Submitted:
24 September 2024
Posted:
25 September 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Genetic Engineering in Bacteria
2.1. Techniques and Tools
2.2. Applications
2.3. Challenges and Future Directions
3. Genetic Engineering in Fungi
3.1. Transformation Techniques
3.2. CRISPR-Cas9
3.3. Applications
3.4. Challenges and Innovations
4. Genetic Engineering in Oomycetes
4.1. Challenges in Oomycete Genetics
4.2. Recent Advances
4.3. Applications
4.4. Future Prospects
5. Comparative Analysis
5.1. Comparison of Techniques
5.2. Case Studies
6. Ethical and Regulatory Considerations
6.1. Safety Concerns
6.2. Regulatory Frameworks
7. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Paul, N.C.; Park, S.-W.; Liu, H.; Choi, S.; Ma, J.; MacCready, J.S.; Chilvers, M.I.; Sang, H. Plant and Fungal Genome Editing to Enhance Plant Disease Resistance Using the CRISPR/Cas9 System. Front. Plant Sci. 2021, 12. [Google Scholar] [CrossRef]
- Wang, L.; Zhao, F.; Liu, H.; Chen, H.; Zhang, F.; Li, S.; Sun, T.; Nekrasov, V.; Huang, S.; Dong, S. A Modified Agrobacterium-Mediated Transformation for Two Oomycete Pathogens. PLOS Pathogens 2023, 19, e1011346. [Google Scholar] [CrossRef]
- Oliveira, L.R.; Gonçalves, A.R.; Quintela, E.D.; Colognese, L.; Cortes, M.V. de C.B.; de Filippi, M.C.C. Genetic Engineering of Filamentous Fungi: Prospects for Obtaining Fourth-Generation Biological Products. Applied Microbiology 2024, 4, 794–810. [Google Scholar] [CrossRef]
- Yang, P.; Bokros, N.; Debolt, S.; Zhao, Z.; Xia, Y. Genome Sequence Source of Bacillus Amyloliquefaciens Strain GD4a, a Bacterial Endophyte Associated with Switchgrass Plants. Phytobiomes Journal 2022, 6, 354–357. [Google Scholar] [CrossRef]
- Malkawi, H.I.; Kapiel, T.Y.S. Microbial Biotechnology: A Key Tool for Addressing Climate Change and Food Insecurity. European Journal of Biology and Biotechnology 2024, 5, 1–15. [Google Scholar] [CrossRef]
- Santos-Beneit, F. What Is the Role of Microbial Biotechnology and Genetic Engineering in Medicine? Microbiologyopen 2024, 13, e1406. [Google Scholar] [CrossRef]
- Roth, M.G.; Westrick, N.M.; Baldwin, T.T. Fungal Biotechnology: From Yesterday to Tomorrow. Front. Fungal Biol. 2023, 4. [Google Scholar] [CrossRef]
- Corbu, V.M.; Gheorghe-Barbu, I.; Dumbravă, A. Ștefania; Vrâncianu, C.O.; Șesan, T.E. Current Insights in Fungal Importance—A Comprehensive Review. Microorganisms 2023, 11, 1384. [Google Scholar] [CrossRef]
- Sharma, S.; Sundaresha, S.; Bhardwaj, V. Biotechnological Approaches in Management of Oomycetes Diseases. 3 Biotech 2021, 11, 274. [Google Scholar] [CrossRef]
- Blakney, A.J.C.; Bainard, L.D.; St-Arnaud, M.; Hijri, M. Soil Chemistry and Soil History Significantly Structure Oomycete Communities in Brassicaceae Crop Rotations. Appl Environ Microbiol 89, e01314-22. [CrossRef]
- Li, T.; Yang, Y.; Qi, H.; Cui, W.; Zhang, L.; Fu, X.; He, X.; Liu, M.; Li, P.; Yu, T. CRISPR/Cas9 Therapeutics: Progress and Prospects. Sig Transduct Target Ther 2023, 8, 1–23. [Google Scholar] [CrossRef]
- Rabaan, A.A.; Al Fares, M.A.; Almaghaslah, M.; Alpakistany, T.; Al Kaabi, N.A.; Alshamrani, S.A.; Alshehri, A.A.; Almazni, I.A.; Saif, A.; Hakami, A.R.; et al. Application of CRISPR-Cas System to Mitigate Superbug Infections. Microorganisms 2023, 11, 2404. [Google Scholar] [CrossRef]
- Vialetto, E.; Miele, S.; Goren, M.G.; Yu, J.; Yu, Y.; Collias, D.; Beamud, B.; Osbelt, L.; Lourenço, M.; Strowig, T.; et al. Systematic Interrogation of CRISPR Antimicrobials in Klebsiella Pneumoniae Reveals Nuclease-, Guide- and Strain-Dependent Features Influencing Antimicrobial Activity. Nucleic Acids Research 2024, 52, 6079–6091. [Google Scholar] [CrossRef]
- Fels, U.; Gevaert, K.; Van Damme, P. Bacterial Genetic Engineering by Means of Recombineering for Reverse Genetics. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef]
- Li, R.; Li, A.; Zhang, Y.; Fu, J. The Emerging Role of Recombineering in Microbiology. Engineering Microbiology 2023, 3, 100097. [Google Scholar] [CrossRef]
- Matuszyńska, A.; Ebenhöh, O.; Zurbriggen, M.D.; Ducat, D.C.; Axmann, I.M. A New Era of Synthetic Biology—Microbial Community Design. Synthetic Biology 2024, 9, ysae011. [Google Scholar] [CrossRef]
- Sudheer, S.; Bai, R.G.; Usmani, Z.; Sharma, M. Insights on Engineered Microbes in Sustainable Agriculture: Biotechnological Developments and Future Prospects. Curr Genomics 2020, 21, 321–333. [Google Scholar] [CrossRef]
- Kamionka, M. Engineering of Therapeutic Proteins Production in Escherichia Coli. Curr Pharm Biotechnol 2011, 12, 268–274. [Google Scholar] [CrossRef]
- Niazi, S.K.; Magoola, M. Advances in Escherichia Coli-Based Therapeutic Protein Expression: Mammalian Conversion, Continuous Manufacturing, and Cell-Free Production. Biologics 2023, 3, 380–401. [Google Scholar] [CrossRef]
- Mugwanda, K.; Hamese, S.; Van Zyl, W.F.; Prinsloo, E.; Du Plessis, M.; Dicks, L.M.T.; Thimiri Govinda Raj, D.B. Recent Advances in Genetic Tools for Engineering Probiotic Lactic Acid Bacteria. Biosci Rep 2023, 43, BSR20211299. [Google Scholar] [CrossRef]
- Dell’ Anno, F.; Rastelli, E.; Sansone, C.; Brunet, C.; Ianora, A.; Dell’ Anno, A. Bacteria, Fungi and Microalgae for the Bioremediation of Marine Sediments Contaminated by Petroleum Hydrocarbons in the Omics Era. Microorganisms 2021, 9, 1695. [Google Scholar] [CrossRef]
- Bala, S.; Garg, D.; Thirumalesh, B.V.; Sharma, M.; Sridhar, K.; Inbaraj, B.S.; Tripathi, M. Recent Strategies for Bioremediation of Emerging Pollutants: A Review for a Green and Sustainable Environment. Toxics 2022, 10, 484. [Google Scholar] [CrossRef] [PubMed]
- Garg, D.; Samota, M.K.; Kontis, N.; Patel, N.; Bala, S.; Rosado, A.S. Revolutionizing Biofuel Generation: Unleashing the Power of CRISPR-Cas Mediated Gene Editing of Extremophiles. Microbiological Research 2023, 274, 127443. [Google Scholar] [CrossRef] [PubMed]
- Yao, R.; Liu, D.; Jia, X.; Zheng, Y.; Liu, W.; Xiao, Y. CRISPR-Cas9/Cas12a Biotechnology and Application in Bacteria. Synthetic and Systems Biotechnology 2018, 3, 135–149. [Google Scholar] [CrossRef]
- Adegboye, M.F.; Ojuederie, O.B.; Talia, P.M.; Babalola, O.O. Bioprospecting of Microbial Strains for Biofuel Production: Metabolic Engineering, Applications, and Challenges. Biotechnology for Biofuels 2021, 14, 5. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Fong, S.S. Challenges and Advances for Genetic Engineering of Non-Model Bacteria and Uses in Consolidated Bioprocessing. Front Microbiol 2017, 8, 2060. [Google Scholar] [CrossRef]
- Egbert, R.; Elmore, J.; Guss, A. Taming Undomesticated Bacteria with a High-Efficiency Genome Engineering Tool Available online: https://www.energy.gov/science/ber/articles/taming-undomesticated-bacteria-high-efficiency-genome-engineering-tool (accessed on 26 August 2024). (accessed on null).
- Zhao, Z.; Bokros, N.; DeBolt, S.; Yang, P.; Xia, Y. Genome Sequence Resource of Bacillus Sp. RRD69, a Beneficial Bacterial Endophyte Isolated from Switchgrass Plants. MPMI 2021, 34, 1320–1323. [Google Scholar] [CrossRef]
- Yang, P.; Yuan, P.; Liu, W.; Zhao, Z.; Bernier, M.C.; Zhang, C.; Adhikari, A.; Opiyo, S.O.; Zhao, L.; Banks, F.; et al. Plant Growth Promotion and Plant Disease Suppression Induced by Bacillus Amyloliquefaciens Strain GD4a. Plants 2024, 13, 672. [Google Scholar] [CrossRef] [PubMed]
- Yang, P. Exploring Plant-Microbe Interactions through the Lens of Beneficial Bacteria, The Ohio State University, 2023.
- Sahib, M.R.; Yang, P.; Bokros, N.; Shapiro, N.; Woyke, T.; Kyrpides, N.C.; Xia, Y.; DeBolt, S. Improved Draft Genome Sequence of Microbacterium Sp. Strain LKL04, a Bacterial Endophyte Associated with Switchgrass Plants. Microbiol Resour Announc 2019, 8. [Google Scholar] [CrossRef]
- Rodriguez-Iglesias, A.; Schmoll, M. Protoplast Transformation for Genome Manipulation in Fungi. In Genetic Transformation Systems in Fungi, Volume 1; van den Berg, M.A., Maruthachalam, K., Eds.; Springer International Publishing: Cham, 2015; pp. 21–40. ISBN 978-3-319-10142-2. [Google Scholar]
- Zhang, Q.; Zhao, L.; Shen, M.; Liu, J.; Li, Y.; Xu, S.; Chen, L.; Shi, G.; Ding, Z. Establishment of an Efficient Polyethylene Glycol (PEG)-Mediated Transformation System in Pleurotus Eryngii Var. Ferulae Using Comprehensive Optimization and Multiple Endogenous Promoters. J Fungi (Basel) 2022, 8, 186. [Google Scholar] [CrossRef]
- Su, Z.-Z.; Dai, M.-D.; Zhu, J.-N.; Zeng, Y.-L.; Lu, X.-J.; Liu, X.-H.; Lin, F.-C. An Efficient Genetic Manipulation Protocol for Dark Septate Endophyte Falciphora Oryzae. Biotechnol Lett 2021, 43, 2045–2052. [Google Scholar] [CrossRef]
- Li, G.; Li, R.; Liu, Q.; Wang, Q.; Chen, M.; Li, B. A Highly Efficient Polyethylene Glycol-Mediated Transformation Method for Mushrooms. FEMS Microbiology Letters 2006, 256, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Sbaraini, N.; Tomazett, M.V.; Penteriche, A.B.; Gonçales, R.A.; Camargo, M. da S.; Bailão, A.M.; Borges, C.L.; Schrank, A.; Soares, C.M. de A.; Staats, C.C. An Efficient Agrobacterium Tumefaciens-Mediated Transformation Method for Simplicillium Subtropicum (Hypocreales: Cordycipitaceae). Genet Mol Biol 2021, 44, e20210073. [Google Scholar] [CrossRef] [PubMed]
- Ropero-Pérez, C.; Manzanares, P.; Marcos, J.F.; Garrigues, S. Agrobacterium Tumefaciens-Mediated Transformation for the Genetic Modification of the Biotechnologically Relevant Fungus Aspergillus Vadensis through Synthetic Biology. Current Research in Biotechnology 2024, 7, 100178. [Google Scholar] [CrossRef]
- Combier, J.-P.; Melayah, D.; Raffier, C.; Gay, G.; Marmeisse, R. Agrobacterium Tumefaciens-Mediated Transformation as a Tool for Insertional Mutagenesis in the Symbiotic Ectomycorrhizal Fungus Hebeloma Cylindrosporum. FEMS Microbiology Letters 2003, 220, 141–148. [Google Scholar] [CrossRef]
- Wei, T.-Y.; Wu, Y.-J.; Xie, Q.-P.; Tang, J.-W.; Yu, Z.-T.; Yang, S.-B.; Chen, S.-X. CRISPR/Cas9-Based Genome Editing in the Filamentous Fungus Glarea Lozoyensis and Its Application in Manipulating gloF. ACS Synth. Biol. 2020, 9, 1968–1977. [Google Scholar] [CrossRef] [PubMed]
- Wang, P. Two Distinct Approaches for CRISPR-Cas9-Mediated Gene Editing in Cryptococcus Neoformans and Related Species. mSphere 2018, 3. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, S.; Yang, L.; Zhang, Q. Application Progress of CRISPR/Cas9 Genome-Editing Technology in Edible Fungi. Front Microbiol 2023, 14, 1169884. [Google Scholar] [CrossRef] [PubMed]
- Weber, J.; Valiante, V.; Nødvig, C.S.; Mattern, D.J.; Slotkowski, R.A.; Mortensen, U.H.; Brakhage, A.A. Functional Reconstitution of a Fungal Natural Product Gene Cluster by Advanced Genome Editing. ACS Synth. Biol. 2017, 6, 62–68. [Google Scholar] [CrossRef]
- Wang, D.; Jin, S.; Lu, Q.; Chen, Y. Advances and Challenges in CRISPR/Cas-Based Fungal Genome Engineering for Secondary Metabolite Production: A Review. J Fungi (Basel) 2023, 9, 362. [Google Scholar] [CrossRef]
- Michaliski, L.F.; Ióca, L.P.; Oliveira, L.S.; Crnkovic, C.M.; Takaki, M.; Freire, V.F.; Berlinck, R.G.S. Improvement of Targeted Fungi Secondary Metabolite Production Using a Systematic Experimental Design and Chemometrics Analysis. Methods and Protocols 2023, 6, 77. [Google Scholar] [CrossRef]
- van Esse, H.P.; Reuber, T.L.; van der Does, D. Genetic Modification to Improve Disease Resistance in Crops. New Phytologist 2020, 225, 70–86. [Google Scholar] [CrossRef] [PubMed]
- Mekapogu, M.; Jung, J.-A.; Kwon, O.-K.; Ahn, M.-S.; Song, H.-Y.; Jang, S. Recent Progress in Enhancing Fungal Disease Resistance in Ornamental Plants. Int J Mol Sci 2021, 22, 7956. [Google Scholar] [CrossRef] [PubMed]
- Maini Rekdal, V.; van der Luijt, C.R.B.; Chen, Y.; Kakumanu, R.; Baidoo, E.E.K.; Petzold, C.J.; Cruz-Morales, P.; Keasling, J.D. Edible Mycelium Bioengineered for Enhanced Nutritional Value and Sensory Appeal Using a Modular Synthetic Biology Toolkit. Nat Commun 2024, 15, 2099. [Google Scholar] [CrossRef]
- Wang, Q.; Zhong, C.; Xiao, H. Genetic Engineering of Filamentous Fungi for Efficient Protein Expression and Secretion. Front. Bioeng. Biotechnol. 2020, 8. [Google Scholar] [CrossRef]
- Ropero-Pérez, C.; Marcos, J.F.; Manzanares, P.; Garrigues, S. Increasing the Efficiency of CRISPR/Cas9-Mediated Genome Editing in the Citrus Postharvest Pathogen Penicillium Digitatum. Fungal Biology and Biotechnology 2024, 11, 8. [Google Scholar] [CrossRef] [PubMed]
- Yin, C.; Hulbert, S.H. Host-Induced Gene Silencing (HIGS) for Elucidating Puccinia Gene Function in Wheat. In Plant Pathogenic Fungi and Oomycetes: Methods and Protocols; Ma, W., Wolpert, T., Eds.; Springer: New York, NY, 2018; pp. 139–150. ISBN 978-1-4939-8724-5. [Google Scholar]
- Feng, H.; Liu, T.; Li, J.; Wan, C.; Ding, F.; Wang, Y.; Zheng, X.; Ye, W. Gene Editing with an Oxathiapiprolin Resistance Selection Marker Reveals That PuLLP, a Loricrin-like Protein, Is Required for Oospore Development in Pythium Ultimum. Phytopathology Research 2023, 5, 34. [Google Scholar] [CrossRef]
- Lamour, K.H.; Win, J.; Kamoun, S. Oomycete Genomics: New Insights and Future Directions. FEMS Microbiol Lett 2007, 274, 1–8. [Google Scholar] [CrossRef]
- Vink, J.N.A.; Hayhurst, M.; Gerth, M.L. Harnessing CRISPR-Cas for Oomycete Genome Editing. Trends Microbiol 2023, 31, 947–958. [Google Scholar] [CrossRef]
- Wang, W.; Xue, Z.; Miao, J.; Cai, M.; Zhang, C.; Li, T.; Zhang, B.; Tyler, B.M.; Liu, X. PcMuORP1, an Oxathiapiprolin-Resistance Gene, Functions as a Novel Selection Marker for Phytophthora Transformation and CRISPR/Cas9 Mediated Genome Editing. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- Ah-Fong, A.M.V.; Boyd, A.M.; Matson, M.E.H.; Judelson, H.S. A Cas12a-Based Gene Editing System for Phytophthora Infestans Reveals Monoallelic Expression of an Elicitor. Molecular Plant Pathology 2021, 22, 737–752. [Google Scholar] [CrossRef]
- Mendoza, C.S.; Findlay, A.; Judelson, H.S. A Variant of LbCas12a and Elevated Incubation Temperatures Enhance the Rate of Gene Editing in the Oomycete Phytophthora Infestans. MPMI 2023, 36, 677–681. [Google Scholar] [CrossRef]
- Verhoeven, T.; Pluis, M.H.; Peippo, M.; Couillaud, G.; Berg, G.C. van den; Evangelisti, E. LbCas12-Mediated Multiplex Gene Editing and 2-Fluoroadenine Counter-Selection in Phytophthora Palmivora 2024, 2024. 02.13.58 0060.
- Langner, T.; Kamoun, S.; Belhaj, K. CRISPR Crops: Plant Genome Editing Toward Disease Resistance. Annual Review of Phytopathology 2018, 56, 479–512. [Google Scholar] [CrossRef]
- Borrelli, V.M.G.; Brambilla, V.; Rogowsky, P.; Marocco, A.; Lanubile, A. The Enhancement of Plant Disease Resistance Using CRISPR/Cas9 Technology. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef]
- Kamoun, S. Molecular Genetics of Pathogenic Oomycetes. Eukaryot Cell 2003, 2, 191–199. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Karlin, S.; Mrázek, J. Predicted Highly Expressed Genes of Diverse Prokaryotic Genomes. Journal of Bacteriology 2000, 182, 5238–5250. [Google Scholar] [CrossRef]
- Jin, F.-J.; Hu, S.; Wang, B.-T.; Jin, L. Advances in Genetic Engineering Technology and Its Application in the Industrial Fungus Aspergillus Oryzae. Front. Microbiol. 2021, 12. [Google Scholar] [CrossRef]
- Yılmaz, Ş.G. Genome Editing Technologies: CRISPR, LEAPER, RESTORE, ARCUT, SATI, and RESCUE. EXCLI Journal 2021, 20, 19–45. [Google Scholar] [CrossRef]
- Capriotti, L.; Baraldi, E.; Mezzetti, B.; Limera, C.; Sabbadini, S. Biotechnological Approaches: Gene Overexpression, Gene Silencing, and Genome Editing to Control Fungal and Oomycete Diseases in Grapevine. International Journal of Molecular Sciences 2020, 21, 5701. [Google Scholar] [CrossRef]
- Ghimire, B.; Saraiva, M.; Andersen, C.B.; Gogoi, A.; Saleh, M.; Zic, N.; van West, P.; Brurberg, M.B. Transformation Systems, Gene Silencing and Gene Editing Technologies in Oomycetes. Fungal Biology Reviews 2022, 40, 37–52. [Google Scholar] [CrossRef]
- Kamoun, S.; Huitema, E.; Vleeshouwers, V.G. Resistance to Oomycetes: A General Role for the Hypersensitive Response? Trends Plant Sci 1999, 4, 196–200. [Google Scholar] [CrossRef]
- Pedrolli, D.B.; Ribeiro, N.V.; Squizato, P.N.; de Jesus, V.N.; Cozetto, D.A.; Tuma, R.B.; Gracindo, A.; Cesar, M.B.; Freire, P.J.C.; da Costa, A.F.M.; et al. Engineering Microbial Living Therapeutics: The Synthetic Biology Toolbox. Trends in Biotechnology 2019, 37, 100–115. [Google Scholar] [CrossRef]
- Peters, J.M.; Koo, B.-M.; Patino, R.; Heussler, G.E.; Hearne, C.C.; Qu, J.; Inclan, Y.F.; Hawkins, J.S.; Lu, C.H.S.; Silvis, M.R.; et al. Enabling Genetic Analysis of Diverse Bacteria with Mobile-CRISPRi. Nature Microbiology 2019, 4, 244–250. [Google Scholar] [CrossRef] [PubMed]
- Arnold, J.; Glazier, J.; Mimee, M. Genetic Engineering of Resident Bacteria in the Gut Microbiome. J Bacteriol 205, e00127-23. [CrossRef]
- Pérez, G.; Lopez-Moya, F.; Chuina, E.; Ibañez-Vea, M.; Garde, E.; López-Llorca, L.V.; Pisabarro, A.G.; Ramírez, L. Strain Degeneration in Pleurotus Ostreatus: A Genotype Dependent Oxidative Stress Process Which Triggers Oxidative Stress, Cellular Detoxifying and Cell Wall Reshaping Genes. Journal of Fungi 2021, 7, 862. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, M.S.; Melo, E.O. Animal Transgenesis and Cloning: Combined Development and Future Perspectives. Methods Mol Biol 2023, 2647, 121–149. [Google Scholar] [CrossRef]
- Yaashikaa, P.R.; Devi, M.K.; Kumar, P.S. Engineering Microbes for Enhancing the Degradation of Environmental Pollutants: A Detailed Review on Synthetic Biology. Environmental Research 2022, 214, 113868. [Google Scholar] [CrossRef]
- Pant, G.; Garlapati, D.; Agrawal, U.; Prasuna, R.G.; Mathimani, T.; Pugazhendhi, A. Biological Approaches Practised Using Genetically Engineered Microbes for a Sustainable Environment: A Review. J Hazard Mater 2021, 405, 124631. [Google Scholar] [CrossRef]
- Bauer-Panskus, A.; Miyazaki, J.; Kawall, K.; Then, C. Risk Assessment of Genetically Engineered Plants That Can Persist and Propagate in the Environment. Environmental Sciences Europe 2020, 32, 32. [Google Scholar] [CrossRef]
- Green Genetic Engineering Law Available online: https://www.leopoldina.org/en/topics/green-genetic-engineering/green-genetic-engineering-law/ (accessed on 27 August 2024). (accessed on null).
- Hanlon, P.; Sewalt, V. GEMs: Genetically Engineered Microorganisms and the Regulatory Oversight of Their Uses in Modern Food Production. Critical Reviews in Food Science and Nutrition 2021, 61, 959–970. [Google Scholar] [CrossRef]
- Sprink, T.; Wilhelm, R. Genome Editing in Biotech Regulations Worldwide. In A Roadmap for Plant Genome Editing; Ricroch, A., Eriksson, D., Miladinović, D., Sweet, J., Van Laere, K., Woźniak-Gientka, E., Eds.; Springer Nature Switzerland: Cham, 2024; pp. 425–435. ISBN 978-3-031-46150-7. [Google Scholar]
- Vos, P.D.; Rossetti, G.; Mantegna, J.L.; Siira, S.J.; Gandadireja, A.P.; Bruce, M.; Raven, S.A.; Khersonsky, O.; Fleishman, S.J.; Filipovska, A.; et al. Computationally Designed Hyperactive Cas9 Enzymes. Nat Commun 2022, 13, 3023. [Google Scholar] [CrossRef] [PubMed]
- Cloud-Based Platform for Biotech R&D | Benchling. Available online: https://www.benchling.com (accessed on 2 September 2024).
- Bae, S.; Park, J.; Kim, J.-S. Cas-OFFinder: A Fast and Versatile Algorithm That Searches for Potential off-Target Sites of Cas9 RNA-Guided Endonucleases. Bioinformatics 2014, 30, 1473–1475. [Google Scholar] [CrossRef] [PubMed]
- Labuhn, M.; Adams, F.F.; Ng, M.; Knoess, S.; Schambach, A.; Charpentier, E.M.; Schwarzer, A.; Mateo, J.L.; Klusmann, J.-H.; Heckl, D. Refined sgRNA Efficacy Prediction Improves Large- and Small-Scale CRISPR–Cas9 Applications. Nucleic Acids Research 2018, 46, 1375–1385. [Google Scholar] [CrossRef] [PubMed]
- Stemmer, M.; Thumberger, T.; Keyer, M. del S.; Wittbrodt, J.; Mateo, J.L. CCTop: An Intuitive, Flexible and Reliable CRISPR/Cas9 Target Prediction Tool. PLOS ONE 2015, 10, e0124633. [Google Scholar] [CrossRef]
- Labun, K.; Montague, T.G.; Krause, M.; Torres Cleuren, Y.N.; Tjeldnes, H.; Valen, E. CHOPCHOP v3: Expanding the CRISPR Web Toolbox beyond Genome Editing. Nucleic Acids Research 2019, 47, W171–W174. [Google Scholar] [CrossRef] [PubMed]
- Concordet, J.-P.; Haeussler, M. CRISPOR: Intuitive Guide Selection for CRISPR/Cas9 Genome Editing Experiments and Screens. Nucleic Acids Research 2018, 46, W242–W245. [Google Scholar] [CrossRef]
- Naito, Y.; Hino, K.; Bono, H.; Ui-Tei, K. CRISPRdirect: Software for Designing CRISPR/Cas Guide RNA with Reduced off-Target Sites. Bioinformatics 2015, 31, 1120–1123. [Google Scholar] [CrossRef]
- Liu, H.; Wei, Z.; Dominguez, A.; Li, Y.; Wang, X.; Qi, L.S. CRISPR-ERA: A Comprehensive Design Tool for CRISPR-Mediated Gene Editing, Repression and Activation. Bioinformatics 2015, 31, 3676–3678. [Google Scholar] [CrossRef]
- CRISPick Available online: https://portals.broadinstitute.org/gppx/crispick/public (accessed on 2 September 2024).
- CRISPR-Cas9 Guide RNA Design Checker | IDT Available online: https://www.idtdna.com/site/order/designtool/index/CRISPR_SEQUENCE (accessed on 2 September 2024).
- Pliatsika, V.; Rigoutsos, I. “Off-Spotter”: Very Fast and Exhaustive Enumeration of Genomic Lookalikes for Designing CRISPR/Cas Guide RNAs. Biology Direct 2015, 10, 4. [Google Scholar] [CrossRef]
- Enzmann, B.L.; Wronski, A. Synthego’s Engineered Cells Allow Scientists to “Cut Out” CRISPR Optimization [SPONSORED]. The CRISPR Journal 2018, 1, 255–257. [Google Scholar] [CrossRef]
- Patel, A.; Iannello, G.; Diaz, A.G.; Sirabella, D.; Thaker, V.; Corneo, B. Efficient Cas9-Based Genome Editing Using CRISPR Analysis Webtools in Severe Early-Onset-Obesity Patient-Derived iPSCs. Current Protocols 2022, 2, e519. [Google Scholar] [CrossRef]
- Yamada, S.; Suzuki, Y.; Kouzuma, A.; Watanabe, K. Development of a CRISPR Interference System for Selective Gene Knockdown in Acidithiobacillus Ferrooxidans. Journal of Bioscience and Bioengineering 2022, 133, 105–109. [Google Scholar] [CrossRef] [PubMed]
- Schultenkämper, K.; Brito, L.F.; López, M.G.; Brautaset, T.; Wendisch, V.F. Establishment and Application of CRISPR Interference to Affect Sporulation, Hydrogen Peroxide Detoxification, and Mannitol Catabolism in the Methylotrophic Thermophile Bacillus Methanolicus. Appl Microbiol Biotechnol 2019, 103, 5879–5889. [Google Scholar] [CrossRef] [PubMed]
- Mougiakos, I.; Mohanraju, P.; Bosma, E.F.; Vrouwe, V.; Finger Bou, M.; Naduthodi, M.I.S.; Gussak, A.; Brinkman, R.B.L.; van Kranenburg, R.; van der Oost, J. Characterizing a Thermostable Cas9 for Bacterial Genome Editing and Silencing. Nat Commun 2017, 8, 1647. [Google Scholar] [CrossRef] [PubMed]
- Mougiakos, I.; Bosma, E.F.; Weenink, K.; Vossen, E.; Goijvaerts, K.; van der Oost, J.; van Kranenburg, R. Efficient Genome Editing of a Facultative Thermophile Using Mesophilic spCas9. ACS Synth. Biol. 2017, 6, 849–861. [Google Scholar] [CrossRef]
- Altenbuchner, J. Editing of the Bacillus Subtilis Genome by the CRISPR-Cas9 System. Applied and Environmental Microbiology 2016, 82, 5421–5427. [Google Scholar] [CrossRef]
- Wasels, F.; Jean-Marie, J.; Collas, F.; López-Contreras, A.M.; Lopes Ferreira, N. A Two-Plasmid Inducible CRISPR/Cas9 Genome Editing Tool for Clostridium Acetobutylicum. Journal of Microbiological Methods 2017, 140, 5–11. [Google Scholar] [CrossRef]
- Nagaraju, S.; Davies, N.K.; Walker, D.J.F.; Köpke, M.; Simpson, S.D. Genome Editing of Clostridium Autoethanogenum Using CRISPR/Cas9. Biotechnol Biofuels 2016, 9, 219. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Z.-T.; Seo, S.-O.; Lynn, P.; Lu, T.; Jin, Y.-S.; Blaschek, H.P. Bacterial Genome Editing with CRISPR-Cas9: Deletion, Integration, Single Nucleotide Modification, and Desirable “Clean” Mutant Selection in Clostridium Beijerinckii as an Example. ACS Synth. Biol. 2016, 5, 721–732. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Li, Y.; Shi, Z.; Hemme, C.L.; Li, Y.; Zhu, Y.; Van Nostrand, J.D.; He, Z.; Zhou, J. Efficient Genome Editing in Clostridium Cellulolyticum via CRISPR-Cas9 Nickase. Applied and Environmental Microbiology 2015, 81, 4423–4431. [Google Scholar] [CrossRef]
- Cleto, S.; Jensen, J.V.; Wendisch, V.F.; Lu, T.K. Corynebacterium Glutamicum Metabolic Engineering with CRISPR Interference (CRISPRi). ACS Synth. Biol. 2016, 5, 375–385. [Google Scholar] [CrossRef]
- Woolston, B.M.; Emerson, D.F.; Currie, D.H.; Stephanopoulos, G. Rediverting Carbon Flux in Clostridium Ljungdahlii Using CRISPR Interference (CRISPRi). Metabolic Engineering 2018, 48, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Pyne, M.E.; Bruder, M.R.; Moo-Young, M.; Chung, D.A.; Chou, C.P. Harnessing Heterologous and Endogenous CRISPR-Cas Machineries for Efficient Markerless Genome Editing in Clostridium. Sci Rep 2016, 6, 25666. [Google Scholar] [CrossRef] [PubMed]
- Walker, J.E.; Lanahan, A.A.; Zheng, T.; Toruno, C.; Lynd, L.R.; Cameron, J.C.; Olson, D.G.; Eckert, C.A. Development of Both Type I–B and Type II CRISPR/Cas Genome Editing Systems in the Cellulolytic Bacterium Clostridium Thermocellum. Metabolic Engineering Communications 2020, 10, e00116. [Google Scholar] [CrossRef]
- Zhang, J.; Zong, W.; Hong, W.; Zhang, Z.-T.; Wang, Y. Exploiting Endogenous CRISPR-Cas System for Multiplex Genome Editing in Clostridium Tyrobutyricum and Engineer the Strain for High-Level Butanol Production. Metabolic Engineering 2018, 47, 49–59. [Google Scholar] [CrossRef]
- Turco, F.; Garavaglia, M.; Van Houdt, R.; Hill, P.; Rawson, F.J.; Kovacs, K. Synthetic Biology Toolbox, Including a Single-Plasmid CRISPR-Cas9 System to Biologically Engineer the Electrogenic, Metal-Resistant Bacterium Cupriavidus Metallidurans CH34. ACS Synthetic Biology 2022, 11, 3617–3628. [Google Scholar] [CrossRef] [PubMed]
- Xiong, B.; Li, Z.; Liu, L.; Zhao, D.; Zhang, X.; Bi, C. Genome Editing of Ralstonia Eutropha Using an Electroporation-Based CRISPR-Cas9 Technique. Biotechnol Biofuels 2018, 11, 172. [Google Scholar] [CrossRef]
- Jiang, W.; Bikard, D.; Cox, D.; Zhang, F.; Marraffini, L.A. RNA-Guided Editing of Bacterial Genomes Using CRISPR-Cas Systems. Nat Biotechnol 2013, 31, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Rath, D.; Amlinger, L.; Hoekzema, M.; Devulapally, P.R.; Lundgren, M. Efficient Programmable Gene Silencing by Cascade. Nucleic Acids Research 2015, 43, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Qin, Q.; Ling, C.; Zhao, Y.; Yang, T.; Yin, J.; Guo, Y.; Chen, G.Q. CRISPR/Cas9 Editing Genome of Extremophile Halomonas Spp. Metabolic Engineering 2018, 47, 219–229. [Google Scholar] [CrossRef]
- Ganguly, J.; Martin-Pascual, M.; van Kranenburg, R. CRISPR Interference (CRISPRi) as Transcriptional Repression Tool for Hungateiclostridium Thermocellum DSM 1313. Microbial Biotechnology 2020, 13, 339–349. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.; Baik, J.; Lim, S.-H. A Model Independent S/W Framework for Search-Based Software Testing. The Scientific World Journal 2014, 2014, 126348. [Google Scholar] [CrossRef] [PubMed]
- Mo, X.-H.; Zhang, H.; Wang, T.-M.; Zhang, C.; Zhang, C.; Xing, X.-H.; Yang, S. Establishment of CRISPR Interference in Methylorubrum Extorquens and Application of Rapidly Mining a New Phytoene Desaturase Involved in Carotenoid Biosynthesis. Appl Microbiol Biotechnol 2020, 104, 4515–4532. [Google Scholar] [CrossRef]
- Batianis, C.; Kozaeva, E.; Damalas, S.G.; Martín-Pascual, M.; Volke, D.C.; Nikel, P.I.; Martins dos Santos, V.A.P. An Expanded CRISPRi Toolbox for Tunable Control of Gene Expression in Pseudomonas Putida. Microbial Biotechnology 2020, 13, 368–385. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yuan, J. CRISPR/Cas12a-Mediated Genome Engineering in the Photosynthetic Bacterium Rhodobacter Capsulatus. Microbial Biotechnology 2021, 14, 2700–2710. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Jiao, S.; Wang, M.; Yu, H.; Shen, Z. A CRISPR/Cas9-Based Genome Editing System for Rhodococcus Ruber TH. Metabolic Engineering 2020, 57, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR Provides Acquired Resistance Against Viruses in Prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef]
- Cobb, R.E.; Wang, Y.; Zhao, H. High-Efficiency Multiplex Genome Editing of Streptomyces Species Using an Engineered CRISPR/Cas System. ACS Synth. Biol. 2015, 4, 723–728. [Google Scholar] [CrossRef]
- Tong, Y.; Charusanti, P.; Zhang, L.; Weber, T.; Lee, S.Y. CRISPR-Cas9 Based Engineering of Actinomycetal Genomes. ACS Synth. Biol. 2015, 4, 1020–1029. [Google Scholar] [CrossRef]
- Choi, S.Y.; Woo, H.M. CRISPRi-dCas12a: A dCas12a-Mediated CRISPR Interference for Repression of Multiple Genes and Metabolic Engineering in Cyanobacteria. ACS Synth. Biol. 2020, 9, 2351–2361. [Google Scholar] [CrossRef]
- Gordon, G.C.; Korosh, T.C.; Cameron, J.C.; Markley, A.L.; Begemann, M.B.; Pfleger, B.F. CRISPR Interference as a Titratable, Trans-Acting Regulatory Tool for Metabolic Engineering in the Cyanobacterium Synechococcus Sp. Strain PCC 7002. Metabolic Engineering 2016, 38, 170–179. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Chen, B.; Duan, C.; Sun, B.; Yang, J.; Yang, S. Multigene Editing in the Escherichia Coli Genome via the CRISPR-Cas9 System. Applied and Environmental Microbiology 2015, 81, 2506–2514. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.H.; Ostrov, N.; Wong, B.G.; Gold, M.A.; Khalil, A.S.; Church, G.M. Functional Genomics of the Rapidly Replicating Bacterium Vibrio Natriegens by CRISPRi. Nat Microbiol 2019, 4, 1105–1113. [Google Scholar] [CrossRef]
- Nødvig, C.S.; Nielsen, J.B.; Kogle, M.E.; Mortensen, U.H. A CRISPR-Cas9 System for Genetic Engineering of Filamentous Fungi. PLOS ONE 2015, 10, e0133085. [Google Scholar] [CrossRef] [PubMed]
- Waltz, E. Gene-Edited CRISPR Mushroom Escapes US Regulation. Nature News 2016, 532, 293. [Google Scholar] [CrossRef]
- Nargesi, S.; Kaboli, S.; Thekkiniath, J.; Heidari, S.; Keramati, F.; Seyedmousavi, S.; Hedayati, M.T. Recent Advances in Genome Editing Tools in Medical Mycology Research. J Fungi (Basel) 2021, 7, 257. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, Y.; Liu, T.; Zhang, H.; Zhang, H.; Li, J. The GlaA Signal Peptide Substantially Increases the Expression and Secretion of α-Galactosidase in Aspergillus Niger. Biotechnol Lett 2018, 40, 949–955. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, D.I.; Olsthoorn, M.M.A.; Maillet, I.; Akeroyd, M.; Breestraat, S.; Donkers, S.; van der Hoeven, R.A.M.; van den Hondel, C.A.M.J.J.; Kooistra, R.; Lapointe, T.; et al. Effective Lead Selection for Improved Protein Production in Aspergillus Niger Based on Integrated Genomics. Fungal Genetics and Biology 2009, 46, S141–S152. [Google Scholar] [CrossRef]
- Wang, Y.; Xue, W.; Sims, A.H.; Zhao, C.; Wang, A.; Tang, G.; Qin, J.; Wang, H. Isolation of Four Pepsin-like Protease Genes from Aspergillus Niger and Analysis of the Effect of Disruptions on Heterologous Laccase Expression. Fungal Genetics and Biology 2008, 45, 17–27. [Google Scholar] [CrossRef]
- Fiedler, M.R.M.; Barthel, L.; Kubisch, C.; Nai, C.; Meyer, V. Construction of an Improved Aspergillus Niger Platform for Enhanced Glucoamylase Secretion. Microbial Cell Factories 2018, 17, 95. [Google Scholar] [CrossRef]
- Ohno, A.; Maruyama, J.; Nemoto, T.; Arioka, M.; Kitamoto, K. A Carrier Fusion Significantly Induces Unfolded Protein Response in Heterologous Protein Production by Aspergillus Oryzae. Appl Microbiol Biotechnol 2011, 92, 1197–1206. [Google Scholar] [CrossRef] [PubMed]
- Hoang, H.-D.; Maruyama, J.; Kitamoto, K. Modulating Endoplasmic Reticulum-Golgi Cargo Receptors for Improving Secretion of Carrier-Fused Heterologous Proteins in the Filamentous Fungus Aspergillus Oryzae. Applied and Environmental Microbiology 2015, 81, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Vyas, V.K.; Barrasa, M.I.; Fink, G.R. A Candida Albicans CRISPR System Permits Genetic Engineering of Essential Genes and Gene Families. Science Advances 2015, 1, e1500248. [Google Scholar] [CrossRef]
- Tsai, S.-C.; Tsai, L.-D.; Li, Y.-K. An Isolated Candida Albicans TL3 Capable of Degrading Phenol at Large Concentration. Bioscience, Biotechnology, and Biochemistry 2005, 69, 2358–2367. [Google Scholar] [CrossRef]
- Min, K.; Ichikawa, Y.; Woolford, C.A.; Mitchell, A.P. Candida Albicans Gene Deletion with a Transient CRISPR-Cas9 System. mSphere 2016, 1. [Google Scholar] [CrossRef]
- Yang, P.; Chen, Y.; Wu, H.; Fang, W.; Liang, Q.; Zheng, Y.; Olsson, S.; Zhang, D.; Zhou, J.; Wang, Z.; et al. The 5-Oxoprolinase Is Required for Conidiation, Sexual Reproduction, Virulence and Deoxynivalenol Production of Fusarium Graminearum. Curr Genet 2018, 64, 285–301. [Google Scholar] [CrossRef] [PubMed]
- Shi, T.-Q.; Gao, J.; Wang, W.-J.; Wang, K.-F.; Xu, G.-Q.; Huang, H.; Ji, X.-J. CRISPR/Cas9-Based Genome Editing in the Filamentous Fungus Fusarium Fujikuroi and Its Application in Strain Engineering for Gibberellic Acid Production. ACS Synth. Biol. 2019, 8, 445–454. [Google Scholar] [CrossRef]
- Horwitz, S.B.; Goldman, I.D. A Conversation with Susan Band Horwitz. Annual Review of Pharmacology and Toxicology 2015, 55, 1–9. [Google Scholar] [CrossRef]
- Löbs, A.-K.; Engel, R.; Schwartz, C.; Flores, A.; Wheeldon, I. CRISPR–Cas9-Enabled Genetic Disruptions for Understanding Ethanol and Ethyl Acetate Biosynthesis in Kluyveromyces Marxianus. Biotechnol Biofuels 2017, 10, 164. [Google Scholar] [CrossRef]
- Chen, X.; Selvaraj, P.; Lin, L.; Fang, W.; Wu, C.; Yang, P.; Zhang, J.; Abubakar, Y.S.; Yang, F.; Lu, G.; et al. Rab7/Retromer-Based Endolysosomal Trafficking Is Essential for Proper Host Invasion in Rice Blast. New Phytologist 2023, 239. [Google Scholar] [CrossRef]
- Liu, W.; An, C.; Shu, X.; Meng, X.; Yao, Y.; Zhang, J.; Chen, F.; Xiang, H.; Yang, S.; Gao, X.; et al. A Dual-Plasmid CRISPR/Cas System for Mycotoxin Elimination in Polykaryotic Industrial Fungi. ACS Synth. Biol. 2020, 9, 2087–2095. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Gao, R.; Li, J.; Lin, L.; Zhao, J.; Sun, W.; Tian, C. Development of a Genome-Editing CRISPR/Cas9 System in Thermophilic Fungal Myceliophthora Species and Its Application to Hyper-Cellulase Production Strain Engineering. Biotechnology for Biofuels 2017, 10, 1. [Google Scholar] [CrossRef] [PubMed]
- Reilly, M.C.; Qin, L.; Craig, J.P.; Starr, T.L.; Glass, N.L. Deletion of Homologs of the SREBP Pathway Results in Hyper-Production of Cellulases in Neurospora Crassa and Trichoderma Reesei. Biotechnology for Biofuels 2015, 8, 121. [Google Scholar] [CrossRef]
- Qin, L.; Wu, V.W.; Glass, N.L. Deciphering the Regulatory Network between the SREBP Pathway and Protein Secretion in Neurospora Crassa. mBio 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Pohl, C.; Kiel, J.A.K.W.; Driessen, A.J.M.; Bovenberg, R.A.L.; Nygård, Y. CRISPR/Cas9 Based Genome Editing of Penicillium Chrysogenum. ACS Synth. Biol. 2016, 5, 754–764. [Google Scholar] [CrossRef] [PubMed]
- Weninger, A.; Hatzl, A.-M.; Schmid, C.; Vogl, T.; Glieder, A. Combinatorial Optimization of CRISPR/Cas9 Expression Enables Precision Genome Engineering in the Methylotrophic Yeast Pichia Pastoris. Journal of Biotechnology 2016, 235, 139–149. [Google Scholar] [CrossRef]
- Zhong, Y.; Liu, X.; Xiao, P.; Wei, S.; Wang, T. Expression and Secretion of the Human Erythropoietin Using an Optimized Cbh1 Promoter and the Native CBH I Signal Sequence in the Industrial Fungus Trichoderma Reesei. Appl Biochem Biotechnol 2011, 165, 1169–1177. [Google Scholar] [CrossRef]
- Wu, Y.; Sun, X.; Xue, X.; Luo, H.; Yao, B.; Xie, X.; Su, X. Overexpressing Key Component Genes of the Secretion Pathway for Enhanced Secretion of an Aspergillus Niger Glucose Oxidase in Trichoderma Reesei. Enzyme and Microbial Technology 2017, 106, 83–87. [Google Scholar] [CrossRef]
- Ronda, C.; Maury, J.; Jakočiu̅nas, T.; Baallal Jacobsen, S.A.; Germann, S.M.; Harrison, S.J.; Borodina, I.; Keasling, J.D.; Jensen, M.K.; Nielsen, A.T. CrEdit: CRISPR Mediated Multi-Loci Gene Integration in Saccharomyces Cerevisiae. Microb Cell Fact 2015, 14, 97. [Google Scholar] [CrossRef]
- Mans, R.; van Rossum, H.M.; Wijsman, M.; Backx, A.; Kuijpers, N.G.A.; van den Broek, M.; Daran-Lapujade, P.; Pronk, J.T.; van Maris, A.J.A.; Daran, J.-M.G. CRISPR/Cas9: A Molecular Swiss Army Knife for Simultaneous Introduction of Multiple Genetic Modifications in Saccharomyces Cerevisiae. FEMS Yeast Research 2015, 15, fov004. [Google Scholar] [CrossRef]
- Gilbert, B.J.; Miller, C.; Corrick, F.; Watson, R.A. Should Trainee Doctors Use the Developing World to Gain Clinical Experience? The Annual Varsity Medical Debate - London, Friday 20th January, 2012. Philosophy, Ethics, and Humanities in Medicine 2013, 8. [CrossRef]
- Li, J.; Zhang, Y.; Zhang, Y.; Yu, P.-L.; Pan, H.; Rollins, J.A. Introduction of Large Sequence Inserts by CRISPR-Cas9 To Create Pathogenicity Mutants in the Multinucleate Filamentous Pathogen Sclerotinia Sclerotiorum. mBio 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, C.; Shabbir-Hussain, M.; Frogue, K.; Blenner, M.; Wheeldon, I. Standardized Markerless Gene Integration for Pathway Engineering in Yarrowia Lipolytica. ACS Synth. Biol. 2017, 6, 402–409. [Google Scholar] [CrossRef] [PubMed]
- Miao, J.; Chi, Y.; Lin, D.; Tyler, B.M.; Liu, X. Mutations in ORP1 Conferring Oxathiapiprolin Resistance Confirmed by Genome Editing Using CRISPR/Cas9 in Phytophthora Capsici and P. Sojae. Phytopathology® 2018, 108, 1412–1419. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Tyler, B.M. Efficient Disruption and Replacement of an Effector Gene in the Oomycete Phytophthora Sojae Using CRISPR/Cas9. Molecular Plant Pathology 2016, 17, 127–139. [Google Scholar] [CrossRef] [PubMed]
- Geiser, E.; Wiebach, V.; Wierckx, N.; Blank, L.M. Prospecting the Biodiversity of the Fungal Family Ustilaginaceae for the Production of Value-Added Chemicals. Fungal Biol Biotechnol 2014, 1, 2. [Google Scholar] [CrossRef]
- Schuster, M.; Schweizer, G.; Reissmann, S.; Kahmann, R. Genome Editing in Ustilago Maydis Using the CRISPR–Cas System. Fungal Genetics and Biology 2016, 89, 3–9. [Google Scholar] [CrossRef]
- Gao, S.; Tong, Y.; Wen, Z.; Zhu, L.; Ge, M.; Chen, D.; Jiang, Y.; Yang, S. Multiplex Gene Editing of the Yarrowia Lipolytica Genome Using the CRISPR-Cas9 System. Journal of Industrial Microbiology and Biotechnology 2016, 43, 1085–1093. [Google Scholar] [CrossRef]
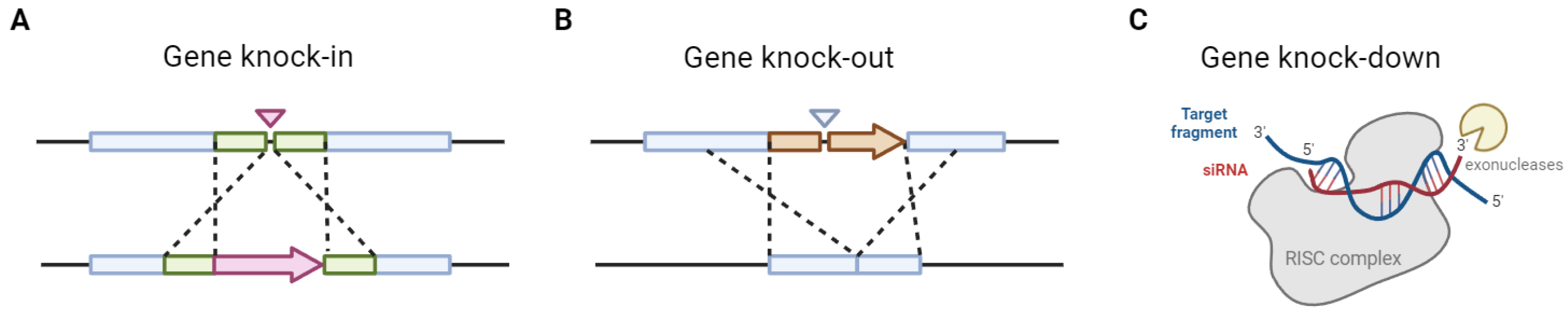
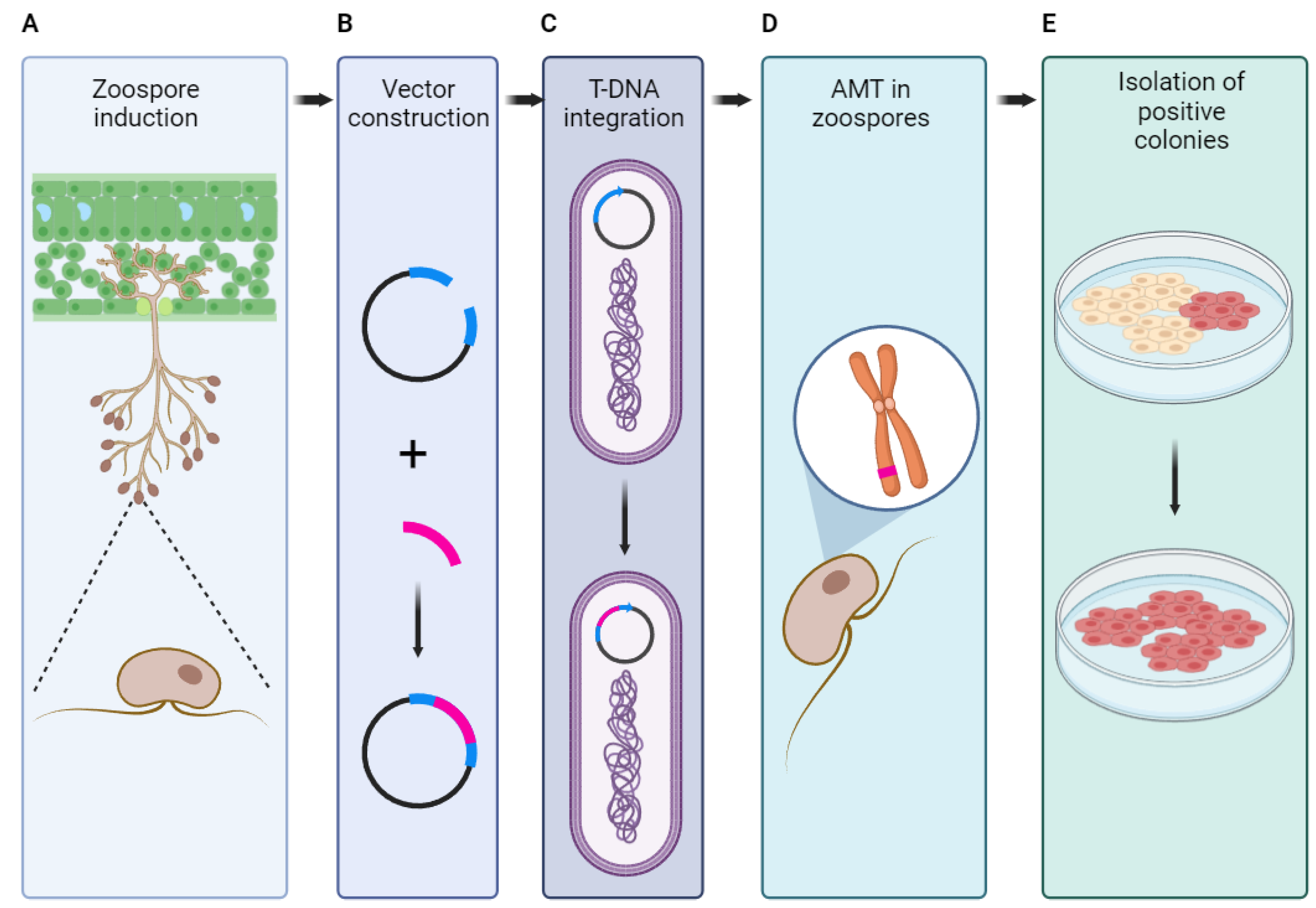
| Tool name | Access link | PAM sequence | Functions | Reference(s) |
|---|---|---|---|---|
| ATUM | https://www.atum.bio/eCommerce/cas9/input | NGG, NAG | Knock-in, knock-out | [79] |
| Benchling | https://www.benchling.com/crispr | Custom PAM | Knock-in, knock-out, knock-down | [80] |
| CasOFFinder | http://www.rgenome.net/cas-offinder/ | NGG, NRG, NNAGAAW, NNNNGMTT, NNGRRT, NNNVRYAC, NNNNRYAC, TTTN, TTTV, NNGTGA, TTN, NNNRRT, KYTV, NGCG, NGA, NGT, NG, TTN, ATTN, DTTN, NAAN, NNNNCC, TYCV, TATV, NNNNGNA, YTTV, NCC, NNNNCNR, NNNNCNAA, NNN, NRN, NGC, TTTR, NTTR, TTCN, NRTH, NNGG, NGGNG, NGGNG, NRTA, NNNA, TTNA, TTNA, NRNH, NGNG, NRCH | Knock-in, knock-out, knock-down | [81] |
| CCTop | https://cctop.cos.uni-heidelberg.de:8043/ | NGG, NRG, NG, NG-NRG, NGA, NGCG, TTTN, TTTV, YTN, TTN, YTTN, NNGRRT, NNNNGATT, NNAGAAW, NAAAAC, NNNNRYAC | Knock-in, knock-out | [82,83] |
| CHOPCHOP | https://chopchop.cbu.uib.no/ | NGG, NRG, NG, NG-NRG, NGA, NGCG, TTTN, TTTV, YTN, TTN, YTTN, NNGRRT, NNNNGATT, NNAGAAW, NAAAAC, NNNNRYAC | Knock-in, knock-out, activation, repression, nanopore enrichment | [84] |
| CRISPOR | http://crispor.gi.ucsc.edu/ | NGG, NNG, NGN, NNGT, NAA, NNG, NNG, NNGRRT, NGK, NNNRRT, NGA, NNNNCC, NGCG, NNAGAA, NGGNG, NNNNGMTT, NNNNACA, NNNNRYAC, TTTV, TTTN, ATTN, NTN, TYCV, TATV, TTTA, TCTA, TCCA, CCCA, GGTT, YTTV, TTYN, NNNNCNAA, NNN, NRN, NYN | Knock-in, knock-out | [85] |
| CRISPRdirect | https://crispr.dbcls.jp/ | NGG, NRG, NNGRRT, NG | Knock-in, knock-out | [86] |
| CRISPR-ERA | http://crispr-era.stanford.edu/ | NGG | Gene editing, gene repression, gene activation | [87] |
| CRISPick | https://portals.broadinstitute.org/gppx/crispick/public | NGG, NNGRR, TTTV | Knock-in, knock-out, knock-down | [88] |
| IDT | https://www.idtdna.com/site/order/designtool/index/CRISPR_SEQUENCE | Custom PAM | Knock-in, knock-out, knock-down | [89] |
| Off-Spotter | https://cm.jefferson.edu/Off-Spotter/ | NGG, NAG, NNNNACA, NNGRRT | Knock-in, knock-out | [90] |
| Synthego | https://design.synthego.com/#/ | Custom PAM | Knock-in, knock-out, knock-down | [91] |
| TrueDesign Genome Editor | https://www.thermofisher.com/us/en/home/life-science/genome-editing/invitrogen-truedesign-genome-editor.html | Custom PAM | Knockout a target gene, add a fluorescent or epitope tag, Insert, delete, or replace up to 30 bases, long insertion up to 10kb, Generate a SNP | [92] |
| Microbial species | Modifications | Importance | Reference(s) |
|---|---|---|---|
| Bacteria | |||
| Acidithiobacillus ferrooxidans | Knock-down | Utilized for the bioleaching of metals | [93] |
| Bacillus methanolicus | Knock-down | Methylotrophic bacteria | [94] |
| Bacillus smithii | Knock-in | Moderate thermophile capable of C5 and C6 sugar metabolism | [95,96] |
| Bacillus subtilis | Knock-in | Producer of industrial enzymes and valuable low-molecular-weight substances | [97] |
| Clostridium acetobutylicum | Knock-in | Isopropanol production | [98] |
| Clostridium autoethanogenum | Knock-in | Capable of fermenting CO, CO2, and H2 into biofuel ethanol and 2,3-butanediol | [99] |
| Clostridium beijerinckii | Knock-in | Production strain for biofuels and biochemical | [100] |
| Clostridium cellulolyticum | Knock-in | Capable of conversion of lignocellulosic biomass to valuable endproducts | [101] |
| Corynebacterium glutamicum | Knock-down | Producer of amino acids | [102] |
| Clostridium ljungdahlii | Knock-down | Capable of producing ethanol from synthesis gas | [103] |
| Clostridium pasteurianum | Knock-in | Capable of converting waste glycerol to butanol | [104] |
| Clostridium thermocellum | Knock-out | Thermophilic bacteria | [105] |
| Clostridium tyrobutyricum | Knock-out | Butanol production | [106] |
| Corynebacterium glutamicum | Knock-down | Producer of amino acids | [102] |
| Cupriavidus metallidurans | Knock-in | Facultative chemolithoautotroph | [107] |
| Cupriavidus necator | Knock-in | Facultative chemolithoautotroph | [108] |
| Escherichia coli | Knock-in, Knock-down, Knock-out | Programmed antimicrobial, recombination, multiplex recombination, CRISPRi, multiplexed CRISPRi, gene circuit, RNA targeting | [109,110] |
| Halomonas bluephagenesis | Knock-out | 3-Hydroxyvalerate production | [111] |
| Hungateiclostridium thermocellum | Knock-down | Thermophilic bacterium | [112] |
| Lactobacillus reuteri | Knock-in | Probiotic strain and producer of biotherapeutics | [113] |
| Methylobacterium extorquens | Knock-down | Methylotrophic bacterium | [114] |
| Pseudomonas putida | Knock-down | Exhibits solvent tolerance and highly versatile metabolism | [115] |
| Rhodobacter capsulatus | Knock-down | Facultative photo- and chemolithoautotroph species | [116] |
| Rhodococcus ruber TH | Knock-out | Acrylamide production | [117] |
| Streptococcus thermophilus | Knock-in | Probiotic and industrial fermentation strains | [118] |
| Streptomyces albus | Knock-in | Producer of heterologous secondary metabolites | [119] |
| Streptomyces coelicolor | Knock-in | Source of pharmacologically active and industrially relevant secondary metabolites | [119,120] |
| Synechococcus elongatus PCC 7942 | Knock-down | Squalene production | [121] |
| Synechococcus sp. PCC 7002 | Knock-down | Lactate production | [122] |
| Tatumella citrea | Knock-in | Producer of vitamin C precursor (2-keto-D-gluconic acid) | [123] |
| Vibrio natriegens | Knock-down | Fast-growing bacteria | [124] |
| Fungi | |||
| Aspergillus aculeatus | Knock-in | Source of and producer of enzymes | [125] |
| Agaricus bisporus | Knock-in | Anti-browning mushroom with extended shelf life and enhanced resistance to blemishes | [126] |
| Aspergillus fumigatus | Knock-in | For studying mechanisms of azole resistance | [127] |
| Aspergillus niger | Knock-in | Enhances the expression and secretion of α-galactosidase; improves protein production; facilitates heterologous laccase expression; and boosts glucoamylase secretion | [128,129,130,131] |
| Aspergillus oryzae | Knock-in | Enhances the expression and secretion of α-galactosidase; improves protein production; facilitates heterologous laccase expression; and boosts glucoamylase secretion | [132,133] |
| Candida albicans | Knock-in | Common production strain, capable of phenol and formaldehyde catabolism | [134,135,136] |
| Fusarium graminearum | Knock-out | Functional study of 5-oxoprolinase | [137] |
| Fusarium fujikuroi | Knock-out | GA4/GA7 mixtures production | [138] |
| Kluyveromyces lactis | Knock-in | Common production strain | [139] |
| Kluyveromyces marxianus | Knock-out | Ethyl acetate production | [140] |
| Magnaporthe oryzae | Knock-out | Study of endolysosomal trafficking mechanisms in rice blast | [141] |
| Monascus purpureus KL-001 | Knock-out | Monascus Red pigment production | [142] |
| Myceliophthora thermophile | Knock-in | Thermophilic strain and producer of cellulases | [143] |
| Neurospora crassa | Knock-in | hyper-production of cellulases | [144,145] |
| Penicillium chrysogenum | Knock-in | Producer of β-lactam antibiotics | [146] |
| Pichia pastoris | Knock-in | Common production strain | [147] |
| Trichoderma reesei | Knock-in | Expression and secretion of human erythropoietin; enhanced secretion of glucose oxidase; and hyper-cellulase production | [143,148,149] |
| Saccharomyces cerevisiae | Knock-in, Knock-down | Common production strain | [150,151,152] |
| Sclerotinia sclerotiorum | Knock-in | Introduction of large sequence inserts | [153] |
| Synechococcus elongatus PCC 7942 | Knock-in, Knock-out | Succinate production | |
| Yarrowia lipolytica | Knock-in | Lycopene production | [154] |
| Oomycetes | |||
| Phytophthora capsici | Knock-out | Conferring resistance to oxathiapiprolin | [155] |
| Phytophthora infestans | Knock-out | Increase in transformation efficiency; a modified genetic transformation and genome editing system | [2] |
| Phytophthora sojae | Knock-out | Efficient disruption and replacement of an effector gene. | [156] |
| Plasmopara viticola | Knock-out | Increase in transformation efficiency; a modified genetic transformation and genome editing system | [2] |
| Ustilago maydis | Knock-out | Natural producer of valuable biochemicals; causative agent of corn smut | [157,158] |
| Yarrowia lipolytica | Knock-in, Knock-out | Natural producer of valuable biochemical | [154,159] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





