Submitted:
23 September 2024
Posted:
23 September 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Datasets
2.2. Immune Infiltration Analysis by ssGSEA
2.3. DNAm Age Calculation
2.4. RNA Age Calculation
2.5. Trajectory Analysis Based on Bulk Sequencing
2.6. Single-Cell RNA-Seq Analysis
2.6.1. Data Preprocess, Cell Clustering, and Annotation
2.6.2. Integration of Various Single-Cell RNA-Seq Datasets Across Multiple Samples
2.6.3. Inference of T Cell Fate by Trajectory Analysis
2.6.4. Cell Type Enrichment of Various Age-Based Subgroups
2.7. Statistical Analysis
3. Results
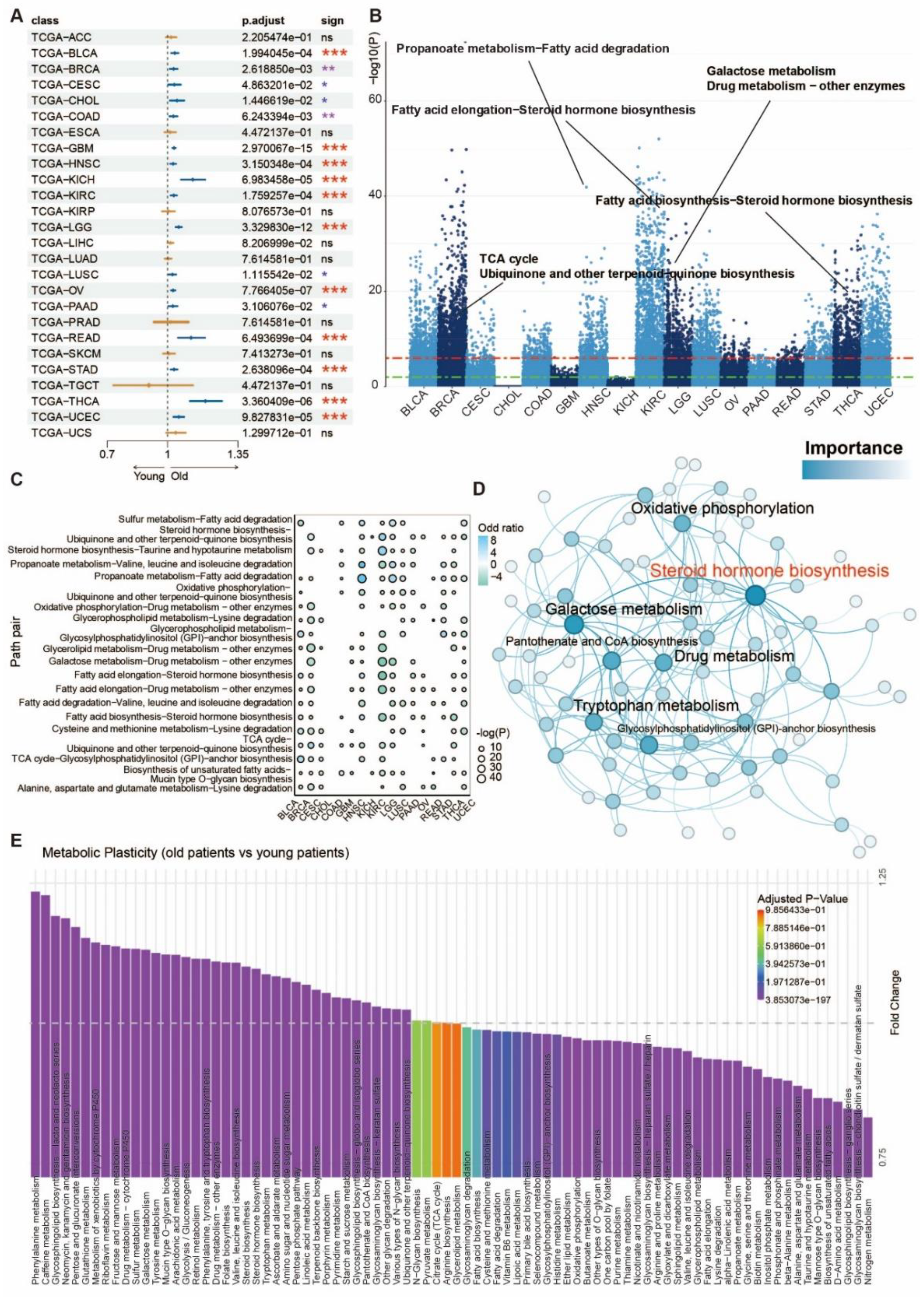
3.1. Aging-Related Metabolic Alterations: Insights from Pan-Cancer Bulk Sequencing Transcriptome Analysis
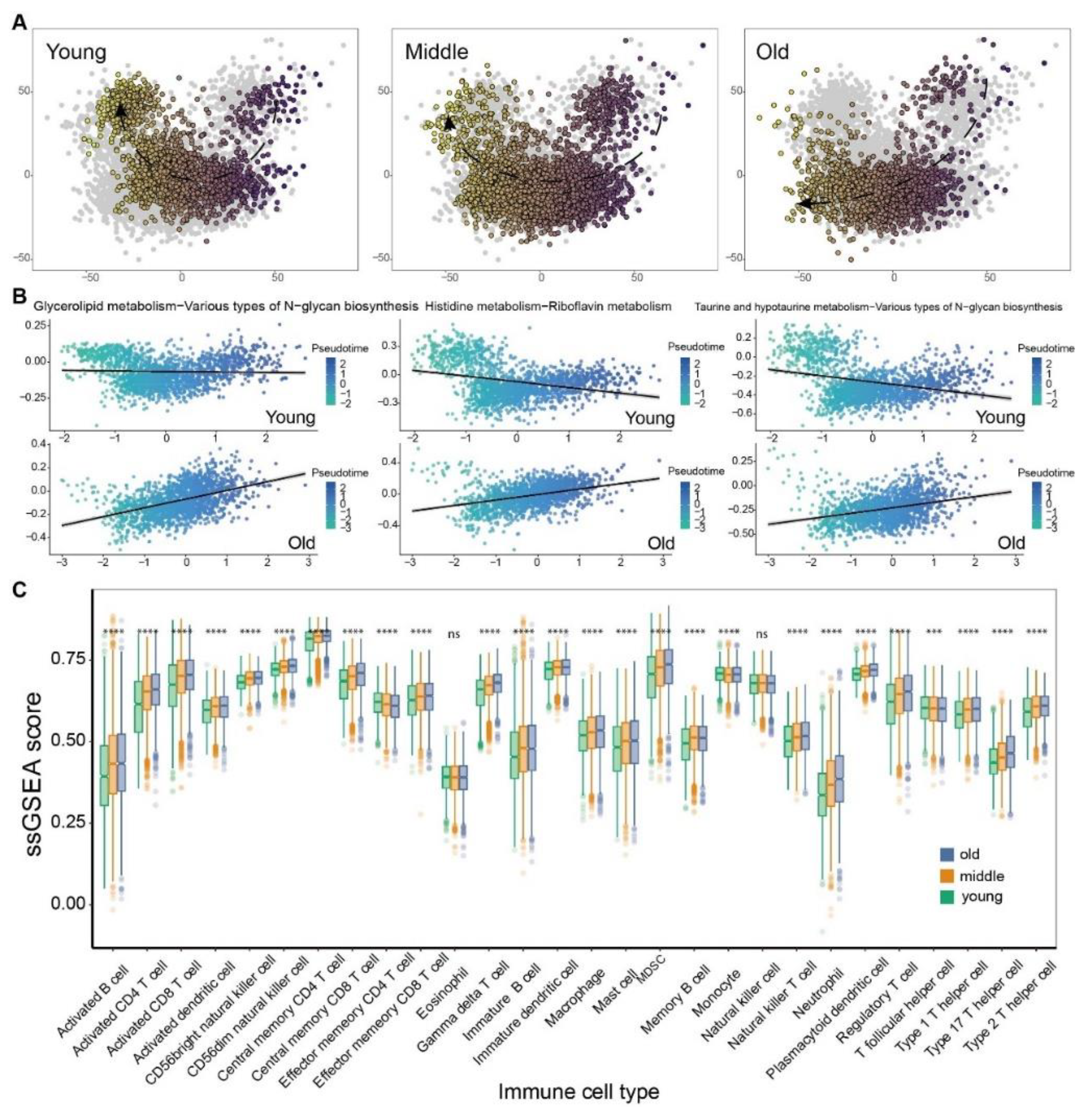
3.2. The metabolic switch correlates to molecular features of senescence in age-related cancers
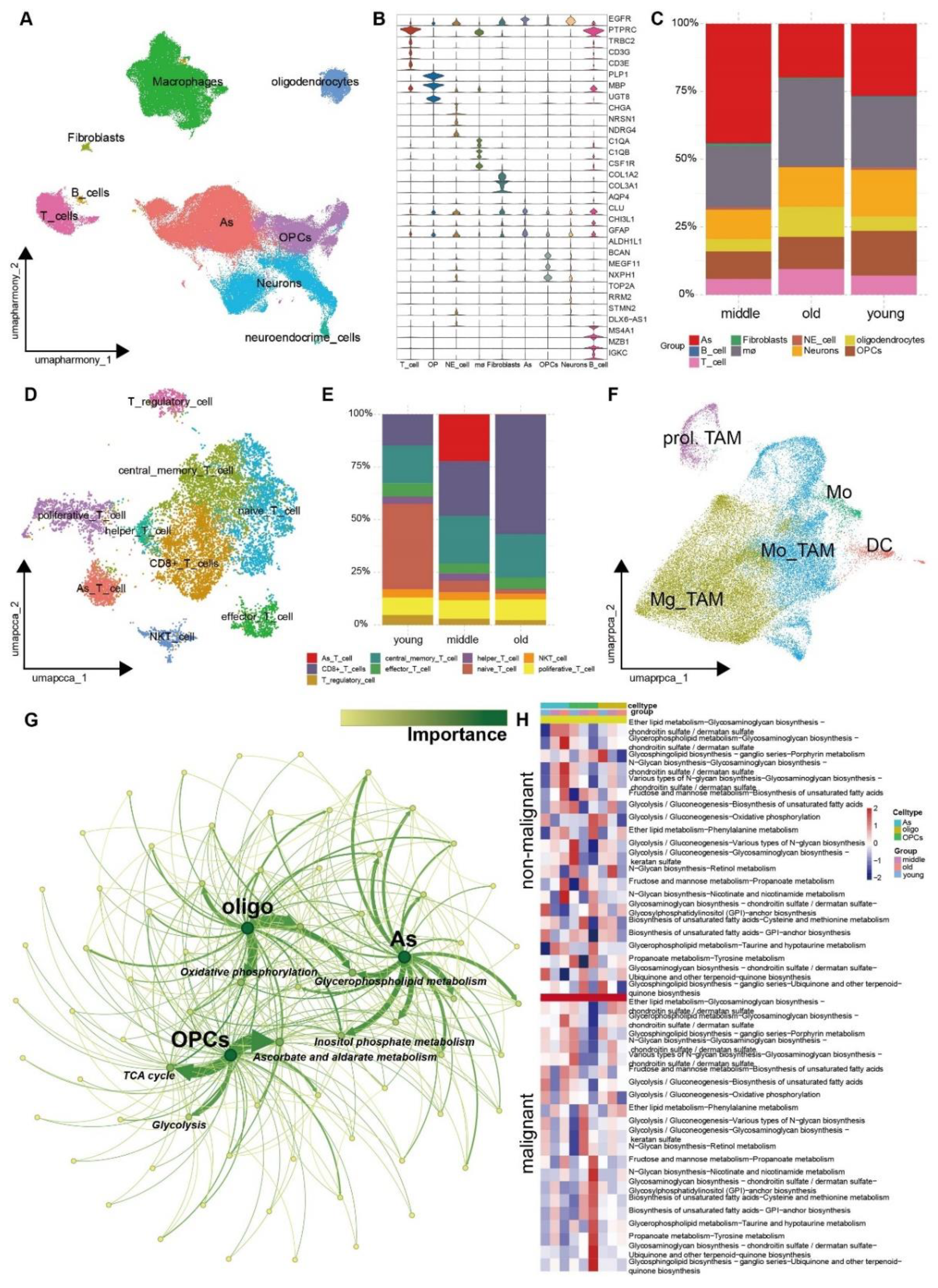
3.3. The Landscape of Tumor Microenvironment Showed Distinct Discrepancy among Aged-Based Subgroups
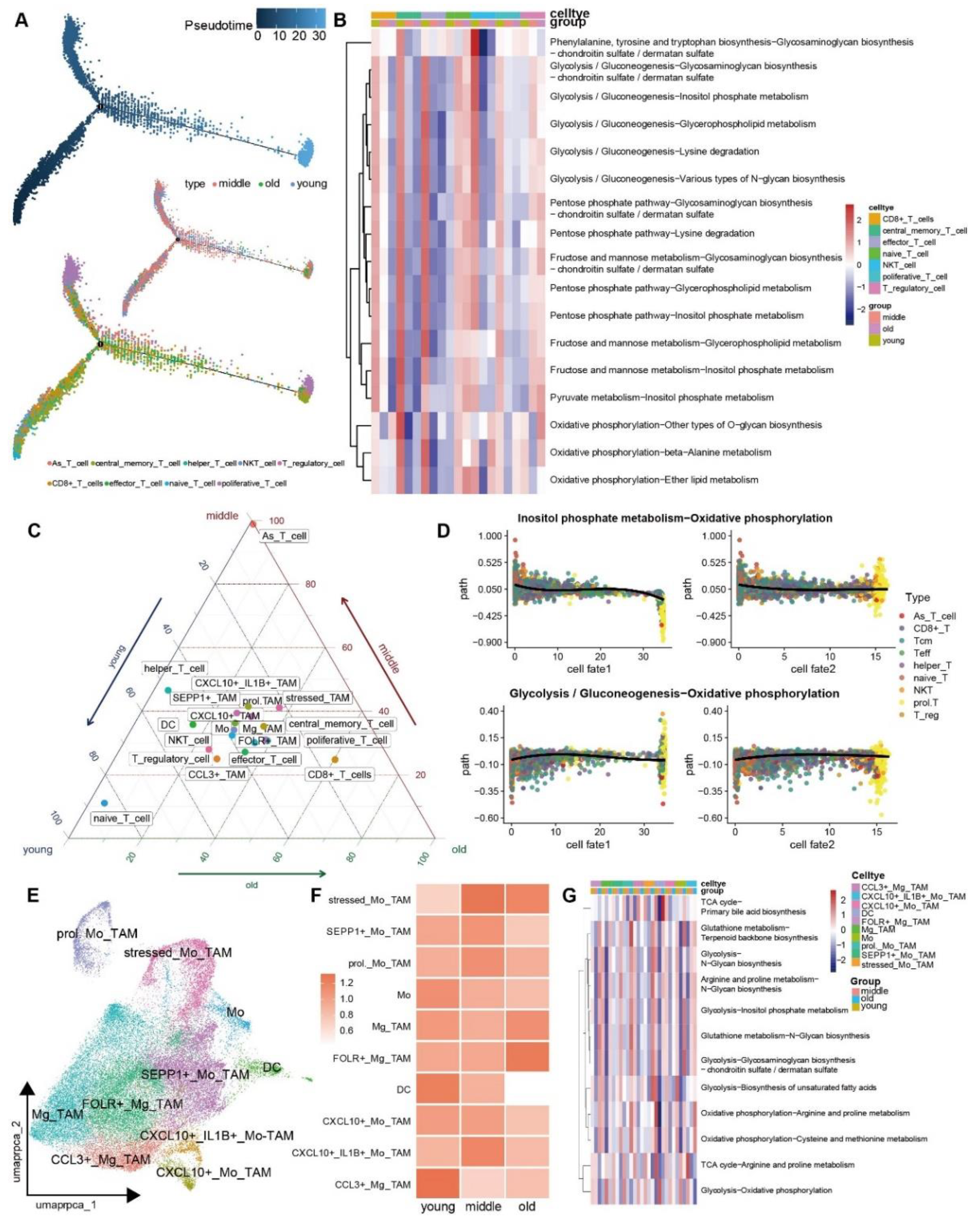
3.4. The Metabolic Reprogramming of Various Cell Types within TME Exhibits Increased Heterogeneity as the Aging Procession
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- White, M.C.; Holman, D.M.; Goodman, R.A.; Richardson, L.C. Cancer Risk Among Older Adults: Time for Cancer Prevention to Go Silver. Gerontologist 2019, 59, S1–S6. [Google Scholar] [CrossRef] [PubMed]
- Lauper, J.M.; Krause, A.; Vaughan, T.L.; Monnat, R.J. Spectrum and Risk of Neoplasia in Werner Syndrome: A Systematic Review. PLoS ONE 2013, 8, e59709. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab 2016, 23, 27–47. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, N.N.; Zhu, J.; Thompson, C.B. The hallmarks of cancer metabolism: Still emerging. Cell Metab 2022, 34, 355–377. [Google Scholar] [CrossRef]
- Han, S.; Georgiev, P.; Ringel, A.E.; Sharpe, A.H.; Haigis, M.C. Age-associated remodeling of T cell immunity and metabolism. Cell Metab 2023, 35, 36–55. [Google Scholar] [CrossRef]
- Weistuch, C.; Mujica-Parodi, L.R.; Razban, R.M.; Antal, B.; van Nieuwenhuizen, H.; Amgalan, A.; Dill, K.A. Metabolism modulates network synchrony in the aging brain. Proc Natl Acad Sci U S A 2021, 118. [Google Scholar] [CrossRef] [PubMed]
- Agnihotri, S.; Zadeh, G. Metabolic reprogramming in glioblastoma: The influence of cancer metabolism on epigenetics and unanswered questions. Neuro Oncol 2016, 18, 160–172. [Google Scholar] [CrossRef]
- Warburg, O. On the origin of cancer cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef]
- Drapela, S.; Ilter, D.; Gomes, A.P. Metabolic reprogramming: A bridge between aging and tumorigenesis. Mol Oncol 2022, 16, 3295–3318. [Google Scholar] [CrossRef]
- Nakajima, Y.; Chamoto, K.; Oura, T.; Honjo, T. Critical role of the CD44(low)CD62L(low) CD8(+) T cell subset in restoring antitumor immunity in aged mice. Proc Natl Acad Sci U S A 2021, 118. [Google Scholar] [CrossRef]
- Ron-Harel, N.; Notarangelo, G.; Ghergurovich, J.M.; Paulo, J.A.; Sage, P.T.; Santos, D.; Satterstrom, F.K.; Gygi, S.P.; Rabinowitz, J.D.; Sharpe, A.H.; et al. Defective respiration and one-carbon metabolism contribute to impaired naive T cell activation in aged mice. Proc Natl Acad Sci U S A 2018, 115, 13347–13352. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Deng, M.; Wang, Z.; Huang, C. MMP3C: An in-silico framework to depict cancer metabolic plasticity using gene expression profiles. Brief Bioinform 2023, 25. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Deng, M.; Leng, D.; Sun, B.; Zheng, P.; Zhang, X.D. MIRS: An AI scoring system for predicting the prognosis and therapy of breast cancer. iScience 2023, 26, 108322. [Google Scholar] [CrossRef]
- Feng, Y.W.; Chen, X.Y.; Zhang, X.D.; Huang, C. Metabolic Pathway Pairwise-Based Signature as a Potential Non-Invasive Diagnostic Marker in Alzheimer's Disease Patients. Genes-Basel 2023, 14. [Google Scholar] [CrossRef]
- Pombo Antunes, A.R.; Scheyltjens, I.; Lodi, F.; Messiaen, J.; Antoranz, A.; Duerinck, J.; Kancheva, D.; Martens, L.; De Vlaminck, K.; Van Hove, H.; et al. Single-cell profiling of myeloid cells in glioblastoma across species and disease stage reveals macrophage competition and specialization. Nat Neurosci 2021, 24, 595–610. [Google Scholar] [CrossRef]
- Charoentong, P.; Finotello, F.; Angelova, M.; Mayer, C.; Efremova, M.; Rieder, D.; Hackl, H.; Trajanoski, Z. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep 2017, 18, 248–262. [Google Scholar] [CrossRef]
- Pelegí-Sisó, D.; de Prado, P.; Ronkainen, J.; Bustamante, M.; González, J.R. : A Bioconductor package to estimate DNA methylation age. Bioinformatics 2021, 37, 1759–1760. [Google Scholar] [CrossRef] [PubMed]
- Horvath, S. DNA methylation age of human tissues and cell types (vol 14, pg R115, 2013). Genome Biology 2015, 16. [Google Scholar] [CrossRef]
- Ren, X.; Kuan, P.F. RNAAgeCalc: A multi-tissue transcriptional age calculator. PLoS ONE 2020, 15. [Google Scholar] [CrossRef]
- Campbell, K.R.; Yau, C. Uncovering pseudotemporal trajectories with covariates from single cell and bulk expression data. Nature Communications 2018, 9. [Google Scholar] [CrossRef]
- R. Core Team, R. R. Core Team, R. R: A language and environment for statistical computing. 2013.
- Hao, Y.H.; Hao, S.; Andersen-Nissen, E.; Mauck, W.M.; Zheng, S.W.; Butler, A.; Lee, M.J.; Wilk, A.J.; Darby, C.; Zager, M.; et al. Integrated analysis of multimodal single-cell data. Cell 2021, 184, 3573. [Google Scholar] [CrossRef] [PubMed]
- Germain, P.L.; Lun, A.; Garcia Meixide, C.; Macnair, W.; Robinson, M.D. Doublet identification in single-cell sequencing data using scDblFinder. F1000Res 2021, 10, 979. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Hill, A.; Packer, J.; Lin, D.; Ma, Y.A.; Trapnell, C. Single-cell mRNA quantification and differential analysis with Census. Nat Methods 2017, 14, 309–315. [Google Scholar] [CrossRef]
- Tomczak, K.; Czerwinska, P.; Wiznerowicz, M. The Cancer Genome Atlas (TCGA): An immeasurable source of knowledge. Contemp Oncol (Pozn) 2015, 19, A68–77. [Google Scholar] [CrossRef] [PubMed]
- Vuononvirta, J.; Marelli-Berg, F.M.; Poobalasingam, T. Metabolic regulation of T lymphocyte motility and migration. Mol Aspects Med 2021, 77. [Google Scholar] [CrossRef]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An Open Source Software for Exploring and Manipulating Networks. Proceedings of the International AAAI Conference on Web and Social Media 2009, 3, 361–362. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015, 43, e47. [Google Scholar] [CrossRef]
- Shah, Y.; Verma, A.; Marderstein, A.R.; White, J.; Bhinder, B.; Medina, J.S.G.; Elemento, O. Pan-cancer analysis reveals molecular patterns associated with age. Cell Rep 2021, 37. [Google Scholar] [CrossRef]
- Johnson, M.; Bell, A.; Lauing, K.L.; Ladomersky, E.; Zhai, L.J.; Penco-Campillo, M.; Shah, Y.J.; Mauer, E.; Xiu, J.; Nicolaides, T.; et al. Advanced Age in Humans and Mouse Models of Glioblastoma Show Decreased Survival from Extratumoral Influence. Clin Cancer Res 2023, 29, 4973–4989. [Google Scholar] [CrossRef]
- Zhao, T.; Zeng, J.; Xu, Y.; Su, Z.; Chong, Y.; Ling, T.; Xu, H.; Shi, H.; Zhu, M.; Mo, Q.; et al. Chitinase-3 like-protein-1 promotes glioma progression via the NF-kappaB signaling pathway and tumor microenvironment reprogramming. Theranostics 2022, 12, 6989–7008. [Google Scholar] [CrossRef]
- Zhang, L.; Yu, X.; Zheng, L.; Zhang, Y.; Li, Y.; Fang, Q.; Gao, R.; Kang, B.; Zhang, Q.; Huang, J.Y.; et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature 2018, 564, 268–272. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.X.; Li, T.Y.; Xu, Y.Q.; Zhang, X.X.; Li, F.; Bai, J.; Chen, J.; Jiang, W.Q.; Yang, K.Y.; Ou, Q.; et al. CellMarker 2.0: An updated database of manually curated cell markers in human/mouse and web tools based on scRNA-seq data. Nucleic Acids Research 2023, 51, D870–D876. [Google Scholar] [CrossRef] [PubMed]
- Bonuccelli, G.; Whitaker-Menezes, D.; Castello-Cros, R.; Pavlides, S.; Pestell, R.G.; Fatatis, A.; Witkiewicz, A.K.; Vander Heiden, M.G.; Migneco, G.; Chiavarina, B.; et al. The reverse Warburg effect: Glycolysis inhibitors prevent the tumor promoting effects of caveolin-1 deficient cancer associated fibroblasts. Cell Cycle 2010, 9, 1960–1971. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, D.A.; Batich, K.A.; Gunn, M.D.; Huang, M.N.; Sanchez-Perez, L.; Nair, S.K.; Congdon, K.L.; Reap, E.A.; Archer, G.E.; Desjardins, A.; et al. Tetanus toxoid and CCL3 improve dendritic cell vaccines in mice and glioblastoma patients. Nature 2015, 519, 366–369. [Google Scholar] [CrossRef]
- Cui, X.; Kang, C. Stressing the Role of CCL3 in Reversing the Immunosuppressive Microenvironment in Gliomas. Cancer Immunol Res 2024, 12, 514. [Google Scholar] [CrossRef]
- Kodama, M.; Nakayama, K.I. A second Warburg-like effect in cancer metabolism: The metabolic shift of glutamine-derived nitrogen A shift in glutamine-derived nitrogen metabolism from glutaminolysis to de novo nucleotide biosynthesis contributes to malignant evolution of cancer. Bioessays 2020, 42. [Google Scholar] [CrossRef]
- Feng, Z.; Hanson, R.W.; Berger, N.A.; Trubitsyn, A. Reprogramming of energy metabolism as a driver of aging. Oncotarget 2016, 7, 15410–15420. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




