Submitted:
30 September 2024
Posted:
01 October 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Experimental Materials
2.2. Experimental Design
2.3. Chromosome Index Determination
2.4. Statistical Analysis
3. Results
3.1. Karyotype Analysis of Diploid Chromosomes in Root Tip during the Mitotic Stage
3.2. Karyotype Analysis of Autotetraploid in Root Tip During the Mitotic Stage
4. Discussion
4.1. Karyotype of Tartary Buckwheat
4.2. Karyotype Changes after Chromosome Doubling
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lin, R.F. The Development and Utilization of Tartary Buckwheat Resources. In Proceedings of the Proceedings of the 9 International Symposium on Buckwheat, 2004, 252-258.
- Morris, M.R. Cytogenetic studies on buckwheat. J. Hered. 1951, 42, 85-89. [CrossRef]
- Chen, Q.F. Karyotype analysis of five Fagopyrum species native to China. Guihaia 2001, 21, 107-110. [CrossRef]
- Du, X.C.; M. Y.; Liu,P.; Xu,G. D.Karyotype Analysis of Two Buckwheat Variety Chromosomes. Subtrop. Plant Sci. 2005, 34, 36-38.
- He, F.F. Karyotype analysis of buckwheat. J. Southwest Agric. Univ. 1992, 14, 522-523.
- Lei, B.J.; R. Z. Studies on the karyotype of chromosomes of buckwheats. J. Sichuan Univ. 2000, 37, 142-143.
- Liu, J.L.; Tang, Y.; Shao,J. R.; Luo,Q.; Sun,J. X. Karyotypic Studies of Two Wild Buckwheat Species in the Fagopyrum Mill. J. Northwest Botany 2009, 29, 1798-1803.
- Sheng, M.Y. Physical mapping of the 45S and 5SrDNA of cultivated buckwheat. J. Plant Genet. Resour. 2013, 14, 317-321. [CrossRef]
- Shi, J.Q.; Li,Y. Q.; Zhang,Z. W.; Wu,B.; Wang,A. H. Analysis of karyotypes and evolutionary features of wild buckwheat species of buckwheat. J. Plant Genet. Resour. 2016, 17, 455-460.
- Suresh, N.; Tamaki, H.; Junichi,W.; Kazumi,T.; Hiroko,K.; Shigeru,S. Karyotype analysis of buckwheat using atomic force microscopy. Microsc. Microanal. 2011, 17, 572-577. [CrossRef]
- Wang, J.S.; Chai, Y.;Zhao,X.T.; JI,W. Q. Karyotype Analysis of Chinese Buckwheat Cultivars. Acta Bot. Boreali-Occident. Sin. 2005, 25, 1114-1117.
- Yang, X.Y.; Wu, Z. F.; Chen,H.; Shao,J. R.; Wu,Q. Karyotype and genetic relationship based on RAPD markers of six wild buckwheat species (Fagopyrum spp.) from southwest of China. Genet. Resour. Crop Evol. 2010, 57, 649-655. [CrossRef]
- Zhang, H.Z.; Guan, Z. X.; Liu,X. Y.; Liu,Y. H. . Karyotype analysis of Fagopyrum exculetum and F. tataricum. J. Inn. Mong Agric. Univ. 2000, 21, 69-74.
- Zhou, M.L.; Bai, D. Q.; Tang,Y.; Zhu, X. M.; Shao,J. R. Genetic diversity of four new species related to southwestern Sichuan buckwheats as revealed by karyotype, ISSR and allozyme characterization. Plant Syst. Evol. 2012, 298, 751-759. [CrossRef]
- Zhu, F.S.; Lin,R. F.; Li,Y. Q.; Niu,D. K. The initial report of studies on different types of chromosomes of Buckwheat. Chin. J. Cell Biol. 1984, 6, 130-131.
- Li, M.X.; Chen, R.Y. A suggestion on the standardization of karyotype analysis in plants. J. Wuhan Bot. Res. 1985, 3, 297-302.
- Levan, A.; Fredya, K.; Sandeberg, A. Nomenclature for centromeric position on chromosomes. Hereditas 1964, 52, 201-220. [CrossRef]
- Wang, W.X.; Li, C. Y.; Xiang,S. Q.; Liang,G. L. . Karyotypes Analysis and 45S rDNA-FISH of the Tetraploid and Diploid in Carica papaya L. Acta Horticulturae Sinica 2007, 34, 345-348.
- Zhang, H.M.; Zhang, S. N.;Yu,X. H.;Hou,X. L. Karyotype Analysis of Autotetraploid Broccoli. Acta Bot. Boreali-Occident. Sin. 2010, 30, 63-67.
- Melo, C.A.F.; Martins, M.I.G.; Oliveira, M.B.M.; Benko-Iseppon, A.M.; Carvalho, R. Karyotype analysis for diploid and polyploid species of the Solanum L. Plant Syst. Evol. 2011, 293, 227-235. [CrossRef]
- Zheng, J.S.; Zhang, S. N.; Sun,C. Z.; Wang,J. J.; Hou,X. L. Karyotype analysis of diploid and autotetraploid non-heading Chinese cabbage. J. Nanjing Agric. Univ. 2012, 35, 131-134.
- Yuan, J.M.; Dang, X. M.; Zhan,Y. F.; Li,Y. R.; Dan,Z.; Su,Y. L.; Mu,W. F.; Yang,C. K. . Karyotype analysis of diploid and autotetraploid mini-watermelon. Nor. Horticul. 2013, 40-43.
- Gao, F.H.; He, L. J.; Chen,H. J.; Liu,J. W.; Shan,Y. M.; Wang,W. J.; Yu,L. X. . Chromosome doubling and polyploid karyotype analysis of wild Lycium ruthenium. Mol. Plant Breed 2020, 18, 7522-7529.
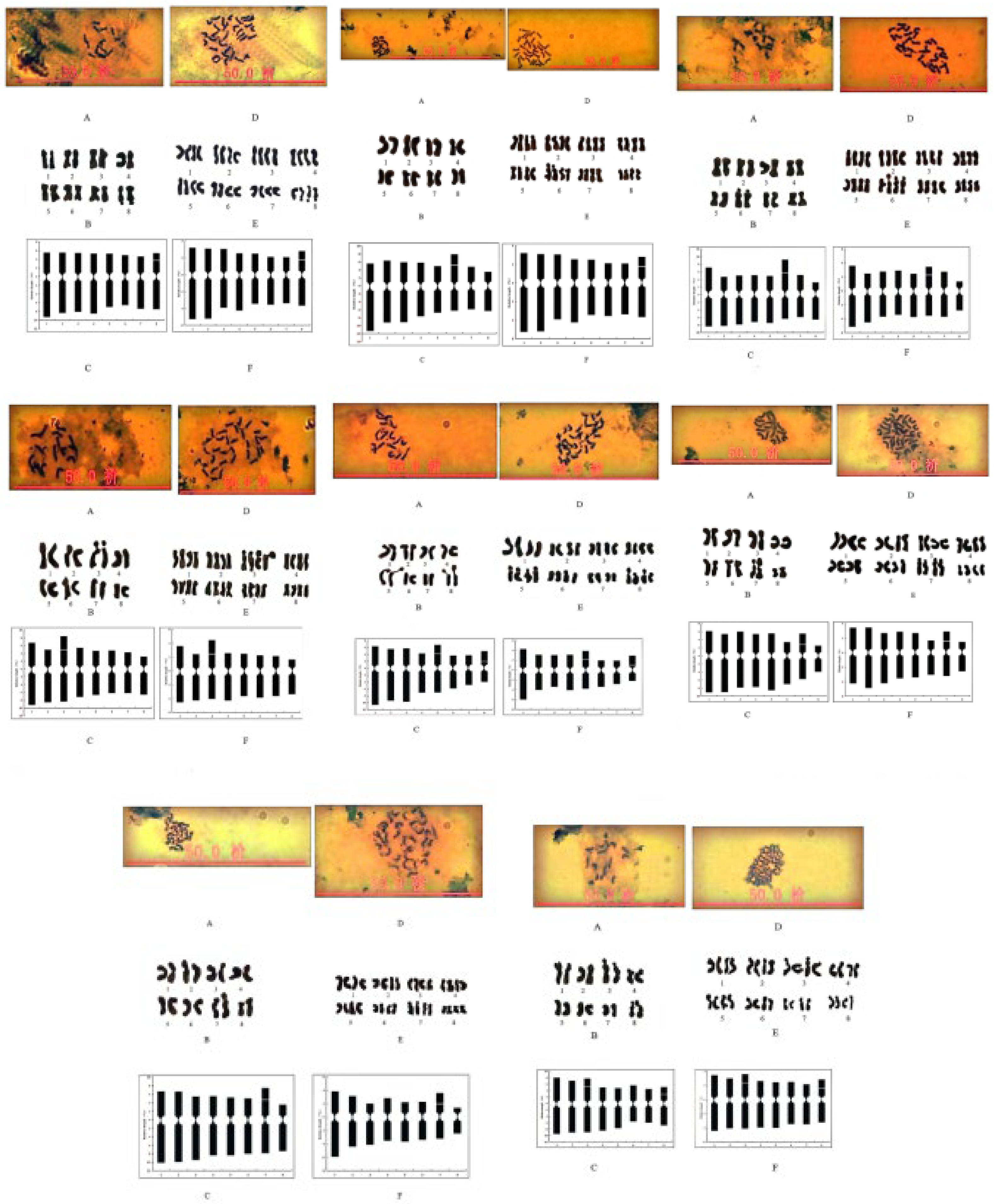
| Accessions (diploids) |
Spices | Sources | Accessions (autotetraploids) |
| T6 | Basu tartary buckwheat | Nala, Basu County, Tibet, China | T6* |
| T9 | WN090 | Weining, Guizhou, China | T9* |
| T18 | Wugang tartary Buckwheat | Wugang, Hunan, China | T18* |
| T22 | WN065 | Weining, Guizhou, China | T22* |
| T25 | Erjizao | Weining, Guizhou, China | T25* |
| T27 | Qianwei No.1 | Weining, Guizhou, China | T27* |
| T36 | Liuqiao No.1 | Liupanshui, Guizhou, China | T36* |
| T39 | Qianwei No.2 | Weining, Guizhou, China | T39* |
| Accessions | Karyotypic formula | A-SAT | P-SAT | Karyotpe type |
| T6 | 2n = 2x = 16 = 12m + 4sm (2SAT) | 2 | 8 | 2A |
| T6* | 2n = 4x = 32 = 24m + 8sm (4SAT) | 4 | 8 | 2A |
| T9 | 2n = 2x = 16 = 12m (2SAT) + 4sm | 2 | 6 | 2A |
| T9* | 2n = 4x = 32 = 24m(4SAT) + 8sm | 4 | 6 | 2A |
| T18 | 2n = 2x = 16 = 12m(2SAT) + 4sm | 2 | 6 | 2A |
| T18* | 2n = 4x = 32 = 24m(4SAT) + 8sm | 4 | 6 | 2A |
| T22 | 2n = 2x = 16 = 12m (2SAT) + 4sm | 2 | 3 | 2A |
| T22* | 2n = 4x = 32 = 24m(4SAT) + 8sm | 4 | 3 | 2A |
| T25 | 2n = 2x = 16 = 12m + 4sm(4SAT) | 4 | 5,8 | 2A |
| T25* | 2n = 4x = 32 = 24m + 8sm(8SAT) | 8 | 5,8 | 2A |
| T27 | 2n = 2x = 16 = 12m + 4sm(2SAT) | 2 | 7 | 2A |
| T27* | 2n = 4x = 32 = 24m + 8sm(4SAT) | 4 | 7 | 2A |
| T36 | 2n = 2x = 16 = 12m(2SAT) + 4sm | 2 | 7 | 2A |
| T36* | 2n = 4x = 32 = 24m(4SAT) + 8sm | 4 | 7 | 2A |
| T39 | 2n = 2x = 16 = 12m + 4sm(4SAT) | 4 | 3,8 | 2A |
| T39* | 2n = 4x = 32 = 24m + 8sm(8SAT) | 8 | 3,8 | 2A |
| Chromosomal code(Ploidy) | Accessions | ||||||||||||||||||||||||
| T6 | T9 | T18 | T22 | T25 | T27 | T36 | T39 | ||||||||||||||||||
| RL | AR | PC | RL | AR | PC | RL | AR | PC | RL | AR | PC | RL | AR | PC | RL | AR | PC | RL | AR | PC | RL | AR | PC | ||
| 1(2x) | 14.29 | 1.67 | m | 15.79 | 2.01 | sm | 15.14 | 1.17 | m | 15.73 | 1.36 | m | 17.06 | 1.66 | m | 15.38 | 1.46 | m | 14.59 | 1.50 | m | 15.90 | 1.19 | m | |
| 2(2x) | 13.37 | 1.51 | m | 14.48 | 1.44 | m | 12.60 | 1.74 | sm | 13.32 | 1.71 | sm | 15.18 | 1.68 | m | 14.57 | 1.65 | m | 14.37 | 1.49 | m | 14.69 | 1.27 | m | |
| 3(2x) | 13.03 | 1.48 | m | 14.03 | 1.53 | m | 12.58 | 1.55 | m | 13.06 | 1.69 | m# | 15.71 | 1.67 | m | 14.03 | 1.30 | m | 12.99 | 1.71 | sm | 12.85 | 1.74 | sm# | |
| 4(2x) | 13.32 | 1.59 | m | 12.55 | 1.30 | m | 12.00 | 1.40 | m | 12.45 | 1.30 | m | 11.55 | 1.64 | m | 13.30 | 1.41 | m | 12.10 | 1.50 | m | 12.21 | 1.70 | m | |
| 5(2x) | 11.83 | 1.31 | m | 10.89 | 1.36 | m | 12.11 | 1.51 | m | 11.19 | 1.42 | m | 11.25 | 1.71 | sm# | 14.24 | 1.52 | m | 11.83 | 1.58 | m | 11.32 | 1.62 | m | |
| 6(2x) | 11.10 | 1.32 | m | 10.55 | 1.22 | m# | 11.75 | 1.12 | m# | 10.81 | 1.29 | m | 9.68 | 1.58 | m | 10.54 | 2.02 | sm | 11.33 | 1.57 | m | 10.24 | 1.00 | m | |
| 7(2x) | 10.63 | 2.02 | sm | 9.93 | 1.22 | m | 10.81 | 1.14 | m | 10.33 | 1.43 | m | 8.74 | 1.29 | m | 8.56 | 2.07 | sm# | 10.83 | 1.68 | m# | 9.57 | 1.33 | m | |
| 8(2x) | 10.75 | 2.03 | sm# | 9.17 | 1.77 | sm | 9.54 | 2.01 | sm | 9.65 | 2.03 | sm | 5.90 | 2.08 | sm# | 6.55 | 1.53 | m | 9.49 | 2.04 | sm | 8.83 | 2.36 | sm# | |
| 1(4x) | 15.72 | 1.63 | m | 14.83 | 2.05 | sm | 16.68 | 1.42 | m | 15.25 | 1.26 | m | 19.02 | 1.39 | m | 15.25 | 1.22 | m | 20.30 | 1.53 | m | 14.76 | 1.27 | m | |
| 2(4x) | 15.41 | 1.66 | m | 13.27 | 1.64 | m | 13.35 | 1.80 | sm | 12.59 | 1.73 | sm | 13.43 | 1.29 | m | 16.54 | 1.39 | m | 15.69 | 1.39 | m | 13.27 | 1.34 | m | |
| 3(4x) | 12.96 | 1.26 | m | 12.93 | 1.70 | m | 12.36 | 1.32 | m | 12.12 | 1.67 | m# | 12.11 | 1.10 | m | 13.75 | 1.54 | m | 12.78 | 2.01 | sm | 12.10 | 1.75 | sm# | |
| 4(4x) | 12.63 | 1.65 | m | 12.40 | 1.49 | m | 12.13 | 1.16 | m | 12.85 | 1.65 | m | 13.51 | 1.30 | m | 12.75 | 1.21 | m | 13.20 | 1.23 | m | 12.33 | 1.50 | m | |
| 5(4x) | 11.07 | 1.36 | m | 12.14 | 1.25 | m | 11.81 | 1.51 | m | 11.52 | 1.45 | m | 11.18 | 1.72 | sm# | 12.13 | 1.25 | m | 12.20 | 1.63 | m | 12.11 | 1.63 | m | |
| 6(4x) | 10.55 | 1.66 | m | 11.76 | 1.47 | m# | 11.15 | 1.55 | m | 11.51 | 1.70 | m | 10.05 | 1.70 | m | 9.39 | 1.79 | sm | 11.81 | 1.46 | m# | 11.15 | 1.38 | m | |
| 7(4x) | 9.60 | 1.86 | sm | 11.18 | 1.61 | m | 12.35 | 1.34 | m | 10.86 | 1.67 | m | 8.91 | 1.45 | m | 9.48 | 2.01 | sm# | 10.73 | 1.63 | m | 10.60 | 1.60 | m | |
| 8(4x) | 10.05 | 2.08 | sm# | 9.57 | 1.78 | sm | 7.93 | 2.04 | sm | 9.44 | 2.02 | sm | 6.04 | 2.07 | sm# | 8.05 | 1.69 | m | 7.94 | 1.73 | sm | 9.01 | 2.02 | sm# | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





