Results and Discussion
The average island length of Rattus
norvegicus in the
X-chromosome is 596.19 and Sus scrofa has an average island length of 580.44 it means Rattus norvegicus has
a greater average island length as compared to Sus scrofa as shown in the table. Variation in island length is more in both
species. The standard error of the CpG island of the X-chromosome of Rattus norvegicus is 3.68 and in Sus scrofa the value is2.02. It means Rattus norvegicus has a
greater variation in their genome. Island number varies in both species.
X-chromosome of Rattus norvegicus island
number is 4465 and in Sus scrofa the value is 13539. It means Sus scrofa has more island number as
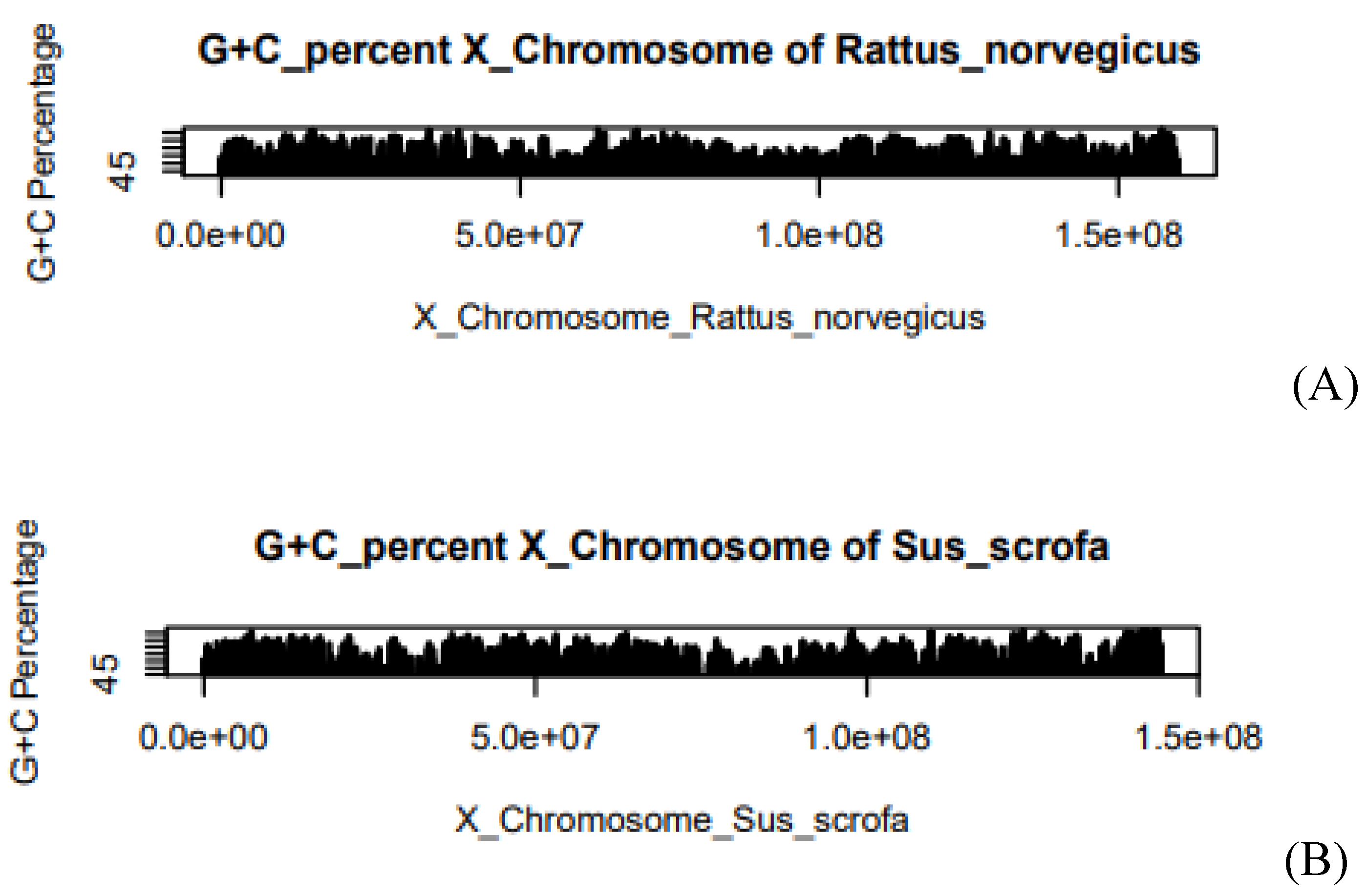
compared to Rattus norvegicus. Average G+C
concentration is close to each other in both species. In the X-chromosome of Sus
scrofa, the value is54.5 and the
case of the Rattus norvegicus value is 53.03. The average CpG concentration is close to each other as shown in Table 1. The value of average
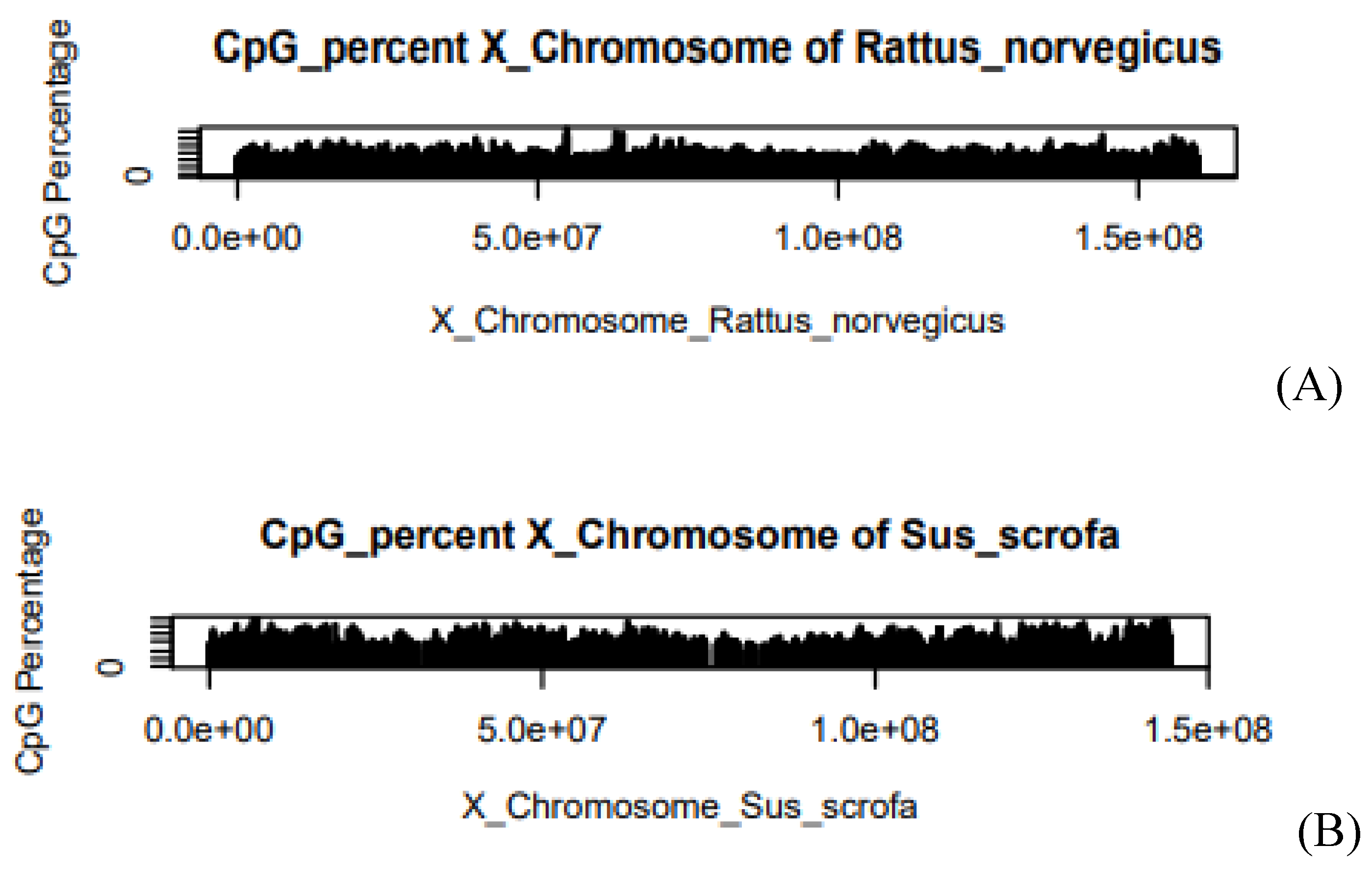
CpG concentration in Rattus norvegicus is 4.94 and in the
case of Sus scrofa it is 5.13. It means there is a small difference in the value of both species.
The average ratio (Observed CpG/Expected CpG) is 0.73 in Rattus
norvegicus in the case of Sus
scrofa 0.7. It means the value
of the average ratio of Rattus norvegicus is
greater than the value of Sus scrofa. We have
got the minimum island length which is the same in both species. Both species
have a similar value that is 500. The maximum island length value of Sus scrofa is 5639 and in the case of Rattus
norvegicus that is 4484. It means Sus
scrofa has a maximum island length as compared to Rattus norvegicus.
Table 1.
Final parameters of CpG island in in X-chromosome of Rattus norvegicus and Sus scrofa.
Table 1.
Final parameters of CpG island in in X-chromosome of Rattus norvegicus and Sus scrofa.
| Features |
Rat |
Swine |
| Average Island Length |
596.19 |
580.44 |
| Standard error (Island Length) |
3.68 |
2.02 |
| Island Number |
4465 |
13539 |
| Average G+ C percent |
53.03 |
54.5 |
| Standard error G+ C percent |
0.07 |
0.05 |
| Average CpG percent |
4.94 |
5.13 |
| Standard error CpG percent |
0.01 |
0.01 |
| Average Ratio |
0.73 |
0.70 |
| Standard error Ratio |
0 |
0 |
| Minimum Island length |
500 |
500 |
| Maximum Island length |
4484 |
5639 |
The average island length of Rattus
norvegicus in Y-chromosome is 560.46 and Sus scrofa has an average island
length of 567.28 it means Sus scrofa has a greater average island length as compared to Rattus
norvegicus as shown in the table. Variation in
island length is more in both species. The standard error of the CpG island of
Y-chromosome of Rattus norvegicus is 12.67 and in Sus scrofa the value is 4.55. It means Rattus norvegicus has a greater variation in their genome. Island number varies in
both species. Y-chromosome of Rattus norvegicus island number is 110 and in Sus
scrofa the value is 1820. It means Sus scrofa has more island number as
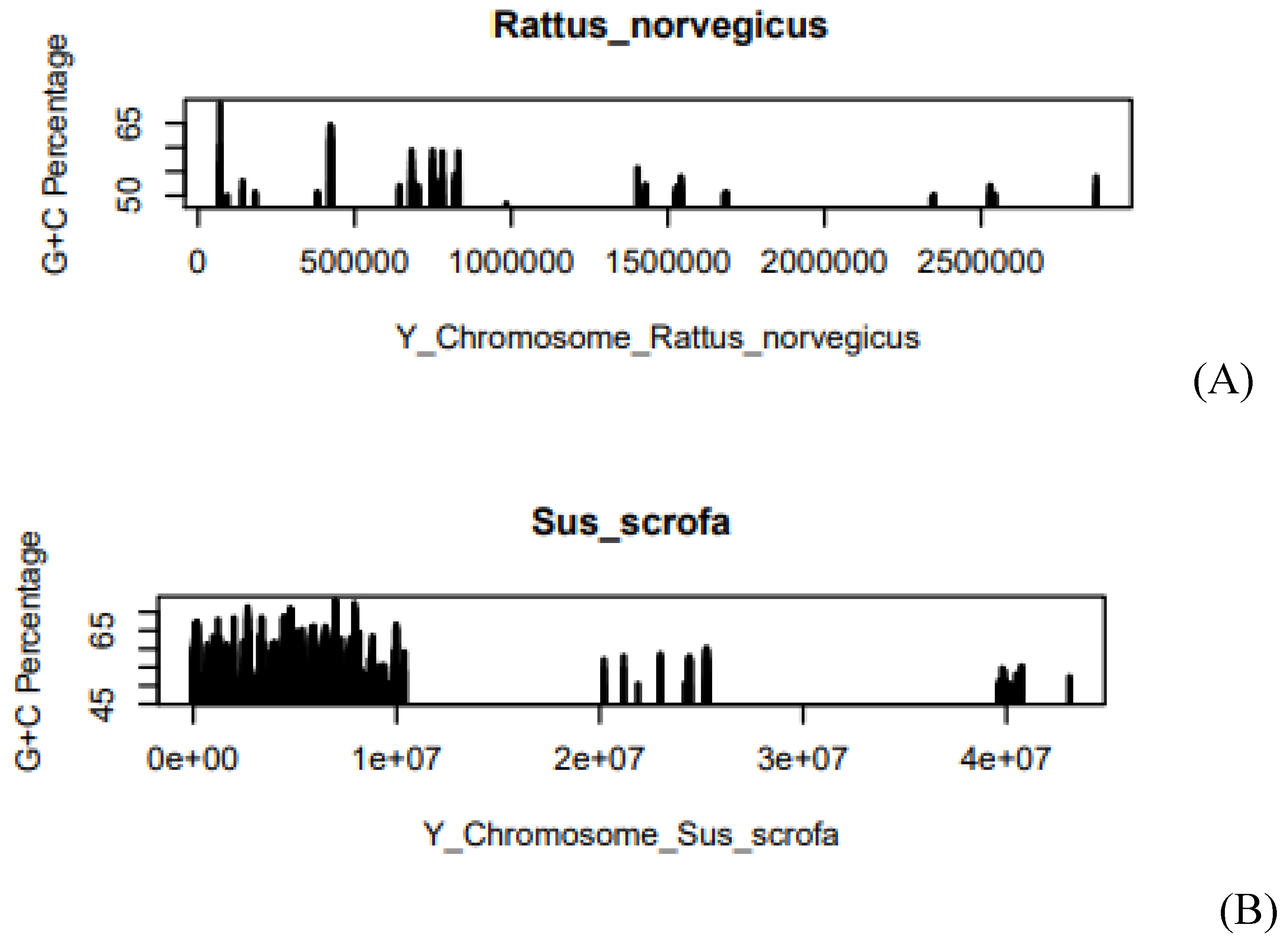
compared to Rattus norvegicus. Average G+C
concentration is close to each other in both species. In the Y-chromosome of Sus
scrofa the value is 53.86 and in
the case of the Rattus norvegicus value is 52.7. The average CpG concentration is close to each other as shown in Table 2. The value of average
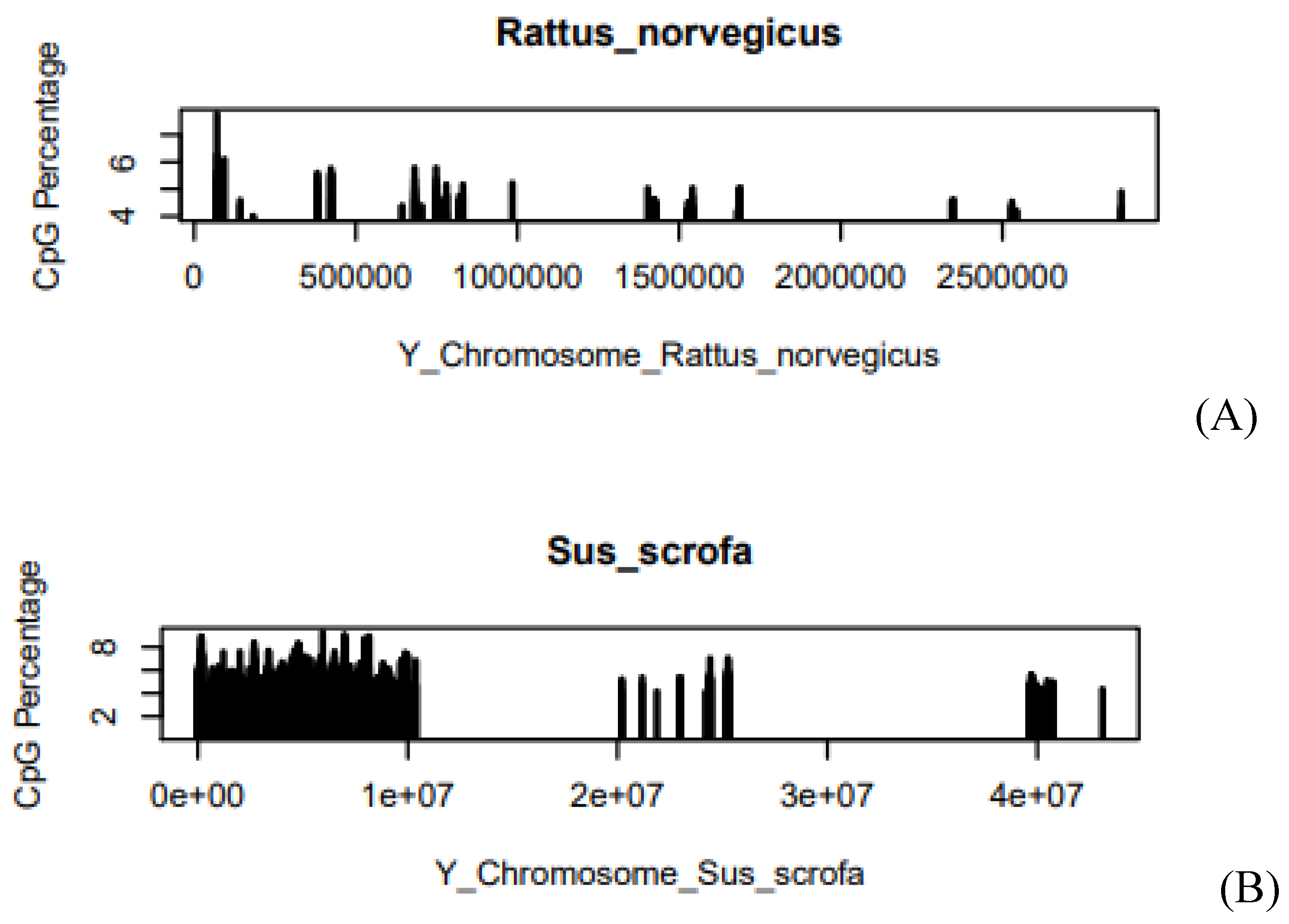
CpG concentration in Rattus norvegicus is 4.73 and in the case of Sus scrofa, it is
4.96. It means there is a small difference in the value
of both species. The average ratio (Observed CpG/Expected CpG) is 0.7 in
Rattus norvegicus in the case of Sus scrofa 0.7. We have got the minimum island length which is the same in
both species. Both species have a similar value that is 500. The maximum island
length value of Sus scrofa is 2650 and in the case of Rattus norvegicus, that is 1059. It means Sus scrofa has a maximum island length as compared to Rattus
norvegicus.
Table 2.
Final parameters of CpG island in in Y-chromosome of Rattus norvegicus and Sus scrofa.
Table 2.
Final parameters of CpG island in in Y-chromosome of Rattus norvegicus and Sus scrofa.
| Features |
Rat |
Swine |
| Average Island Length |
560.46 |
567.28 |
| Standard error Island Length |
12.67 |
4.55 |
| Island Number |
110 |
1820 |
| Average G+ C percent |
52.7 |
53.86 |
| Standard error G+ C percent |
0.37 |
0.11 |
| Average CpG percent |
4.73 |
4.96 |
| Standard error CpG percent |
0.06 |
0.02 |
| Average Ratio |
0.7 |
0.7 |
| Standard error Ratio |
0.01 |
0 |
| Minimum Island length |
500 |
500 |
| Maximum Island length |
1059 |
2650 |
Figure 1.
G+C% of Rattus norvegicus in X-chromosome, (B) G+C% of Sus scrofa in X-chromosome. The X-axis of the graph describes the length of the chromosome. Where e is the exponential constant, for example, 5.0e+07 can be written as 5*107. On the Y-axis, the graph shows the G+C%. At some positions, we have obtained the higher G+C% whereas at some positions we have obtained the lower G+C%.
Figure 1.
G+C% of Rattus norvegicus in X-chromosome, (B) G+C% of Sus scrofa in X-chromosome. The X-axis of the graph describes the length of the chromosome. Where e is the exponential constant, for example, 5.0e+07 can be written as 5*107. On the Y-axis, the graph shows the G+C%. At some positions, we have obtained the higher G+C% whereas at some positions we have obtained the lower G+C%.
Figure 2.
(A) G+C% of Rattus norvegicus in Y-chromosome, (B) G+C% of Sus scrofa in Y-chromosome. The X-axis of the graph describes the length of the chromosome. Where e is the exponential constant, for example, 5.0e+07 can be written as 5*107. On the Y-axis, the graph shows the G+C%. At some positions, we have obtained the higher G+C% whereas at some positions we have obtained the lower G+C%.
Figure 2.
(A) G+C% of Rattus norvegicus in Y-chromosome, (B) G+C% of Sus scrofa in Y-chromosome. The X-axis of the graph describes the length of the chromosome. Where e is the exponential constant, for example, 5.0e+07 can be written as 5*107. On the Y-axis, the graph shows the G+C%. At some positions, we have obtained the higher G+C% whereas at some positions we have obtained the lower G+C%.
Figure 3.
(A) CpG Percentage of Rattus norvegicus in X-chromosome, (B) CpG Percentage of Sus scrofa in X-chromosome. The X-axis of the graph describes the length of the chromosome. Where e is the exponential constant, for example, 5.0e+07 can be written as 5*107. On the Y-axis, the graph shows the CpG Percentage.
Figure 3.
(A) CpG Percentage of Rattus norvegicus in X-chromosome, (B) CpG Percentage of Sus scrofa in X-chromosome. The X-axis of the graph describes the length of the chromosome. Where e is the exponential constant, for example, 5.0e+07 can be written as 5*107. On the Y-axis, the graph shows the CpG Percentage.
Figure 4.
(A) CpG Percentage of Rattus norvegicus in Y-chromosome, (B) CpG Percentage of Sus scrofa in Y-chromosome. The X-axis of the graph describes the length of the chromosome. Where e is the exponential constant, for example, 5.0e+07 can be written as 5*107. On the Y-axis, the graph shows the CpG Percentage.
Figure 4.
(A) CpG Percentage of Rattus norvegicus in Y-chromosome, (B) CpG Percentage of Sus scrofa in Y-chromosome. The X-axis of the graph describes the length of the chromosome. Where e is the exponential constant, for example, 5.0e+07 can be written as 5*107. On the Y-axis, the graph shows the CpG Percentage.
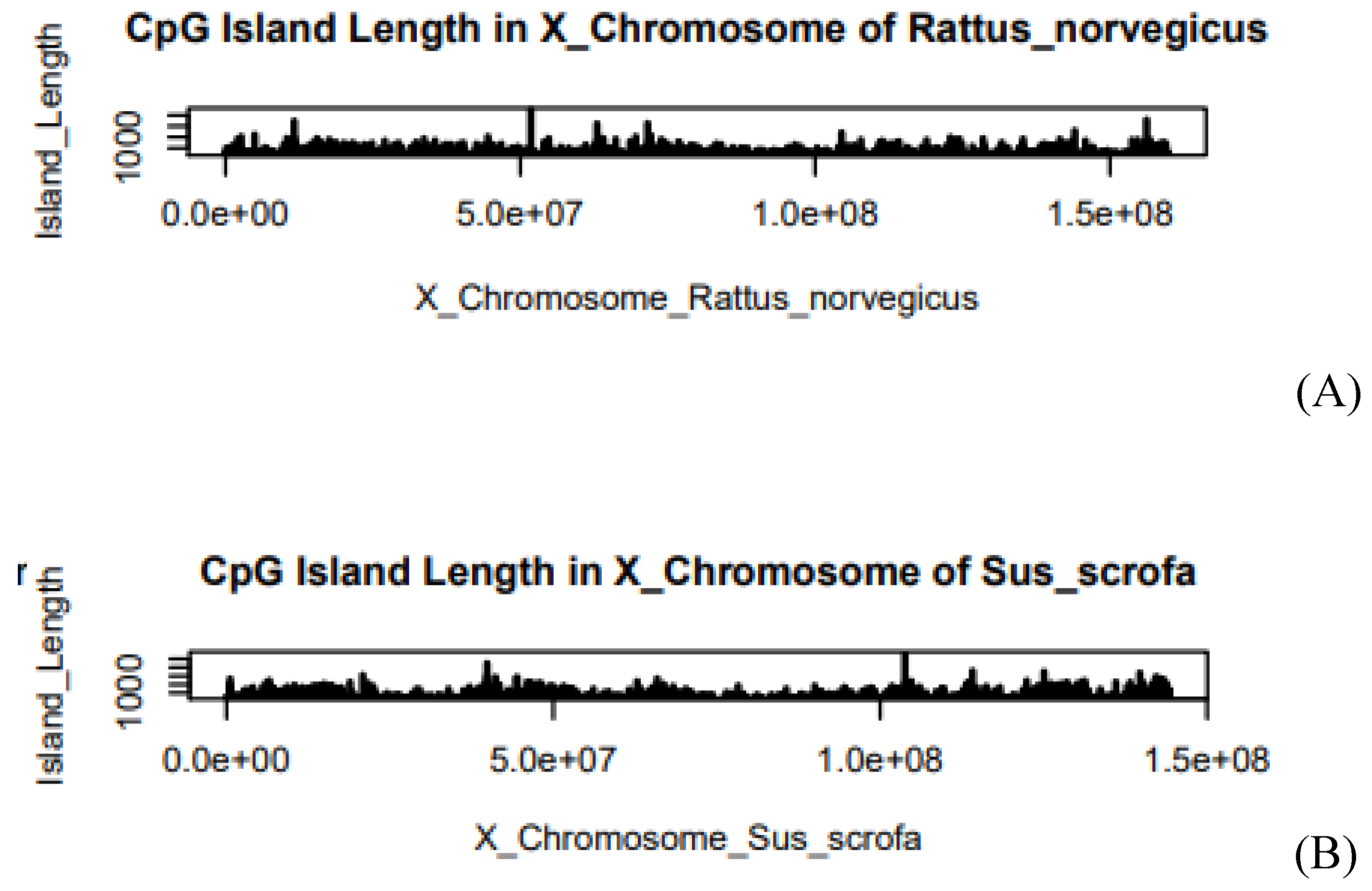
Figure 5.
(A) Island Length of Rattus norvegicus in X-chromosome, (B) Island Length of Sus scrofa in X-chromosome. The X-axis of the graph describes the length of the chromosome. Where e is the exponential constant, for example, 5.0e+07 can be written as 5*107. On the Y-axis, the graph shows the Island Length.
Figure 5.
(A) Island Length of Rattus norvegicus in X-chromosome, (B) Island Length of Sus scrofa in X-chromosome. The X-axis of the graph describes the length of the chromosome. Where e is the exponential constant, for example, 5.0e+07 can be written as 5*107. On the Y-axis, the graph shows the Island Length.
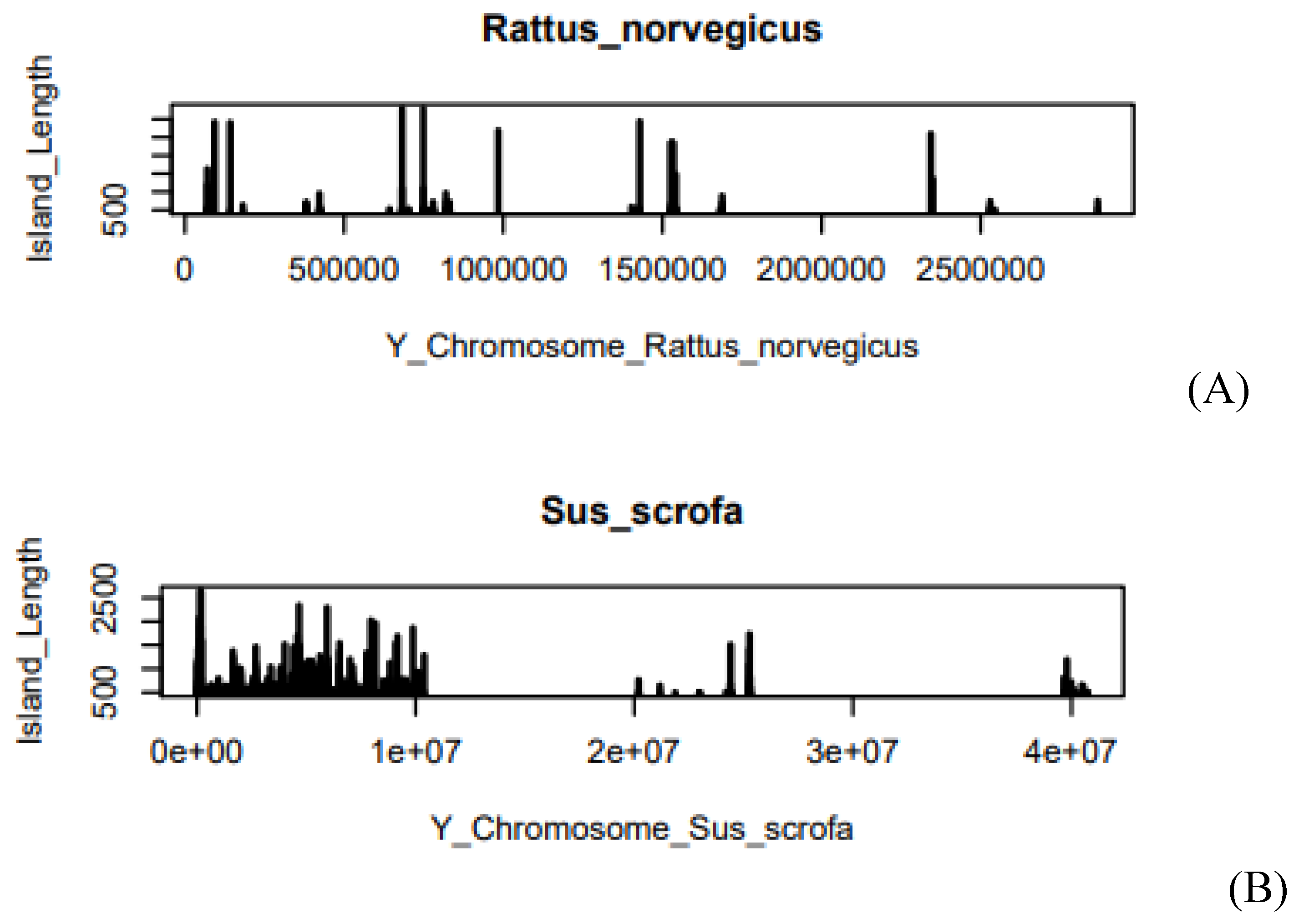
Figure 6.
(A) Island Length of Rattus norvegicus in the Y-chromosome, (B) Island Length of Sus scrofa in the Y-chromosome. The x-axis of the graph describes the length of the chromosome. Where e is the exponential constant, for example, 5.0e+07 can be written as 5*107. On the Y-axis, the graph shows the Island Length.
Figure 6.
(A) Island Length of Rattus norvegicus in the Y-chromosome, (B) Island Length of Sus scrofa in the Y-chromosome. The x-axis of the graph describes the length of the chromosome. Where e is the exponential constant, for example, 5.0e+07 can be written as 5*107. On the Y-axis, the graph shows the Island Length.
In prokaryotes it is associated with restriction-modification systems, targeting short palindromic or nearly palindromic sites. DNA methylation is used as a signal for the regulation of a particular DNA protein interaction. Methylation systems typically comprise of a DNA methylase and one or more DNA binding proteins which will overlap the target methylation site on DNA, subsequently blocking methylation of that site.The laboratory rat (Rattus norvegicus) have incalculable benefits to human health it is mostly used in experimental medicine and drug production. The diploid number chromosome is 42 in Rattus norvegicus and has an autosomal complement consisting of seven pairs of metacentrics, three pairs of submetacentric, and 10 pairs of acrocentrics. The X-chromosome is an acrocentric of medium size and the Y is the smallest acrocentric of the complement (Duncan and Van Peenen 1971). Rattus norvegicus, has multiple copies of Sry Y-chromosome while the maximum mammals have only a single copy (Turner et al. 2007). Pigs (Sus scrofa) have been domesticated in the old world since antiquity. Pig provides valuable products to humans; including fertilizer, leather, pork, and a variety of medicines an indigenous pig has a diploid number of chromosomes (2n) of 38, which includes 18 pairs of autosomes and one pair of allosomes or sex-chromosomes. First, 5 pairs of autosomes are submetacentric, the next 2 pairs are subtelocentric, a subsequent 5 pairs are metacentrics, the remaining six pairs are telocentric and sex-chromosomes are metacentrics in nature. The first chromosome was the longest pair and a thirteenth pair was the second largest, while a Y-chromosome was the smallest in the karyotype of the pig (Vishnu et al. 2015).