Submitted:
10 September 2024
Posted:
11 September 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
3. Results
3.1. Sequencing Statistics
3.2. Most Transcribed Genes in Each Time Point
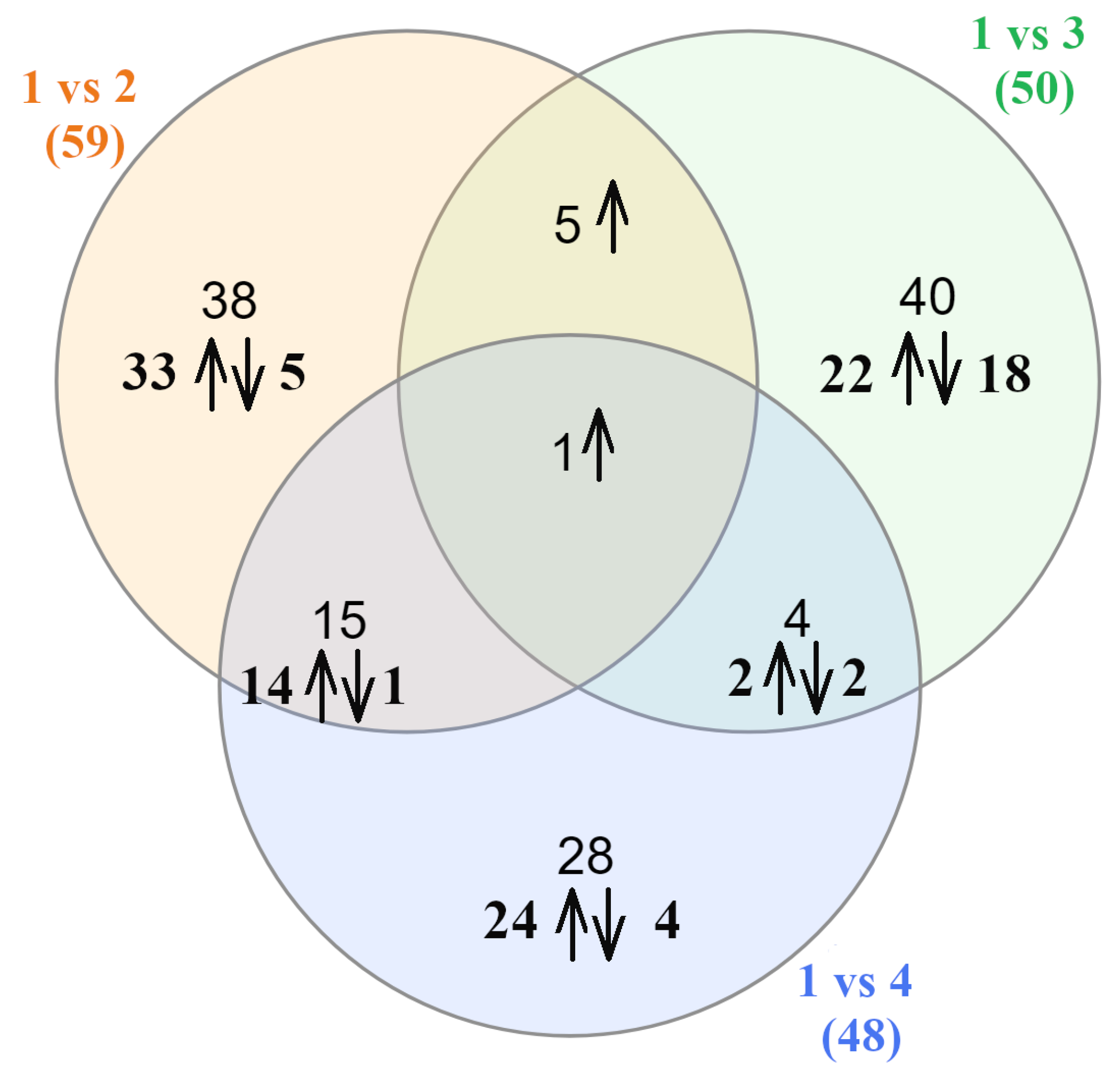
3.3. Differential Gene Expression
| Mapping Reference ID | Annotation | Fold change | P-value | |
|---|---|---|---|---|
| Upregulated | ||||
| CCPA1999.b1 | Hypothetical protein HETIRDRAFT_426980 | 181.85 | 4.72E-05 | |
| CCPB2345.b1 | Carotenoid ester lipase precursor | 145.16 | 2.55E-04 | |
| CCOZ2064.b1 | ATP-utilizing phosphoenolpyruvate carboxykinase | 64.23 | 9.74E-05 | |
| CCPA2867.g1 | Aldo/keto reductase | 45.65 | 1.76E-04 | |
| CCPB993.g1 | Terpenoid cyclases/protein prenyltransferase alpha-alpha toroid | 43.52 | 5.23E-03 | |
| CCPC5739.g1 | NAD-P-binding protein | 34.14 | 1.76E-03 | |
| CCPC2435.b1 | ---Na---* | 33.93 | 2.87E-04 | |
| CCPA4098.g1 | GPI mannosyltransferase 3 | 29.59 | 2.71E-03 | |
| CCPB1601.b1 | ---Na--- | 29.53 | 8.77E-04 | |
| CCOZ1600.b1 | Methionine adenosyltransferase | 28.24 | 4.23E-03 | |
| CCPC3360.b1 | Isocitrate lyase | 27.32 | 9.33E-05 | |
| CCPA4929.b1 | Alpha/beta hydrolase | 25.73 | 5.18E-04 | |
| Downregulated | ||||
| CCPC8078.b1 | Transcription regulator | –23.45 | 7.99E-03 | |
| CCOZ5192.g1 | Dnaj-domain-containing protein | –20.35 | 3.12E-03 | |
| CCPB3914.b1 | Hypothetical protein HETIRDRAFT_426907 | –18.18 | 4.34E-03 | |
| CCOZ3764.b1 | Negative regulator of differentiation 1 | –12.93 | 9.40E-03 | |
| CCPC2832.b1 | Ornithine decarboxylase antizyme-domain-containing protein | –12.24 | 8.22E-03 | |
| CCPA3492.b1 | Hypothetical protein HETIRDRAFT_477666 | –8.36 | 3.06E-03 | |
| Mapping Reference ID | Annotation | Fold change | P-value |
|---|---|---|---|
| Upregulated | |||
| CCPC2187.b1 | Malic enzyme | 61.69 | 2.06E-04 |
| CCOZ3444.b1 | Heat shock protein 70 | 55.59 | 3.80E-03 |
| CCPB993.g1 | Terpenoid cyclases/protein prenyltransferase alpha-alpha toroid | 44.46 | 1.40E-03 |
| CCPC2829.b1 | Pali-domain-containing protein | 32.82 | 2.45E-04 |
| CCPA4010.b1 | Fatty acid desaturase-domain-containing protein | 27.64 | 4.38E-03 |
| CCPC4213.b1 | Hypothetical protein HETIRDRAFT_325943 | 25.72 | 5.16E-03 |
| 11E44-04-08 | Predicted protein | 24.54 | 8.11E-04 |
| CCPC4213.g1 | Hypothetical protein HETIRDRAFT_325943 | 24.37 | 6.08E-03 |
| CCPA4569.b1 | Hypothetical protein HETIRDRAFT_441917 | 23.75 | 3.45E-04 |
| CCPC6772.g1 | Putative BAG domain containing protein | 23.30 | 7.21E-03 |
| Downregulated | |||
| CCPC3479.g1 | Groes-like protein | –41.47 | 1.64E-03 |
| CCPA5017.g1 | Protein arginine N-methyltransferase | –39.95 | 3.01E-04 |
| CCOZ3601.b1 | Secy protein | –27.45 | 6.26E-03 |
| 16D10 | HSP20-like chaperone | –25.79 | 2.78E-03 |
| CCOZ4082.b1 | Glucoamylase | –23.26 | 2.19E-03 |
| CCPA3011.b1 | Cell division control/GTP binding protein | –20.94 | 8.14E-03 |
| CCPA3961.g1 | Glutamate decarboxylase | –18.18 | 7.47E-03 |
| CCPC7984.b1 | ---Na--- | –17.48 | 5.70E-03 |
| D69E9 | 40S ribosomal protein S26 | –16.73 | 3.76E-03 |
| CCPB4097.b1 | Hypothetical protein HETIRDRAFT_439855 | –14.76 | 4.80E-03 |
| CCPC6286.b1 | Leucine aminopeptidase | –13.02 | 7.79E-03 |
| Mapping Reference ID | Annotation | Fold change | P-value |
|---|---|---|---|
| Upregulated | |||
| CCPA575.b1 | Delta-12 fatty acid desaturase | 48.12 | 7.94E-05 |
| CCPA1686.b1 | Polysaccharide lyase family 1 protein | 47.55 | 4.24E-03 |
| CCPA1999.b1 | Hypothetical protein HETIRDRAFT_426980 | 46.41 | 1.60E-04 |
| 10F24-03-16 | Elongase of fatty acids ELO | 45.14 | 9.47E-03 |
| CCPC993.b1 | Erylysin B | 38.75 | 5.07E-03 |
| CCPC1268.b1 | Delta-12 fatty acid desaturase protein | 38.60 | 1.89E-03 |
| CCPA3999.b1 | Fatty acid desaturase-domain-containing protein | 35.32 | 4.51E-03 |
| CCPC5436.b1 | Hypothetical protein EW146_g3762 | 33.84 | 4.25E-03 |
| CCPA5235.g1 | Hypothetical protein HETIRDRAFT_468348 | 33.44 | 3.52E-03 |
| CCPC8046.b1 | Delta-12 fatty acid desaturase | 33.02 | 1.59E-04 |
| Downregulated | |||
| D128H9 | RS27A protein | –128.69 | 2.57E-04 |
| CCPA2234.b1 | Hypothetical protein HETIRDRAFT_409605 | –50.89 | 7.84E-04 |
| CCOZ3606.b1 | Glycoside hydrolase superfamily | –21.73 | 5.62E-03 |
| CCPB3914.b1 | Hypothetical protein HETIRDRAFT_426907 | –20.09 | 3.36E-03 |
| CCPA5017.g1 | Protein arginine N-methyltransferase | –18.89 | 5.61E-03 |
| CCPA2400.b1 | General substrate transporter | –13.93 | 8.68E-03 |
| CCPB4930.b1 | Family 43 glycosylhydrolase | –11.52 | 3.40E-03 |
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- San-Miguel-Ayanz, J.; de Rigo, D.; Caudullo, G.; Houston Durrant, T.; Mauri, A. European Atlas of Forest Tree Species. Publication Office of the European Union, 2016, 202. ISSN: 9279528335.
- Farjon, A. A Handbook of the World’s Conifers. Brill, 2010, 1–1111. [CrossRef]
- Gonzalez Diaz, P. Development and Maintenance of Genetic Diversity in Scots Pine, Pinus sylvestris (L.); Ph.D. Thesis, University of Stirling, Stirling, U.K., 2018.
- Tyrmi, J.S.; Vuosku, J.; Acosta, J.J.; Li, Z.; Sterck, L.; Cervera, M.T.; Savolainen, O.; Pyhäjärvi, T. Genomics of Clinal Local Adaptation in Pinus Sylvestris under Continuous Environmental and Spatial Genetic Setting. G3 Genes, Genomes, Genet. 2020, 10, 2683–2696. [Google Scholar] [CrossRef]
- Hsieh, S.; Uchman, A. Spatially Associated or Composite Life Traces from Holocene Paleosols and Dune Sands Provide Evidence for Past Biotic Interactions. Sci. Nat. 2023, 110. [Google Scholar] [CrossRef]
- Siitonen, J. Ips Acuminatus Kills Pines in Southern Finland. Silva Fenn. 2014, 48. [Google Scholar] [CrossRef]
- Hlávková, D.; Doležal, P. Cambioxylophagous Pests of Scots Pine: Ecological Physiology of European Populations—A Review. Front. For. Glob. Chang. 2022, 5, 864651. [Google Scholar] [CrossRef]
- Garbelotto, M.; Gonthier, P. Biology, Epidemiology, and Control of Heterobasidion Species Worldwide. Annu. Rev. Phytopathol. 2013, 51, 39–59. [Google Scholar] [CrossRef]
- Asiegbu, F.O.; Adomas, A.; Stenlid, J. Conifer Root and Butt Rot Caused by Heterobasidion Annosum (Fr.) Bref. s.L. Mol. Plant Pathol. 2005, 6, 395–409. [Google Scholar] [CrossRef]
- Samils, N. Monitoring the Control Methods of Heterobasidion Annosum s.l. Root Rot. Ph.D. Thesis, Swedish University of Agricultural Sciences, Uppsala, Sweden, 2008, ISBN: 9789185913800.
- Piri, T. Silvicultural Control of Heterobasidion Root Rot in Norway Spruce Forests in Southern Finland : Regeneration and Vitality Fertilization of Infected Stands; Ph.D. Thesis, University of Helsinki, Finnish Forest Research Institute, Vantaa Research Centre, Finland, 2003.
- Piri, T. Response of Compensatory-Fertilized Pinus Sylvestris to Infection by Heterobasidion Annosum. Scand. J. For. Res. 2010, 15, 218–224. [Google Scholar] [CrossRef]
- Rönnberg, J.; Berglund, M.; Johansson, U.; Cleary, M. Incidence of Heterobasidion Spp. Following Different Thinning Regimes in Norway Spruce in Southern Sweden. For. Ecol. Manage. 2013, 289, 409–415. [Google Scholar] [CrossRef]
- Blomquist, M.; Cleary, M.; Sherwood, P.; Pinto, W.; Herrera, S.L.; Marčiulynienė, D.; Elsafy, M.; Bakal, I.; Nilsson, A.; Rönnberg, J. The Potential of Biological Control against Heterobasidion Root Rot Is Not Realized in Practical Forestry. For. Ecol. Manage. 2023, 531, 120778. [Google Scholar] [CrossRef]
- Kenigsvalde, K.; Brauners, I.; Zaļuma, A.; Jansons, J.; Gaitnieks, T. Biological Protection of Conifers against Heterobasidion Infection – Interaction between Root-Rot Fungus and Phlebiopsis Gigantea. Research for Rural Development 2017. [Google Scholar] [CrossRef]
- Piri, T.; Saarinen, M.; Hamberg, L.; Hantula, J.; Gaitnieks, T. Efficacy of Biological and Chemical Control Agents against Heterobasidion Spore Infections of Norway Spruce and Scots Pine Stumps on Drained Peatland. J. Fungi 2023, 9, 346. [Google Scholar] [CrossRef]
- Korshikov, I.I.; Demkovich, A.E. Genetic Features of Root Fungus-Resistant Scotch Pine Trees in Artificial Stands in the Steppe Zone of Ukraine. Cytol. Genet. 2008, 42, 323–328. [Google Scholar] [CrossRef]
- Vasiliauskas, A. Šakninės Pinties (Heterobasidion Annosum (Fr.) Bref.) Paplitimas Pušynuose, Įveistuose Žemės Ūkio Naudmenuose, Kovos Su Ja Priemonės Ir Kovos Rezultatai [Root Rot Caused by Heterobasidion Annosum in Pinus Sylvestris Plantations on Former Agricultural Land]. Miskininkyste 2001, 2, 42–59. [Google Scholar]
- Rieksts-Riekstiņš, R.; Zeltiņš, P.; Baliuckas, V.; Bruna, L.; Zaluma, A.; Kapostiņš, R. Pinus Sylvestris Breeding for Resistance against Natural Infection of the Fungus Heterobasidion Annosum. Forests 2020, 11, 23. [Google Scholar] [CrossRef]
- Adomas, A.; Heller, G.; Li, G.; Olson, Å.; Chu, T.M.; Osborne, J.; Craig, D.; Van Zyl, L.; Wolfinger, R.; Sederoff, R.; et al. Transcript Profiling of a Conifer Pathosystem: Response of Pinus Sylvestris Root Tissues to Pathogen (Heterobasidion Annosum) Invasion. Tree Physiol. 2007, 27, 1441–1458. [Google Scholar] [CrossRef]
- Mukrimin, M.; Kovalchuk, A.; Ghimire, R.P.; Kivimäenpää, M.; Sun, H.; Holopainen, J.K.; Asiegbu, F.O. Evaluation of Potential Genetic and Chemical Markers for Scots Pine Tolerance against Heterobasidion Annosum Infection. Planta 2019, 250, 1881–1895. [Google Scholar] [CrossRef]
- Hernandez-Escribano, L.; Visser, E.A.; Iturritxa, E.; Raposo, R.; Naidoo, S. The Transcriptome of Pinus Pinaster under Fusarium Circinatum Challenge. BMC Genomics 2020, 21, 1–18. [Google Scholar] [CrossRef]
- Šķipars, V.; Ruņģis, D. Transcript Dynamics in Wounded and Inoculated Scots Pine. Int. J. Mol. Sci. 2021, 22, 1–20. [Google Scholar] [CrossRef]
- Liu, M.; Wang, K.; Haapanen, M.; Ghimire, R.P.; Kivimäenpää, M.; Asiegbu, F.O. Analysis of Transcriptome and Terpene Constituents of Scots Pine Genotypes Inherently Resistant or Susceptible to Heterobasidion Annosum. Front. Plant Sci. 2022, 13, 947734. [Google Scholar] [CrossRef]
- Dalman, K.; Himmelstrand, K.; Olson, Å.; Lind, M.; Brandström-Durling, M.; Stenlid, J. A Genome-Wide Association Study Identifies Genomic Regions for Virulence in the Non-Model Organism Heterobasidion Annosum s.s. PLoS One 2013, 8, e53525. [Google Scholar] [CrossRef]
- Adomas, A.; Eklund, M.; Johansson, M.; Asiegbu, F.O. Identification and Analysis of Differentially Expressed CDNAs during Nonself-Competitive Interaction between Phlebiopsis Gigantea and Heterobasidion Parviporum. FEMS Microbiol. Ecol. 2006, 57, 26–39. [Google Scholar] [CrossRef]
- Zeng, Z.; Sun, H.; Vainio, E.J.; Raffaello, T.; Kovalchuk, A.; Morin, E.; Duplessis, S.; Asiegbu, F.O. Intraspecific Comparative Genomics of Isolates of the Norway Spruce Pathogen (Heterobasidion Parviporum) and Identification of Its Potential Virulence Factors. BMC Genomics 2018, 19, 1–21. [Google Scholar] [CrossRef]
- Urban, M.; Cuzick, A.; Seager, J.; Wood, V.; Rutherford, K.; Venkatesh, S.Y.; De Silva, N.; Martinez, M.C.; Pedro, H.; Yates, A.D.; et al. PHI-Base: The Pathogen–Host Interactions Database. Nucleic Acids Res. 2020, 48, D613–D620. [Google Scholar] [CrossRef]
- Shin, J.; Kim, J.E.; Lee, Y.W.; Son, H. Fungal Cytochrome P450s and the P450 Complement (CYPome) of Fusarium Graminearum. Toxins 2018, 10. [Google Scholar] [CrossRef]
- Kovalchuk, A.; Zeng, Z.; Ghimire, R.P.; Kivimäenpää, M.; Raffaello, T.; Liu, M.; Mukrimin, M.; Kasanen, R.; Sun, H.; Julkunen-Tiitto, R.; et al. Dual RNA-Seq Analysis Provides New Insights into Interactions between Norway Spruce and Necrotrophic Pathogen Heterobasidion Annosum s.l. BMC Plant Biol. 2019, 19, 1–17. [Google Scholar] [CrossRef]
- Wen, Z.; Zeng, Z.; Ren, F.; Asiegbu, F.O. The Conifer Root and Stem Rot Pathogen (Heterobasidion Parviporum): Effectome Analysis and Roles in Interspecific Fungal Interactions. Microorg. 2019, 7, 658. [Google Scholar] [CrossRef]
- Zamora-Ballesteros, C.; Pinto, G.; Amaral, J.; Valledor, L.; Alves, A.; Diez, J.J.; Martín-García, J. Dual Rna-Sequencing Analysis of Resistant (Pinus Pinea) and Susceptible (Pinus Radiata) Hosts during Fusarium Circinatum Challenge. Int. J. Mol. Sci. 2021, 22, 5231. [Google Scholar] [CrossRef]
- Lundén, K.; Danielsson, M.; Durling, M.B.; Ihrmark, K.; Nemesio Gorriz, M.; Stenlid, J.; Asiegbu, F.O.; Elfstrand, M. Transcriptional Responses Associated with Virulence and Defence in the Interaction between Heterobasidion Annosum s.s. and Norway Spruce. PLoS One 2015, 10, e0131182. [Google Scholar] [CrossRef]
- Asiegbu, F.O.; Johansson, M.; Woodward, S.; Hüttermann, A. Biochemistry of the Host - Parasite Interaction; In Heterobasidion Annosum : Biology, Ecology, Impact and Control; Woodward, S.; Stenlid, J.; Karjalainen, R.; Hüttermann, A., Eds.; CABI Publishing: Wallingford, U.K., 1998; pp 176-180.
- Doyle, J.J.; Doyle, J.L. Isolation of Plant DNA from Fresh Tissue. Focus 1987, 12, 13–15. [Google Scholar]
- Asiegbu, F.; Daniel, G.; Johansson, M. Studies on the Infection of Norway Spruce Roots by Heterobasidion Annosum. Canad. J. Bot. 1993, 71, 1552–1561. [Google Scholar] [CrossRef]
- Karlsson, M.; Hietala, A.M.; Kvaalen, H.; Solheim, H.; Olson, Å.; Stenlid, J.; Fossdal, C.G. Quantification of Host and Pathogen DNA and RNA Transcripts in the Interaction of Norway Spruce with Heterobasidion Parviporum. Physiol. Mol. Plant Pathol. 2007, 70, 99–109. [Google Scholar] [CrossRef]
- Olson, Å.; Aerts, A.; Asiegbu, F.; Belbahri, L.; Bouzid, O.; Broberg, A.; Canbäck, B.; Coutinho, P.M.; Cullen, D.; Dalman, K.; et al. Insight into Trade-off between Wood Decay and Parasitism from the Genome of a Fungal Forest Pathogen. New Phytol. 2012, 194, 1001–1013. [Google Scholar] [CrossRef]
- https://genome.jgi.doe.gov/portal/Hetan2/Hetan2.download.html (accessed on 28 June 2023).
- Wachowiak, W.; Trivedi, U.; Perry, A.; Cavers, S. Comparative Transcriptomics of a Complex of Four European Pine Species. BMC Genomics 2015, 16, 234. [Google Scholar] [CrossRef]
- CLC Genomics Workbench User Manual for CLC Genomics Workbench 23.0.5 2023, 1165.
- Heberle, H.; Meirelles, V.G.; da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A Web-Based Tool for the Analysis of Sets through Venn Diagrams. BMC Bioinformatics 2015, 16, 1–7. [Google Scholar] [CrossRef]
- Brutyn, M.; D’Herde, K.; Dhaenens, M.; Rooij, P. Van; Verbrugghe, E.; Hyatt, A.D.; Croubels, S.; Deforce, D.; Ducatelle, R.; Haesebrouck, F.; et al. Batrachochytrium Dendrobatidis Zoospore Secretions Rapidly Disturb Intercellular Junctions in Frog Skin. Fungal Genet. Biol. 2012, 49, 830–837. [Google Scholar] [CrossRef]
- Panevska, A.; Hodnik, V.; Skočaj, M.; Novak, M.; Modic, Š.; Pavlic, I.; Podržaj, S.; Zarić, M.; Resnik, N.; Maček, P.; et al. Pore-Forming Protein Complexes from Pleurotus Mushrooms Kill Western Corn Rootworm and Colorado Potato Beetle through Targeting Membrane Ceramide Phosphoethanolamine. Sci. Reports 2019, 9, 1–14. [Google Scholar] [CrossRef]
- Wynn, J.P.; Hamid, A.B.A.; Ratledge, C. The Role of Malic Enzyme in the Regulation of Lipid Accumulation in Filamentous Fungi. Microbiology 1999, 145, 1911–1917. [Google Scholar] [CrossRef]
- Chang, Q.; Griest, T.A.; Harter, T.M.; Mark Petrash, J. Functional Studies of Aldo-Keto Reductases in Saccharomyces Cerevisiae. Biochim. Biophys. Acta 2007, 1773, 321–329. [Google Scholar] [CrossRef]
- Xu, Y.; Zhou, H.; Zhao, G.; Yang, J.; Luo, Y.; Sun, S.; Wang, Z.; Li, S.; Jin, C. Genetical and O-Glycoproteomic Analyses Reveal the Roles of Three Protein O-Mannosyltransferases in Phytopathogen Fusarium Oxysporum f.sp. Cucumerinum. Fungal Genet. Biol. 2020, 134. [Google Scholar] [CrossRef]
- Gerke, J.; Bayram, Ö.; Braus, G.H. Fungal S-Adenosylmethionine Synthetase and the Control of Development and Secondary Metabolism in Aspergillus Nidulans. Fungal Genet. Biol. 2012, 49, 443–454. [Google Scholar] [CrossRef]
- Watanabe, T.; Tsuda, S.; Nishimura, H.; Honda, Y.; Watanabe, T. Characterization of a Delta12-Fatty Acid Desaturase Gene from Ceriporiopsis Subvermispora, a Selective Lignin-Degrading Fungus. Appl. Microbiol. Biotechnol. 2010, 87, 215–224. [Google Scholar] [CrossRef]
- Van Den Brink, J.; De Vries, R.P. Fungal Enzyme Sets for Plant Polysaccharide Degradation. Appl. Microbiol. Biotechnol. 2011, 91, 1477. [Google Scholar] [CrossRef]
- Huang, Z.Y.; Ye, R.Y.; Yu, H.L.; Li, A.T.; Xu, J.H. Mining Methods and Typical Structural Mechanisms of Terpene Cyclases. Bioresour. Bioprocess. 2021, 8, 1–27. [Google Scholar] [CrossRef]
- González-Hernández, R.A.; Valdez-Cruz, N.A.; Macías-Rubalcava, M.L.; Trujillo-Roldán, M.A. Overview of Fungal Terpene Synthases and Their Regulation. World J. Microbiol. Biotechnol. 2023, 39, 194. [Google Scholar] [CrossRef]
- Palanimurugan, R.; Scheel, H.; Hofmann, K.; Dohmen, R.J. Polyamines Regulate Their Synthesis by Inducing Expression and Blocking Degradation of ODC Antizyme. EMBO J. 2004, 23, 4857–4867. [Google Scholar] [CrossRef]
- Beccaccioli, M.; Reverberi, M.; Scala, V. Fungal Lipids: Biosynthesis and Signalling during Plant-Pathogen Interaction. Front. Biosci. - Landmark 2019, 24, 172–185. [Google Scholar] [CrossRef]
- Cardoza, R.E.; McCormick, S.P.; Lindo, L.; Mayo-Prieto, S.; González-Cazón, D.; Martínez-Reyes, N.; Carro-Huerga, G.; Rodríguez-González, Á.; Proctor, R.H.; Casquero, P.A.; et al. Effect of Farnesol in Trichoderma Physiology and in Fungal-Plant Interaction. J. fungi 2022, 8. [Google Scholar] [CrossRef]
- Vorapreeda, T.; Thammarongtham, C.; Cheevadhanarak, S.; Laoteng, K. Repertoire of Malic Enzymes in Yeast and Fungi: Insight into Their Evolutionary Functional and Structural Significance. Microbiology 2013, 159. [Google Scholar] [CrossRef]
- Hao, G.; Chen, H.; Wang, L.; Gu, Z.; Song, Y.; Zhang, H.; Chen, W.; Chen, Y.Q. Role of Malic Enzyme during Fatty Acid Synthesis in the Oleaginous Fungus Mortierella Alpina. Appl. Environ. Microbiol. 2014, 80, 2672. [Google Scholar] [CrossRef]
- Raffaello, T.; Chen, H.; Kohler, A.; Asiegbu, F.O. Transcriptomic Profiles of Heterobasidion Annosum under Abiotic Stresses and during Saprotrophic Growth in Bark, Sapwood and Heartwood. Environ. Microbiol. 2014, 16, 1654–1667. [Google Scholar] [CrossRef]
- Looi, H.K.; Toh, Y.F.; Yew, S.M.; Na, S.L.; Tan, Y.C.; Chong, P.S.; Khoo, J.S.; Yee, W.Y.; Ng, K.P.; Kuan, C.S. Genomic Insight into Pathogenicity of Dematiaceous Fungus Corynespora Cassiicola. PeerJ 2017, 5. [Google Scholar] [CrossRef]
- Gong, L.; Liu, Y.; Xiong, Y.; Li, T.; Yin, C.; Zhao, J.; Yu, J.; Yin, Q.; Gupta, V.K.; Jiang, Y.; et al. New Insights into the Evolution of Host Specificity of Three Penicillium Species and the Pathogenicity of P. Italicum Involving the Infection of Valencia Orange (Citrus Sinensis). Virulence 2020, 11, 748–768. [Google Scholar] [CrossRef]
- Mewis, K.; Lenfant, N.; Lombard, V.; Henrissat, B. Dividing the Large Glycoside Hydrolase Family 43 into Subfamilies: A Motivation for Detailed Enzyme Characterization. Appl. Environ. Microbiol. 2016, 82, 1686. [Google Scholar] [CrossRef]
- Sahu, P.K.; Jayalakshmi, K.; Tilgam, J.; Gupta, A.; Nagaraju, Y.; Kumar, A.; Hamid, S.; Singh, H.V.; Minkina, T.; Rajput, V.D.; et al. ROS Generated from Biotic Stress: Effects on Plants and Alleviation by Endophytic Microbes. Front. Plant Sci. 2022, 13. [Google Scholar] [CrossRef]
- Heller, J.; Tudzynski, P. Reactive Oxygen Species in Phytopathogenic Fungi: Signaling, Development, and Disease. Annu. Rev. Phytopathol. 2011, 49, 369–390. [Google Scholar] [CrossRef]
- Ahn, S.; Jung, J.; Jang, I.A.; Madsen, E.L.; Park, W. Role of Glyoxylate Shunt in Oxidative Stress Response. J. Biol. Chem. 2016, 291, 11928. [Google Scholar] [CrossRef]
| Sample name1 | Total reads | Percentage of reads mapping to H. annosum transcriptome | Percentage of reads mapping to P. sylvestris transcriptome |
|---|---|---|---|
| 1_5 | 330514752 | 0.32 | 26.69 |
| 1_9 | 148181900 | 0.47 | 69.69 |
| 1_10 | 127532184 | 0.73 | 44.92 |
| 1_14 | 265534720 | 0.44 | 45.81 |
| 2_4 | 302911430 | 0.17 | 26.15 |
| 2_10 | 200927014 | 0.36 | 54.40 |
| 2_14 | 138048364 | 0.12 | 23.87 |
| 2_15 | 297177502 | 0.38 | 42.04 |
| 3_3 | 183571350 | 0.65 | 55.76 |
| 3_5 | 175591306 | 0.38 | 57.62 |
| 3_7 | 214847606 | 0.28 | 41.78 |
| 3_12 | 176541044 | 0.32 | 29.99 |
| 4_1 | 163295084 | 0.27 | 56.09 |
| 4_3 | 123870986 | 0.52 | 49.52 |
| 4_10 | 400583236 | 0.11 | 23.70 |
| 4_13 | 115595890 | 0.24 | 32.19 |
| Biological process | Unique for timepoint |
|---|---|
| Histidine biosynthetic process | 1 WPI |
| Arginine biosynthetic process | 1 WPI |
| Coenzyme A metabolic process | 1 WPI |
| Response to heat | 1 WPI |
| Response to oxygen-containing compound | 1 WPI |
| Isoprenoid biosynthetic process | 1 WPI |
| Response to osmotic stress | 1 WPI |
| Response to oxidative stress | 1 WPI |
| Glyoxylate cycle | 1 WPI |
| Carboxylic acid metabolic process | 1 WPI |
| Protein-containing complex assembly | 1 WPI |
| S-adenosylmethionine biosynthetic process | 1 WPI |
| Proton transmembrane transport | 2 WPI |
| Negative regulation of protein modification process | 2 WPI |
| Negative regulation of phosphate metabolic process | 2 WPI |
| DNA-templated transcription | 2 WPI |
| Macroautophagy | 2 WPI |
| Regulation of translation | 2 WPI |
| Cellular response to amino acid starvation | 2 WPI |
| Regulation of protein dephosphorylation | 2 WPI |
| Positive regulation of transcription by RNA polymerase II | 2 WPI |
| Acetyl-coa biosynthetic process | 3 WPI |
| Citrate metabolic process | 3 WPI |
| Cellular biosynthetic process | 4 WPI |
| Protein import into mitochondrial matrix | 4 WPI |
| Signal transduction | 4 WPI |
| Ergosterol biosynthetic process | 4 WPI |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





