056 ± 6.29 - |
498.833 ± 23.20a 569.130 ± 11.85b 535.684 ± 13.04b |
577.517 ± 10.58 576.256 ± 7.34 579.110 ± 8.77 |
| Test daily gain [g/day] |
AA AC CC |
883.968 ± 58.63 858.058 ± 39.69 866.307 ± 40.44 |
924.189 ± 16.93 913.801 ± 12.60 - |
721.167 ± 27.90Aa 829.30 ± 14.25B 805.158 ± 15.64b |
873.856 ± 17.88 877.083 ± 12.41 884.28 ± 14.82 |
| Feed conversion [kg/kg gain] |
AA AC CC |
2.708 ± 0.11 2.710 ± 0.08 2.737 ± 0.08 |
2.762 ± 0.03 2.724 ± 0.02 - |
2.988 ± 0.07a 2.750 ± 0.04b 2.798 ± 0.04a |
2.811 ± 0.03 2.749 ± 0.02 2.774 ± 0.03 |
| Age at slaughter [days] |
AA AC CC |
169.980 ± 11.07 180.444 ± 7.49 176.930 ± 7.64 |
168.592 ± 2.55 170.854 ± 1.90 - |
204.833 ± 7.98a 178.304 ± 4.08b 189.105 ± 4.48b |
177.185 ± 3.25 177.056 ± 2.26 175.537 ± 2.69 |
| Trait | Genotype | Polish Large White | Polish Landrace | Pulawska | Whole population |
|---|---|---|---|---|---|
| Carcass yield [%] |
AA AC CC |
75.973 ± 0.40 76.031 ± 0.27 76.097 ± 0.28 |
76.885 ± 0.13 76.861 ± 0.10 - |
76.230 ± 0.30 76.420 ± 0.15 76.393 ± 0.17 |
76.479 ± 0.13 76.450 ± 0.09 76.516 ± 0.11 |
| Middle length of carcass [cm] |
AA AC CC |
77.603 ± 1.04 78.764 ± 0.70 78.491 ± 0.71 |
79.209 ± 0.74 79.665 ± 0.55 - |
78.625 ± 0.85 78.452 ± 0.44 78.203 ± 0.48 |
79.133 ± 0.57 79.464 ± 0.39 79.220 ± 0.47 |
| Loin weight [kg] |
AA AC CC |
7.439 ± 0.27 7.327 ± 0.18 7.355 ± 0.19 |
7.765 ± 0.09 7.648 ± 0.07 - |
7.272 ± 0.23 7.700 ± 0.12 7.545 ± 0.13 |
7.681 ± 0.09 7.611 ± 0.06 7.617 ± 0.08 |
| Loin weight without skin and backfat [kg] |
AA AC CC |
5.882 ± 0.26 5.865 ± 0.17 5.855 ± 0.18 |
6.121 ± 0.08 6.041 ± 0.06 - |
5.736 ± 0.20 6.079 ± 0.10 5.840 ± 0.11 |
6.019 ± 0.09 5.987 ± 0.06 5.939 ± 0.07 |
| Ham weight without skin and backfat [kg] |
AA AC CC |
9.538 ± 0.26 9.294 ± 0.17 9.310 ± 0.18 |
9.272 ± 0.09 9.302 ± 0.07 - |
9.184 ± 0.22 9.214 ± 0.11 8.823 ± 0.12 |
9.223 ± 0.09 9.201 ± 0.06 9.119 ± 0.07 |
| Loin eye area [cm2] |
AA AC CC |
53.631 ± 2.35 51.196 ± 1.58 51.518 ± 1.61 |
49.993 ± 0.70a 51.661 ± 0.52b - |
52.217 ± 2.36a 53.972 ± 1.20a 49.587 ± 1.32b |
51.042 ± 0.78 51.971 ± 0.54 51.114 ± 0.65 |
| Width of loin eye [cm] |
AA AC CC |
10.188 ± 0.35 10.195 ± 0.23 10.295 ± 0.24 |
10.091 ± 0.11 10.307 ± 0.08 - |
10.431 ± 0.32A 10.554 ± 0.16A 9.778 ± 0.18B |
10.239 ± 0.12 10.357 ± 0.08 10.262 ± 0.10 |
| Height of loin eye [cm] |
AA AC CC |
6.741 ± 0.28 6.562 ± 0.18 6.608 ± 0.19 |
6.894 ± 0.09 6.851 ± 0.07 - |
6.977 ± 0.25 6.924 ± 0.13 6.772 ± 0.14 |
6.898 ± 0.09 6.834 ± 0.06 6.837 ± 0.08 |
| Average backfat thickness of five measurements [cm] |
AA AC CC |
1.123 ± 0.14 1.122 ± 0.09 1.177 ± 0.10 |
1.341 ± 0.04 1.292 ± 0.03 - |
1.295 ± 0.13 1.371 ± 0.07 1.527 ± 0.07 |
1.339 ± 0.05 1.311 ± 0.03 1.396 ± 0.04 |
| Carcass meat content [%] |
AA AC CC |
63.155 ± 1.35 61.939 ± 0.90 62.117 ± 0.92 |
61.766 ± 0.44 61.890 ± 0.33 - |
61.339 ± 1.18A 62.276 ± 0.60A 59.254 ± 0.66B |
61.798 ± 0.46 61.687 ± 0.32 61.155 ± 0.38 |
| Weight of primary cuts [kg] |
AA AC CC |
24.261 ± 0.53 23.797 ± 0.35 23.859 ± 0.36 |
23.995 ± 0.17 24.049 ± 0.13 - |
23.621 ± 0.45A 23.993 ± 0.23A 22.838 ± 0.25B |
23.862 ± 0.18 23.828 ± 0.12 23.618 ± 0.15 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





