Submitted:
23 September 2024
Posted:
23 September 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Plant Material and DNA Extraction
2.2. DNA Amplification
2.3. Data Analysis
3. Results
3.1. SSR Analysis
| Province | Accession | Common name | Unique alleles (primer name and base pair) | N° unique alleles |
|---|---|---|---|---|
| Sucre | S5 | Quinua | KAAT007_315 | 17 |
| S6 | Quinua | QGAA1_143 | ||
| S7 | Quinua | KAAT007_261, KAAT007_276, KGA16_194, QGA028_196 | ||
| S8 | Atoqpa Quinua | QAAT012_189, QGA028_214, QGAA1_236 | ||
| S9 | Quinua | KAAT007_294, QAAT84_167 | ||
| S22 | Quinua | KCAA011_205 | ||
| S39 | Quinua | QCA14_195 | ||
| S71 | Quinua | KCAA011_245 | ||
| S75 | Quinua | QGAA1_275 | ||
| S83 | Quinua | QGA17_193 | ||
| S92 | Quinua | KAAT007_335 | ||
| La Mar | LM26 | Quinua | KAAT007_300, KAAT007_345 | 3 |
| LM64 | Quinua | KAAT007_350 | ||
| Vilcashuaman | V60 | Quinua | KAAT007_368 | 2 |
| V61 | Quinua | KAAT007_329 | ||
| Victor Fajardo | VF41 | Quinua | KGA20_230 | 3 |
| VF73 | Quinua | KAAT007_326 | ||
| VF79 | Quinua | KCAA011_224 | ||
| Huanca Sancos | HS16 | Quinua | QGA17_182 | 2 |
| HS98 | Quinua | QAAT84_188 | ||
| Lucanas | L115 | Quinua | KAAT007_293 | 7 |
| L116 | Quinua | KGA20_182 | ||
| L117 | Quinua | QGAA1_230 | ||
| L119 | Quinua | KAAT007_299, QAAT84_206 | ||
| L120 | Quinua | QAAT012_240, QGA028_230 | ||
| Parinacochas | P124 | Quinua | KCAA011_230 | 5 |
| P125 | Quinua | QAAT84_215 | ||
| P128 | Quinua | KCAA011_263, QAAT081_ 216, QGAA1_317 | ||
| Cangallo | C34 | Quinua | KAAT007_282 | 3 |
| C186 | Utuloñawi | QAAT050_206 | ||
| C187 | yuraq quinua | QGAA1_430 | ||
| Huamanga | Ha10 | Quinua | QGAA1_245, QGAA1_266 | 28 |
| Ha14 | Quinua | KAAT007_222 | ||
| Ha20 | Quinua | KGA20_226, QGA028_208 | ||
| Ha21 | Quinua | KAAT007_210, KCAA011_238 | ||
| Ha24 | Quinua | QAAT012_216 | ||
| Ha47 | Quinua | KAAT007_297, QAAT84_182 | ||
| Ha50 | Quinua | KAAT007_231, KAAT007_303 | ||
| Ha56 | Quinua | QGA17_206 | ||
| Ha58 | Quinua | KGA16_192 | ||
| Ha62 | Quinua | KAAT007_371 | ||
| Ha67 | Quinua | KGA16_193 | ||
| Ha70 | Quinua | KAAT007_374 | ||
| Ha76 | Quinua | QGA028_212 | ||
| Ha81 | Quinua | QGAA1_239 | ||
| Ha82 | Quinua | QAAT071_188 | ||
| Ha85 | Quinua | QAAT071_146 | ||
| Ha94 | Quinua | KAAT007_296, KAAT007_344 | ||
| Ha96 | Quinua | KAAT007_227 | ||
| Ha189 | yuraq quinua | QGA17_150 | ||
| Ha190 | qello quinua | QAAT050_210 | ||
| Ha194 | Realquinua | QAAT050_256 | ||
| Ha197 | Yuraq salcedo | QGAA1_380 | ||
| Huanta | H28 | Quinua | KAAT007_336, QGAA1_362 | 4 |
| H36 | Quinua | KAAT007_252 | ||
| H203 | yuraq quinua | QCA14_245 | ||
| Total | 74 |
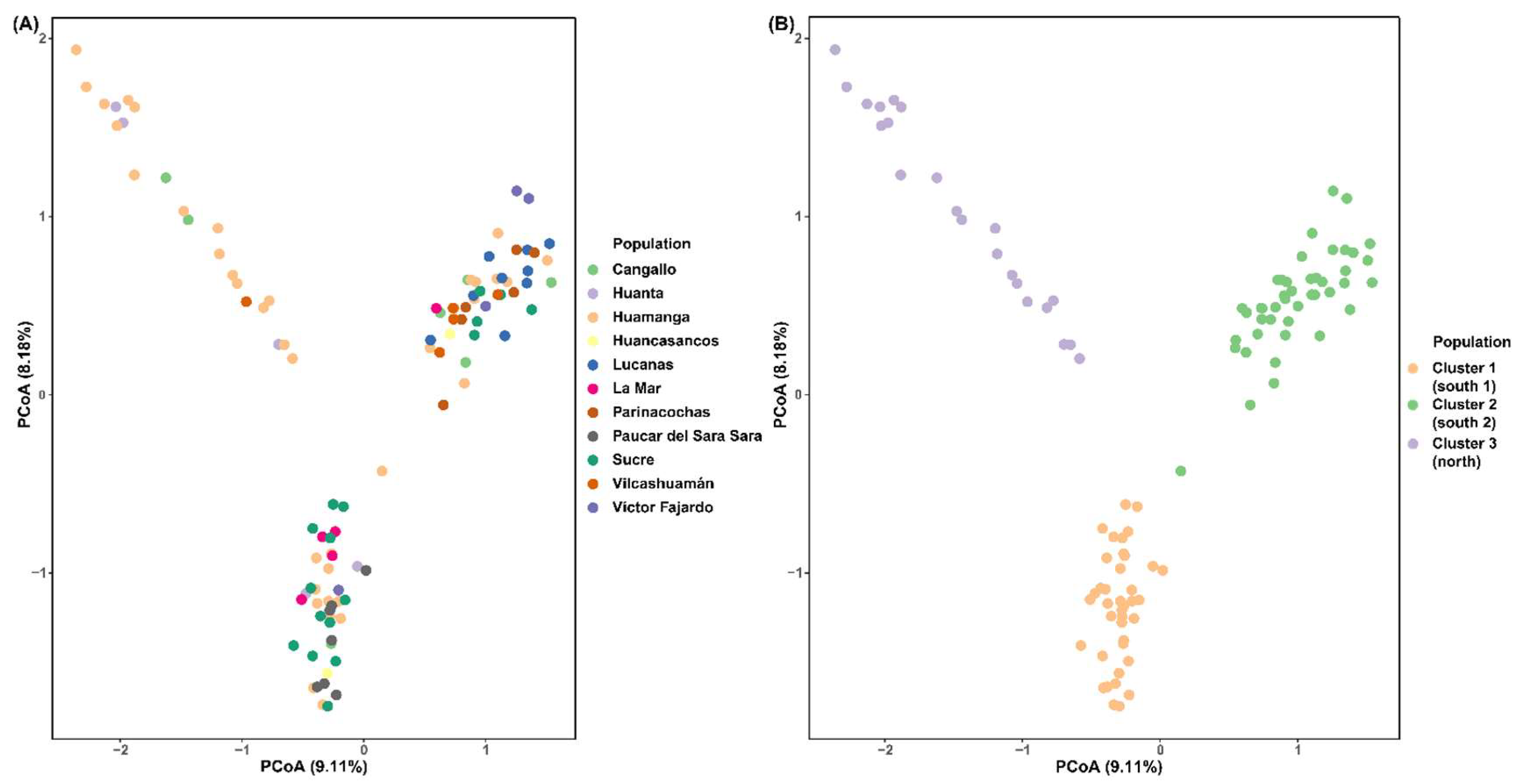
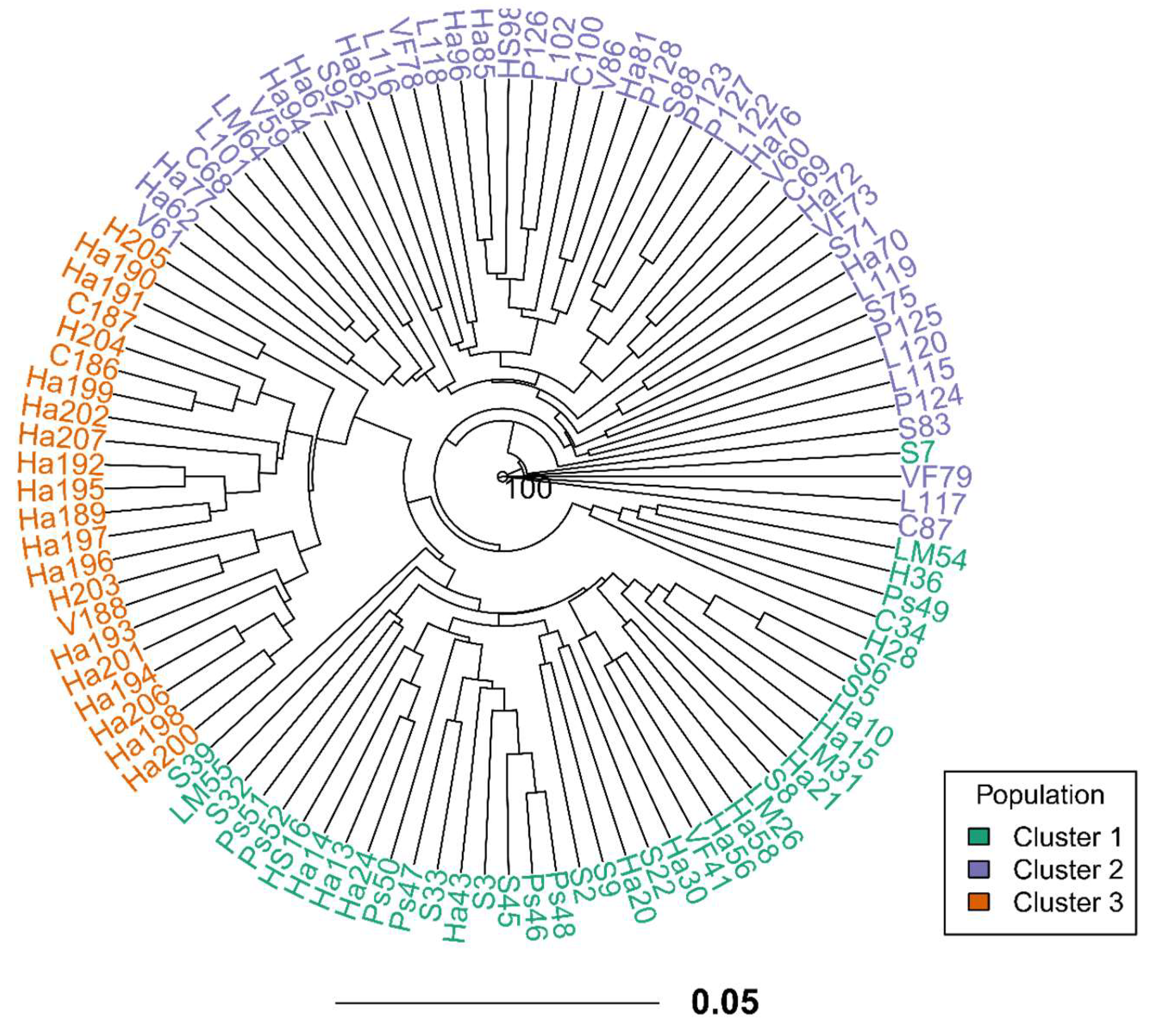
3.2. Genetic Diversity and Population Structure
| Province | Cluster 1 | Cluster 2 | Cluster 3 |
|---|---|---|---|
| Cangallo | 1 | 4 | 2 |
| Huamanga | 11 | 11 | 16 |
| La Mar | 4 | 1 | 0 |
| Huanta | 2 | 0 | 3 |
| Vilcashuaman | 0 | 4 | 1 |
| Huancasancos | 1 | 1 | 0 |
| Victor Fajardo | 1 | 3 | 0 |
| Sucre | 12 | 5 | 0 |
| Lucanas | 0 | 9 | 0 |
| Parinacochas | 0 | 6 | 0 |
| Paucar del Sara Sara | 7 | 0 | 0 |
4. Discussion
4.1. Microsatellite Diversity
4.2. Population Structure
4.3. Genetic Diversity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Heitkam, T.; Weber, B.; Walter, I.; Liedtke, S.; Ost, C.; Schmidt, T. Satellite DNA Landscapes after Allotetraploidization of Quinoa (Chenopodium Quinoa) Reveal Unique A and B Subgenomes. Plant Journal 2020, 103, 32–52. [Google Scholar] [CrossRef] [PubMed]
- Maughan, P.J.; Kolano, B.A.; Maluszynska, J.; Coles, N.D.; Bonifacio, A.; Rojas, J.; Coleman, C.E.; Stevens, M.R.; Fairbanks, D.J.; Parkinson, S.E.; et al. Molecular and Cytological Characterization of Ribosomal RNA Genes in Chenopodium Quinoa and Chenopodium Berlandieri. Genome 2006, 49, 825–839. [Google Scholar] [CrossRef] [PubMed]
- Jarvis, D.E.; Ho, Y.S.; Lightfoot, D.J.; Schmöckel, S.M.; Li, B.; Borm, T.J.A.; Ohyanagi, H.; Mineta, K.; Michell, C.T.; Saber, N.; et al. The Genome of Chenopodium Quinoa. Nature 2017, 542, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Maughan, P.J.; Turner, T.B.; Coleman, C.E.; Elzinga, D.B.; Jellen, E.N.; Morales, J.A.; Udall, J.A.; Fairbanks, D.J.; Bonifacio, A. Characterization of Salt Overly Sensitive 1 (SOS1) Gene Homoeologs in Quinoa (Chenopodium Quinoa Willd.). Genome 2009, 52, 647–657. [Google Scholar] [CrossRef]
- Brown, D.C.; Cepeda-Cornejo, V.; Maughan, P.J.; Jellen, E.N. Characterization of the Granule-Bound Starch Synthase I Gene in Chenopodium. Plant Genome 2015, 8. [Google Scholar] [CrossRef]
- Štorchová, H.; Drabešová, J.; Cháb, D.; Kolář, J.; Jellen, E.N. The Introns in FLOWERING LOCUS T-LIKE (FTL) Genes Are Useful Markers for Tracking Paternity in Tetraploid Chenopodium Quinoa Willd. Genet Resour Crop Evol 2015, 62, 913–925. [Google Scholar] [CrossRef]
- Uhle, M. Fundamentos Etnicos y Arqueologías de Arica y Tacna; Universidad Central: Quito, 1922. [Google Scholar]
- Lumbreras, L.G.; Kaulicke, P.; Santillana, J.I.; Espinoza, W. Economía Prehispanica. In Compendio de historia economica del Peru; Contreras, C., Ed.; Banco Central de Reserva del Perú: Lima, 2020. [Google Scholar]
- Bazile, D.; Fuentes, F.; Mujica, Á. Historical Perspectives and Domestication. In Quinoa: botany, production and uses; CABI: Wallingford, 2013; pp. 16–35. [Google Scholar]
- Bhargava, A.; Shukla, S.; Ohri, D. Chenopodium Quinoa - An Indian Perspective. Ind Crops Prod 2006, 23, 73–87. [Google Scholar] [CrossRef]
- Vargas Zambrano, P.; Arteaga Solorzano, R.; Cruz Viera, L. Análisis Bibliográfico Sobre El Potencial Nutricional de La Quinua (Chenopodium Quinoa) Como Alimento Funcional. Centro azúcar 2019, 46, 89–100. [Google Scholar]
- Melini, V.; Melini, F. Functional Components and Anti-Nutritional Factors in Gluten-Free Grains: A Focus on Quinoa Seeds. Foods 2021, 10, 351. [Google Scholar] [CrossRef]
- Abellán Ruiz, M.S.; Barnuevo Espinosa, M.D.; García Santamaría, C.; Contreras Fernández, C.J.; Aldeguer García, M.; Soto Méndez, F.; Guillén Guillén, I.; Luque Rubia, A.J.; Quinde Ràzuri, F.J.; Martínez Garrido, A.; et al. Efecto Del Consumo de Quinua (Chenopodium Quinoa) Como Coadyuvante En La Intervención Nutricional En Sujetos Prediabéticos. Nutr Hosp 2017, 34, 1163–1169. [Google Scholar] [CrossRef]
- Grados Torrez, R.E.; Trino, R.D.; Perez Gonzales, J.; Gonzales Dávalos, E. Determinación Del Índice Glucémico de Un Producto Elaborado a Base de Amaranto (Amaranthus Caudatus Linnaeus), Quinua (Chenopodium Quinoa Willd) y Tarwi (Lupinus Mutabilis Sweet) Para Tratamiento Coadyuvante de Diabetes Tipo 2 y Obesidad. Revista Con-Ciencia 2018, 6, 73–82. [Google Scholar]
- Abugoch, L.; Castro, E.; Tapia, C.; Añón, M.C.; Gajardo, P.; Villarroel, A. Stability of Quinoa Flour Proteins (Chenopodium Quinoa Willd.) during Storage. Int J Food Sci Technol 2009, 44, 2013–2020. [Google Scholar] [CrossRef]
- Bioversity International; FAO; PROINPA; INIAF; FICA Descriptores Para Quinua (Chenopodium Quinoa Willd.) y Sus Parientes Silvestres; Bioversity International: Rome, 2013;
- Jacobsen, S.-E. The Worldwide Potential for Quinoa ( Chenopodium Quinoa Willd.). Food Reviews International 2003, 19, 167–177. [Google Scholar] [CrossRef]
- Bhargava, A.; Srivastava, S. Quinoa: Botany, Production and Uses; CABI, 2013.
- Kumar, A.; Bhargava, A.; Shukla, S.; Singh, H.B.; Ohri, D. Screening of Exotic Chenopodium Quinoa Accessions for Downy Mildew Resistance under Mid-Eastern Conditions of India. Crop Protection 2006, 25, 879–889. [Google Scholar] [CrossRef]
- Isobe, K.; Sugiyama, H.; Okuda, D.; Murase, Y.; Harada, H.; Miyamoto, M.; Koide, S.; Higo, M.; Torigoe, Y. Effects of Sowing Time on the Seed Yield of Quinoa (Chenopodium Quinoa Willd) in South Kanto, Japan. Agricultural Sciences 2016, 07, 146–153. [Google Scholar] [CrossRef]
- Zohry, A.E.-H. Prospects of Quinoa Cultivation in Marginal Lands of Egypt. Moroccan Journal of Agricultural Sciences 2020, 1, 132–137. [Google Scholar]
- Bertero, H.D.; De La Vega, A.J.; Correa, G.; Jacobsen, S.E.; Mujica, A. Genotype and Genotype-by-Environment Interaction Effects for Grain Yield and Grain Size of Quinoa (Chenopodium Quinoa Willd.) as Revealed by Pattern Analysis of International Multi-Environment Trials. Field Crops Res 2004, 89, 299–318. [Google Scholar] [CrossRef]
- Bhargava, A.; Shukla, S.; Rajan, S.; Ohri, D. Genetic Diversity for Morphological and Quality Traits in Quinoa (Chenopodium Quinoa Willd.) Germplasm. Genet Resour Crop Evol 2007, 54, 167–173. [Google Scholar] [CrossRef]
- Nunez Carrasco, L.; Bazile, D.; Chia, E.; Hocdé, H.; Negrete Sepulveda, J.; Martinez, E.A. Representaciones Sociales Acerca de La Conservacion de La Biodiversidad En El Caso de Productores Tradicionales de Chenopodium Quinoa Willd Del Secano Costero En Las Regiones de O’Higgins y El Maule. In Proceedings of the XXX Congreso Nacional y XV Internacional de Geografia; Talca, 2009; p. 36.
- Morillo, E. Estudio de La Diversidad Genética Del Banco de Germoplasma de Quinua (Chenopodium Quinoa W.) Utilizando Marcadores Moleculares; Quito, 2002.
- Rana, T.S.; Narzary, D.; Ohri, D. Genetic Diversity and Relationships among Some Wild and Cultivated Species of Chenopodium L. (Amaranthaceae) Using RAPD and DAMD Methods. Curr Sci 2010, 98, 840–846. [Google Scholar]
- Del Castillo, C.; Winkel, T.; Mahy, G.; Bizoux, J.P. Genetic Structure of Quinoa (Chenopodium Quinoa Willd.) from the Bolivian Altiplano as Revealed by RAPD Markers. Genet Resour Crop Evol 2007, 54, 897–905. [Google Scholar] [CrossRef]
- Anabalón Rodríguez, L.; Thomet Isla, M. Comparative Analysis of Genetic and Morphologic Diversity among Quinoa Accessions (Chenopodium Quinoa Willd.) of the South of Chile and Highland Accessions. African Journal of Crop Science 2018, 6, 001–007. [Google Scholar]
- García-Godos, P.; Cueva-Castillo, J.M. Genetic Variability of 29 Peruvian Quinoa (Chenopodium Quinoa Willd) Accessions Using AFLP Markers and Multivariate Analysis. Scientia Agropecuaria 2021, 12, 57–64. [Google Scholar] [CrossRef]
- Morillo Coronado, A.-C.; Manjarres, E.H.; Morillo Coronado, Y. Molecular Characterization of Chenopodium Quinoa Willd. Using Inter-Simple Sequence Repeat (ISSR) Markers. Afr J Biotechnol 2017, 16, 483–489. [Google Scholar] [CrossRef]
- Zhang, T.; Gu, M.; Liu, Y.; Lv, Y.; Zhou, L.; Lu, H.; Liang, S.; Bao, H.; Zhao, H. Development of Novel InDel Markers and Genetic Diversity in Chenopodium Quinoa through Whole-Genome Re-Sequencing. BMC Genomics 2017, 18. [Google Scholar] [CrossRef] [PubMed]
- Laosatit, K.; Taytragool, S.; Pimsaythong, K.; Somta, P.; Tanadul, O.U.M. Genetic Diversity of Quinoa (Chenopodium Quinoa Willd.) Germplasm as Revealed by Sequence-Related Amplified Polymorphism Markers. Agriculture and Natural Resources 2021, 55, 341–348. [Google Scholar] [CrossRef]
- El-Harty, E.H.; Ghazy, A.; Alateeq, T.K.; Al-Faifi, S.A.; Khan, M.A.; Afzal, M.; Alghamdi, S.S.; Migdadi, H.M. Morphological and Molecular Characterization of Quinoa Genotypes. 2021. [CrossRef]
- Abd El-Moneim, D.; ELsarag, E.; Aloufi, S.; El-Azraq, A.; ALshamrani, S.; Safhi, F.; Ibrahim, A. Quinoa (Chenopodium Quinoa Willd.): Genetic Diversity According to ISSR and SCoT Markers, Relative Gene Expression, and Morpho-Physiological Variation under Salinity Stress. Plants 2021, 10, 2802. [Google Scholar] [CrossRef]
- Christensen, S.A.; Pratt, D.B.; Pratt, C.; Nelson, P.T.; Stevens, M.R.; Jellen, E.N.; Coleman, C.E.; Fairbanks, D.J.; Bonifacio, A.; Maughan, P.J. Assessment of Genetic Diversity in the USDA and CIP-FAO International Nursery Collections of Quinoa (Chenopodium Quinoa Willd.) Using Microsatellite Markers. Plant Genetic Resources: Characterisation and Utilisation 2007, 5, 82–95. [Google Scholar] [CrossRef]
- Costa Tártara, S.M.; Manifesto, M.M.; Bramardi, S.J.; Bertero, H.D. Genetic Structure in Cultivated Quinoa (Chenopodium Quinoa Willd.), a Reflection of Landscape Structure in Northwest Argentina. Conservation Genetics 2012, 13, 1027–1038. [Google Scholar] [CrossRef]
- Manjarres-Hernández, E.H.; Morillo-Coronado, A.C. Genetic Diversity of Colombian Quinoa (Chenopodium Quinoa Willd.): Implications for Breeding Programs. Genet Resour Crop Evol 2022, 69, 2447–2458. [Google Scholar] [CrossRef]
- Kandy, A.; Choque, H.; Huanca Alanoca, N.; Mamani, F.M.; Torrico, S.V. Diversidad Fenotípica y Genotípica de 50 Nuevas Accesiones de Quinua (Chenopodium Quinoa Willd.), Del Banco de Germoplasma de Granos Altoandinos.
- Vía y Rada Fernández, R.N. Determinación de La Diversidad Genética de 172 Accesiones de La Colección Nacional de Chenopodium Quinoa Willd. “Quinua” Mediante Marcadores Microsatélites, Universidad Ricardo Palma: Lima, 2015.
- Cárdenas Córdova, R.G. Caracterización Molecular de 129 Accesiones de Quinua(Chenopodium Quinoa Willd.) de La Región Puno Mediante Marcadores Microsatélites, Universidad Nacional Mayor de San Marcos: Lima, 2017.
- Romero, M.; Mujica, A.; Pineda, E.; Ccamapaza, Y.; Zavalla, N.; de Juli, B.; Collana, N.; Inia, I.; Blanca de Juli, A.; Blanca de Arequipa, A.; et al. Genetic Identity Based on Simple Sequence Repeat (SSR) Markers for Quinoa (Chenopodium Quinoa Willd.) Chenopodium Petiolare from the CIP-Camacani. Cien. Inv. Agr 2019, 46, 166–178. [Google Scholar] [CrossRef]
- Salazar, J.; de Lourdes Torres, M.; Gutierrez, B.; Torres, A.F. Molecular Characterization of Ecuadorian Quinoa (Chenopodium Quinoa Willd.) Diversity: Implications for Conservation and Breeding. Euphytica 2019, 215. [Google Scholar] [CrossRef]
- Arenas Morales, V.E. Análisis de La Diversidad Genética y Estructura Poblacional En Genotipos de Quínoa Chilena (Chenopodium Quinoa Willd.) Usando Microsatélites, Universidad Andrés Bello: Santiago de Chile, 2018.
- Alfaro Mendivil, E. Establecimiento Del Banco Regional de Germoplasma y Caracterización Fenotípica de Accesiones de Quinua (Chenopodium Quinoa Willd.) Ayacucho, Universidad Nacional San Cristóbal de Huamanga: Huamanga, 2013.
- Menéndez Burns, F.M. Estudio Poblacional y Diversidad Genética de Los Cultivos Primarios de Chenopodium Quinoa En Ayacucho, Perú, Universidad Peruana Cayetano Heredia: Lima, 2016.
- Pedersen, D.; Tremblay, J.; Errázuriz, C.; Gamarra, J. The Sequelae of Political Violence: Assessing Trauma, Suffering and Dislocation in the Peruvian Highlands. Soc Sci Med 2008, 67, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Truth and Reconciliation Commission Final Report.
- Doyle, J.J.; Doyle, J.L. A Rapid DNA Isolation Procedure for Small Quantities of Fresh Leaf Tissue. Phytochemical Bulletin 1987, 19, 11–15. [Google Scholar]
- Mason, S.L.; Stevens, M.R.; Jellen, E.N.; Bonifacio, A.; Fairbanks, D.J.; Coleman, C.E.; McCarty, R.R.; Rasmussen, A.G.; Maughan, P.J. Development and Use of Microsatellite Markers for Germplasm Characterization in Quinoa (Chenopodium Quinoa Willd.). Crop Sci 2005, 45, 1618–1630. [Google Scholar] [CrossRef]
- Jarvis, D.E.; Kopp, O.R.; Jellen, E.N.; Mallory, M.A.; Pattee, J.; Bonifacio, A.; Coleman, C.E.; Stevens, M.R.; Fairbanks, D.J.; Maughan, P.J. Simple Sequence Repeat Marker Development and Genetic Mapping in Quinoa (Chenopodium Quinoa Willd.). J Genet 2008, 87, 39–51. [Google Scholar] [CrossRef]
- Ward, S.M. Response to Selection for Reduced Grain Saponin Content in Quinoa (Chenopodium Quinoa Willd.). Field Crops Res 2000, 68, 157–163. [Google Scholar] [CrossRef]
- Nei, M.; Maruyama, T.; Chakraborty, R. The Bottleneck Effect and Genetic Variability in Populations. Evolution (N Y) 1975, 29, 1–10. [Google Scholar]
- Chesnokov, Yu.V.; Artemyeva, A.M. Evaluation of the Measure of Polymorphism Information of Genetic Diversity. Sel’skokhozyaistvennaya Biologiya 2015, 50, 571–578. [Google Scholar] [CrossRef]
- Prevosti, A.; Ocaña, J.; Alonso, G. Distances between Populations of Drosophila Subobscura, Based on Chromosome Arrangement Frequencies. Theoretical and Applied Genetics 1975, 45, 231–241. [Google Scholar] [CrossRef]
- R Core Team R: A Language and Environment for Statistical Computing 2022.
- Kamvar, Z.N.; Tabima, J.F.; Gr̈unwald, N.J. Poppr: An R Package for Genetic Analysis of Populations with Clonal, Partially Clonal, and/or Sexual Reproduction. PeerJ 2014, 2014, 1–14. [Google Scholar] [CrossRef]
- Dray, S.; Dufour, A.-B. The Ade4 Package: Implementing the Duality Diagram for Ecologists. J Stat Softw 2007, 22. [Google Scholar] [CrossRef]
- Jombart, T. Adegenet : A R Package for the Multivariate Analysis of Genetic Markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the Number of Clusters of Individuals Using the Software STRUCTURE: A Simulation Study. Mol Ecol 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Francis, R.M. Pophelper: An R Package and Web App to Analyse and Visualize Population Structure. In Proceedings of the Molecular Ecology Resources; Blackwell Publishing Ltd, January 1 2017; Vol. 17, pp. 27–32.
- Ott, J. Strategies for Characterizing Highly Polymorphic Markers in Human Gene Mapping. The American Society of Human Genetics 1992, 51, 283–290. [Google Scholar]
- Yataco Capcha, J.; Nami, H.G. A New View on the Late Pleistocene Lithic Remains from Pikimachay Cave, South Central Peru. Archaeological Discovery 2022, 10, 282–334. [Google Scholar] [CrossRef]
- Fuentes, F.F.; Martinez, E.A.; Hinrichsen, P. V.; Jellen, E.N.; Maughan, P.J. Assessment of Genetic Diversity Patterns in Chilean Quinoa (Chenopodium Quinoa Willd.) Germplasm Using Multiplex Fluorescent Microsatellite Markers. Conservation Genetics 2009, 10, 369–377. [Google Scholar] [CrossRef]
- Allende Ciballero, M.J. Caracterización Morfológica y Molecular de Accesiones de Quinua (Chenopodium Quinoa Willd.) Para Estimar Variabilidad Genética, Universidad Nacional Agraria la Molina: Lima, 2017.
- Stepputat, F.; Nyberg Sørensen, N. The Rise and Fall of ‘Internally Displaced People’ in the Central Peruvian Andes. Dev Change 2001, 32, 769–791. [Google Scholar] [CrossRef]
- FAO Evaluación Del Año Internacional de La Quinua (2013); Roma, 2014.
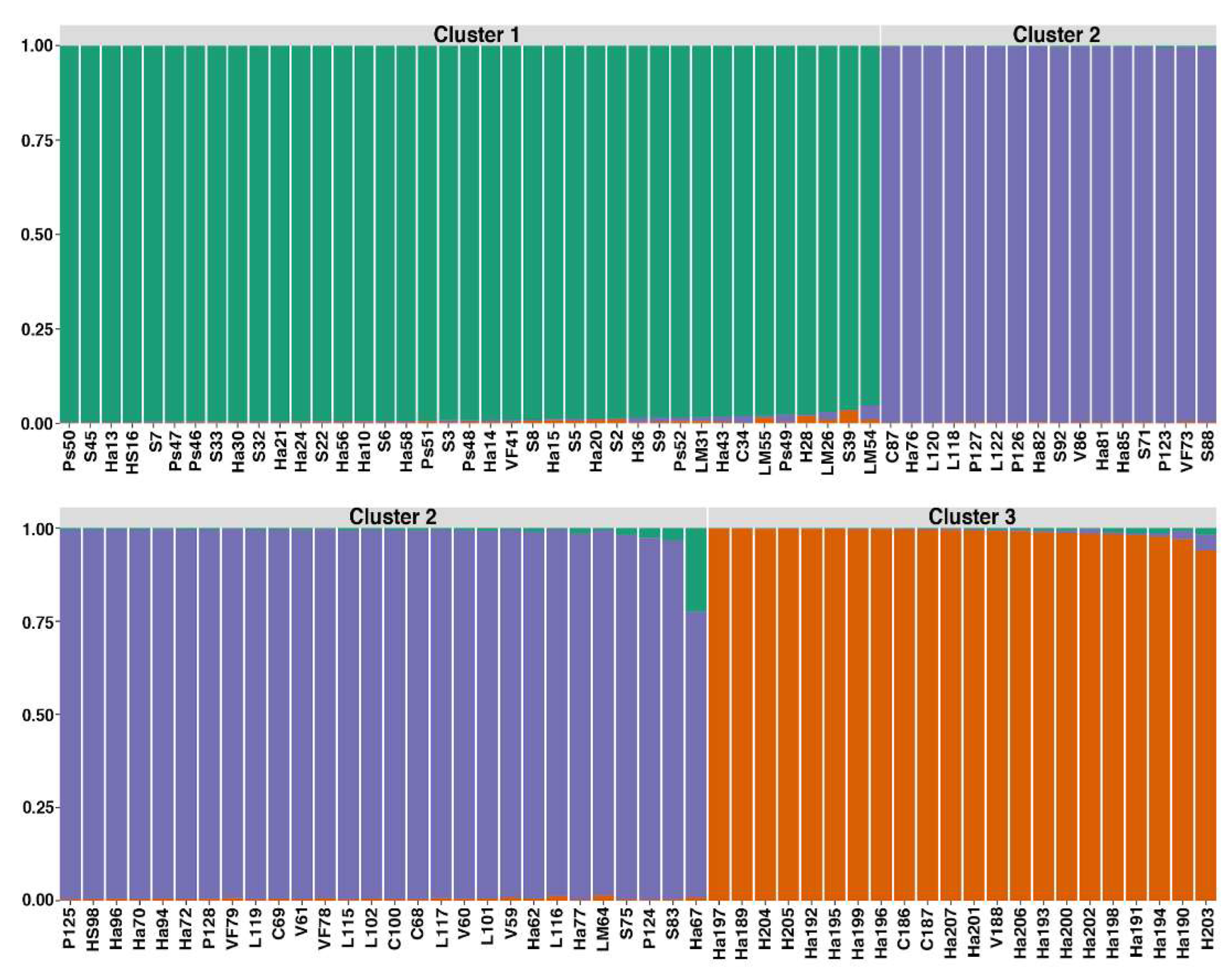
| Cluster | N | H | Lambda | HE | PIC | PPL |
|---|---|---|---|---|---|---|
| C1 (south 1) | 39 | 3.66 | 0.974 | 0.0838 | 0.095 | 73.05 |
| C2 (south 2) | 44 | 3.78 | 0.977 | 0.095 | 0.087 | 68.44 |
| C3 (north) | 22 | 3.09 | 0.955 | 0.0594 | 0.093 | 52.48 |
| Cluster (C) | C1 | C2 | |
|---|---|---|---|
| C2 | 0.023 | ||
| C3 | 0.022 | 0.034 | |
| Source | Df | SS | MS | Est. Var. | % |
|---|---|---|---|---|---|
| Between clusters | 2 | 221.27 | 110.64 | 2.93 | 19.93 |
| Within clusters | 102 | 1200.77 | 11.77 | 11.77 | 80.07 |
| Total | 104 | 1422.00 | 13.67 | 14.70 | 100.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





