Submitted:
30 August 2024
Posted:
03 September 2024
You are already at the latest version
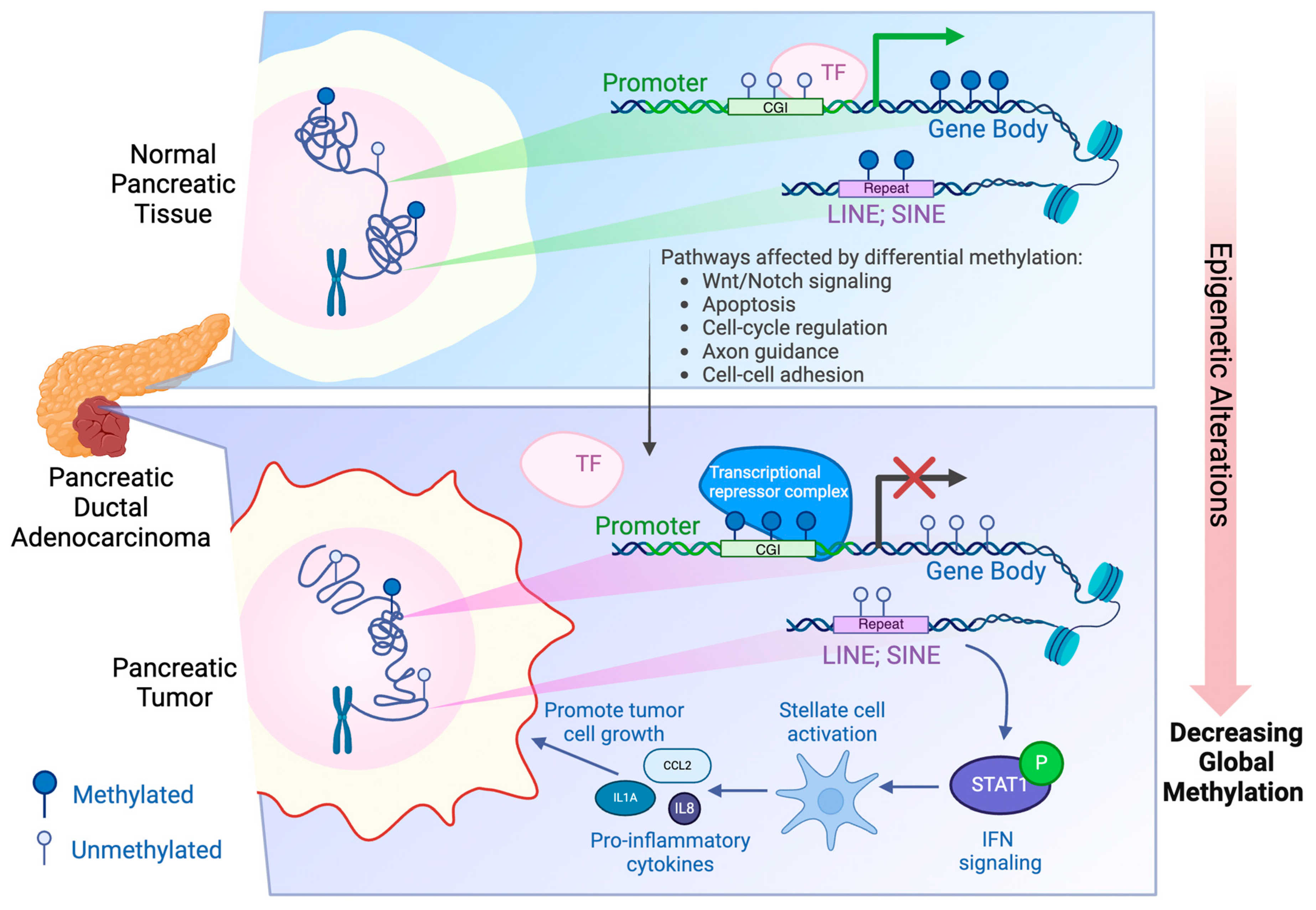
Abstract
Keywords:
Introduction
Regulation of DNA Methylation
Aberrant DNA Methylation in PDAC
Recent WGBS studies on PDAC patients
Conclusion
| Gene Promoter | Hypermethylated or Hypomethylated in PDAC | Affected Gene Expression Level | Molecular Function of the Gene | Functional Phenotype in PDAC | Method | Reference |
| PD-L1 promoter | Hypomethylated | PD-L1 ↑ | Programmed cell death ligand | Aggressive clinical phenotypes ↑, immune cell infiltration ↓, expression of immune checkpoint genes ↓, overall survival ↓ | Microarray | [71] |
| HNF4A promoter | Hypermethylated |
HNF4A ↓ | Controls genes that are especially important for development and function of beta cells in the pancreas; regulate cell proliferation through the Wnt/beta-Catenin Pathway | Patient survival ↓, cell growth, colony formation, and invasiveness ↑, expression decreases during early PDAC stages | Targeted bisulfite sequencing | [72] |
| BEND4 promoter | Hypermethylated | BEND4 ↓ | BEN domain-containing protein 4 involves in non-homologous end-joining signaling by interacting with Ku80 and promotes DNA damage repair; inhibits cell growth, migration and invasion | Prognosis ↓, synthetic lethality towards ATM inhibitor | MSP | [73] |
|
NPY and FAIM2 promoters |
Hypermethylated | NPY ↓, FAIM2 ↓ | NPY is involved in cell motion, invasion, and proliferation, and its hypermethylation is observed in certain cancers. FAIM2 is associated with apoptosis inhibition. | Prognosis ↓, progress to later stages ↑ | Microarray | [55] |
| CDH13 promoter | Hypermethylated | CDH13 ↓ | Cadherin, involved in cell-cell adhesion and contact inhibition of cell growth | Invasion and metastasis ↑ | MSP | [74] |
| Cyclin D2 promoter | Hypermethylated | Cyclin D2 ↓ | Involved in cell cycle regulation, cell differentiation, and neoplastic transformation | Unknown in PDAC context | MSP | [75] |
| SPARC promoter | Hypermethylated | SPARC ↓ | A multifunctional glycoprotein, involved in cell proliferation, cell spreading, adhesion, motility, and invasion | Proliferation ↑ | MSP, microarray | [76] |
| DKK1 promoter | Hypermethylated | DKK1 ↓ | Regulates Wnt signaling by binding to the Wnt coreceptor lipoprotein-related protein-5 (LRP5)/Arrow | Proliferation ↑, migration & invasion ↑ | MSP | [77] |
| MUC Promoter | Hypomethylated | Not tested | Mucins, bind to pathogens as part of the immune system; renewal and differentiation of the epithelium; involved in cell adhesion and signaling | Unknown in PDAC context | Microarray | [78] |
Acknowledgments
References
- Siegel, R.L., A.N. Giaquinto, and A. Jemal, Cancer statistics, 2024. CA Cancer J Clin, 2024. 74(1): p. 12-49.
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zheng, Y.; Yang, F.; Zhu, L.; Zhu, X.-Q.; Wang, Z.-F.; Wu, X.-L.; Zhou, C.-H.; Yan, J.-Y.; Hu, B.-Y.; et al. The Molecular Biology of Pancreatic Adenocarcinoma: Translational Challenges and Clinical Perspectives. Signal Transduct. Target. Ther. 2021, 6, 1–23. [Google Scholar] [CrossRef]
- Zeng, S., et al., Chemoresistance in Pancreatic Cancer. Int J Mol Sci, 2019. 20(18).
- Waddell, N.; Pajic, M.; Patch, A.-M.; Chang, D.K.; Kassahn, K.S.; Bailey, P.; Johns, A.L.; Miller, D.; Nones, K.; Quek, K.; et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015, 518, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Kanda, M.; Matthaei, H.; Wu, J.; Hong, S.M.; Yu, J.; Borges, M.; Hruban, R.H.; Maitra, A.; Kinzler, K.; Vogelstein, B.; et al. Presence of somatic mutations in most early-stage pancreatic intraepithelial neoplasia. Gastroenterology 2012, 142, 730–733. [Google Scholar] [CrossRef] [PubMed]
- Waters, A.M.; Der, C.J. KRAS: The Critical Driver and Therapeutic Target for Pancreatic Cancer. Cold Spring Harb. Perspect. Med. 2018, 8, a031435. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Electronic address, a.a.d.h.e. and N. Cancer Genome Atlas Research, Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell, 2017. 32(2): p. 185-203 e13.
- Halbrook, C.J.; Lyssiotis, C.A.; di Magliano, M.P.; Maitra, A. Pancreatic cancer: Advances and challenges. Cell 2023, 186, 1729–1754. [Google Scholar] [CrossRef]
- Jones, S.; Zhang, X.; Parsons, D.W.; Lin, J.C.-H.; Leary, R.J.; Angenendt, P.; Mankoo, P.; Carter, H.; Kamiyama, H.; Jimeno, A.; et al. Core Signaling Pathways in Human Pancreatic Cancers Revealed by Global Genomic Analyses. Science 2008, 321, 1801–1806. [Google Scholar] [CrossRef]
- Xu, J.; Roe, J.; Lee, E.; Tonelli, C.; Ji, K.Y.; Younis, O.W.; Somervile, T.D.; Yao, M.; Milazzo, J.P.; Tiriac, H.; et al. Engrailed-1 Promotes Pancreatic Cancer Metastasis. Adv. Sci. 2023, 11, e2308537. [Google Scholar] [CrossRef]
- Roe, J.-S.; Hwang, C.-I.; Somerville, T.D.; Milazzo, J.P.; Lee, E.J.; Da Silva, B.; Maiorino, L.; Tiriac, H.; Young, C.M.; Miyabayashi, K.; et al. Enhancer Reprogramming Promotes Pancreatic Cancer Metastasis. Cell 2017, 170, 875–888.e20. [Google Scholar] [CrossRef]
- Kim, H.-R.; Yim, J.; Yoo, H.-B.; Lee, S.E.; Oh, S.; Jung, S.; Hwang, C.-I.; Shin, D.-M.; Kim, T.; Yoo, K.H.; et al. EVI1 activates tumor-promoting transcriptional enhancers in pancreatic cancer. NAR Cancer 2021, 3, zcab023. [Google Scholar] [CrossRef]
- Wang, S.S.; Xu, J.; Ji, K.Y.; Hwang, C.-I. Epigenetic Alterations in Pancreatic Cancer Metastasis. Biomolecules 2021, 11, 1082. [Google Scholar] [CrossRef] [PubMed]
- Saghafinia, S.; Mina, M.; Riggi, N.; Hanahan, D.; Ciriello, G. Pan-Cancer Landscape of Aberrant DNA Methylation across Human Tumors. Cell Rep. 2018, 25, 1066–1080.e8. [Google Scholar] [CrossRef] [PubMed]
- Paz, M.F.; Fraga, M.F.; Avila, S.; Guo, M.; Pollan, M.; Herman, J.G.; Esteller, M. A systematic profile of DNA methylation in human cancer cell lines. Cancer Res. 2003, 63, 1114–1121. [Google Scholar] [PubMed]
- Bergman, Y. and H. Cedar, DNA methylation dynamics in health and disease. Nat Struct Mol Biol, 2013. 20(3): p. 274-81.
- Lomberk, G.; Blum, Y.; Nicolle, R.; Nair, A.; Gaonkar, K.S.; Marisa, L.; Mathison, A.; Sun, Z.; Yan, H.; Elarouci, N.; et al. Distinct epigenetic landscapes underlie the pathobiology of pancreatic cancer subtypes. Nat. Commun. 2018, 9, 1978. [Google Scholar] [CrossRef]
- Spainhour, J.C.; Lim, H.S.; Yi, S.V.; Qiu, P. Correlation Patterns Between DNA Methylation and Gene Expression in The Cancer Genome Atlas. Cancer Informatics 2019, 18. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.; Sen, S.; Weeks, R.J.; Eccles, M.R.; Chatterjee, A. Promoter DNA Hypermethylation and Paradoxical Gene Activation. Trends Cancer 2020, 6, 392–406. [Google Scholar] [CrossRef]
- Wang, S.S.; Hall, M.L.; Lee, E.; Kim, S.-C.; Ramesh, N.; Lee, S.H.; Jang, J.-Y.; Bold, R.J.; Ku, J.-L.; Hwang, C.-I. Whole-genome bisulfite sequencing identifies stage- and subtype-specific DNA methylation signatures in pancreatic cancer. iScience 2024, 27, 109414. [Google Scholar] [CrossRef] [PubMed]
- Espinet, E.; Gu, Z.; Imbusch, C.D.; Giese, N.A.; Büscher, M.; Safavi, M.; Weisenburger, S.; Klein, C.; Vogel, V.; Falcone, M.; et al. Aggressive PDACs Show Hypomethylation of Repetitive Elements and the Execution of an Intrinsic IFN Program Linked to a Ductal Cell of Origin. Cancer Discov. 2020, 11, 638–659. [Google Scholar] [CrossRef]
- Robertson, K.D. DNA methylation and human disease. Nat. Rev. Genet. 2005, 6, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Jabbari, K.; Bernardi, G. Cytosine methylation and CpG, TpG (CpA) and TpA frequencies. Gene 2004, 333, 143–149. [Google Scholar] [CrossRef]
- Li, E. and Y. Zhang, DNA methylation in mammals. Cold Spring Harb Perspect Biol, 2014. 6(5): p. a019133.
- Takahashi, Y.; Valencia, M.M.; Yu, Y.; Ouchi, Y.; Takahashi, K.; Shokhirev, M.N.; Lande, K.; Williams, A.E.; Fresia, C.; Kurita, M.; et al. Transgenerational inheritance of acquired epigenetic signatures at CpG islands in mice. Cell 2023, 186, 715–731.e19. [Google Scholar] [CrossRef]
- Biswas, S.; Rao, C.M. Epigenetic tools (The Writers, The Readers and The Erasers) and their implications in cancer therapy. Eur. J. Pharmacol. 2018, 837, 8–24. [Google Scholar] [CrossRef] [PubMed]
- Okano, M.; Bell, D.W.; Haber, D.A.; Li, E. DNA Methyltransferases Dnmt3a and Dnmt3b Are Essential for De Novo Methylation and Mammalian Development. Cell 1999, 99, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Bestor, T.H.; Ingram, V.M. Two DNA methyltransferases from murine erythroleukemia cells: purification, sequence specificity, and mode of interaction with DNA. Proc. Natl. Acad. Sci. 1983, 80, 5559–5563. [Google Scholar] [CrossRef]
- Probst, A.V.; Dunleavy, E.; Almouzni, G. Epigenetic inheritance during the cell cycle. Nat. Rev. Mol. Cell Biol. 2009, 10, 192–206. [Google Scholar] [CrossRef]
- Wu, S.C. and Y. Zhang, Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol, 2010. 11(9): p. 607-20.
- Zhou, X.; Zhuang, Z.; Wang, W.; He, L.; Wu, H.; Cao, Y.; Pan, F.; Zhao, J.; Hu, Z.; Sekhar, C.; et al. OGG1 is essential in oxidative stress induced DNA demethylation. Cell. Signal. 2016, 28, 1163–1171. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Yang, M.; Wang, S.; Abbas, S.J.; Zhang, J.; Li, Y.; Shao, R.; Liu, Y. An Overview of Epigenetic Methylation in Pancreatic Cancer Progression. Front. Oncol. 2022, 12, 854773. [Google Scholar] [CrossRef]
- Angeloni, A.; Bogdanovic, O. Enhancer DNA methylation: implications for gene regulation. Essays Biochem. 2019, 63, 707–715. [Google Scholar] [CrossRef] [PubMed]
- Brenet, F.; Moh, M.; Funk, P.; Feierstein, E.; Viale, A.J.; Socci, N.D.; Scandura, J.M. DNA Methylation of the First Exon Is Tightly Linked to Transcriptional Silencing. PLOS ONE 2011, 6, e14524. [Google Scholar] [CrossRef]
- Du, Q. , et al., Methyl-CpG-binding domain proteins: readers of the epigenome. Epigenomics, 2015. 7(6): p. 1051-73.
- Skoulidis, F.; Cassidy, L.D.; Pisupati, V.; Jonasson, J.G.; Bjarnason, H.; Eyfjord, J.E.; Karreth, F.A.; Lim, M.; Barber, L.M.; Clatworthy, S.A.; et al. Germline Brca2 Heterozygosity Promotes KrasG12D -Driven Carcinogenesis in a Murine Model of Familial Pancreatic Cancer. Cancer Cell 2010, 18, 499–509. [Google Scholar] [CrossRef]
- Watt, F.; Molloy, P.L. Cytosine methylation prevents binding to DNA of a HeLa cell transcription factor required for optimal expression of the adenovirus major late promoter. Genes Dev. 1988, 2, 1136–1143. [Google Scholar] [CrossRef]
- Ball, M.P. , et al., Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol, 2009. 27(4): p. 361-8.
- Jones, P.A. The DNA methylation paradox. Trends Genet. 1999, 15, 34–37. [Google Scholar] [CrossRef]
- Neri, F.; Rapelli, S.; Krepelova, A.; Incarnato, D.; Parlato, C.; Basile, G.; Maldotti, M.; Anselmi, F.; Oliviero, S. Intragenic DNA methylation prevents spurious transcription initiation. Nature 2017, 543, 72–77. [Google Scholar] [CrossRef]
- Yoder, J.A.; Walsh, C.P.; Bestor, T.H. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997, 13, 335–340. [Google Scholar] [CrossRef]
- Ibrahim, J. , et al., Genome-wide DNA methylation profiling and identification of potential pan-cancer and tumor-specific biomarkers. Mol Oncol, 2022. 16(12): p. 2432-2447.
- Frommer, M.; E McDonald, L.; Millar, D.S.; Collis, C.M.; Watt, F.; Grigg, G.W.; Molloy, P.L.; Paul, C.L. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. 1992, 89, 1827–1831. [Google Scholar] [CrossRef]
- Wang, R.Y.-H.; Gehrke, C.W.; Ehrlich, M. Comparison of bisulfite modification of 5-methyldeoxycytidine and deoxycytidine residues. Nucleic Acids Res. 1980, 8, 4777–4790. [Google Scholar] [CrossRef] [PubMed]
- Umer, M.; Herceg, Z. Deciphering the Epigenetic Code: An Overview of DNA Methylation Analysis Methods. Antioxidants Redox Signal. 2013, 18, 1972–1986. [Google Scholar] [CrossRef] [PubMed]
- Warnecke, P.M.; Stirzaker, C.; Melki, J.R.; Millar, D.S.; Paul, C.L.; Clark, S.J. Detection and measurement of PCR bias in quantitative methylation analysis of bisulphite-treated DNA. Nucleic Acids Res. 1997, 25, 4422–4426. [Google Scholar] [CrossRef]
- Ehrich, M.; Zoll, S.; Sur, S.; Boom, D.v.D. A new method for accurate assessment of DNA quality after bisulfite treatment. Nucleic Acids Res. 2007, 35, e29–e29. [Google Scholar] [CrossRef] [PubMed]
- Meissner, A.; Gnirke, A.; Bell, G.W.; Ramsahoye, B.; Lander, E.S.; Jaenisch, R. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005, 33, 5868–5877. [Google Scholar] [CrossRef]
- Harrison, A.; Parle-McDermott, A. DNA Methylation: A Timeline of Methods and Applications. Front. Genet. 2011, 2, 74. [Google Scholar] [CrossRef] [PubMed]
- Weichenhan, D. , et al., Tagmentation-Based Library Preparation for Low DNA Input Whole Genome Bisulfite Sequencing. Methods Mol Biol, 2018. 1708: p. 105-122.
- Jin, B. and K.D. Robertson, DNA methyltransferases, DNA damage repair, and cancer. Adv Exp Med Biol, 2013. 754: p. 3-29.
- Das, P.M. and R. Singal, DNA methylation and cancer. J Clin Oncol, 2004. 22(22): p. 4632-42.
- Mishra, N.K.; Guda, C. Genome-wide DNA methylation analysis reveals molecular subtypes of pancreatic cancer. Oncotarget 2017, 8, 28990–29012. [Google Scholar] [CrossRef]
- Chatterjee, A. , et al., DNA methylome in pancreatic cancer identified novel promoter hyper-methylation in NPY and FAIM2 genes associated with poor prognosis in Indian patient cohort. Cancer Cell Int, 2022. 22(1): p. 334.
- Bailey, P.; Chang, D.K.; Nones, K.; Johns, A.L.; Patch, A.-M.; Gingras, M.-C.; Miller, D.K.; Christ, A.N.; Bruxner, T.J.C.; Quinn, M.C.; et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016, 531, 47–52. [Google Scholar] [CrossRef]
- Boj, S.F.; Hwang, C.-I.; Baker, L.A.; Chio, I.I.C.; Engle, D.D.; Corbo, V.; Jager, M.; Ponz-Sarvise, M.; Tiriac, H.; Spector, M.S.; et al. Organoid Models of Human and Mouse Ductal Pancreatic Cancer. Cell 2015, 160, 324–338. [Google Scholar] [CrossRef] [PubMed]
- Boj, S.F.; Hwang, C.-I.; A Baker, L.; Engle, D.D.; A Tuveson, D.; Clevers, H. Model organoids provide new research opportunities for ductal pancreatic cancer. Mol. Cell. Oncol. 2015, 3, e1014757. [Google Scholar] [CrossRef]
- Hwang, C.; Boj, S.F.; Clevers, H.; A Tuveson, D. Preclinical models of pancreatic ductal adenocarcinoma. J. Pathol. 2015, 238, 197–204. [Google Scholar] [CrossRef]
- Romero-Calvo, I.; Weber, C.R.; Ray, M.; Brown, M.; Kirby, K.; Nandi, R.K.; Long, T.M.; Sparrow, S.M.; Ugolkov, A.; Qiang, W.; et al. Human Organoids Share Structural and Genetic Features with Primary Pancreatic Adenocarcinoma Tumors. Mol. Cancer Res. 2019, 17, 70–83. [Google Scholar] [CrossRef] [PubMed]
- Sharick, J.T.; Jeffery, J.J.; Karim, M.R.; Walsh, C.M.; Esbona, K.; Cook, R.S.; Skala, M.C. Cellular Metabolic Heterogeneity In Vivo Is Recapitulated in Tumor Organoids. Neoplasia 2019, 21, 615–626. [Google Scholar] [CrossRef] [PubMed]
- Tiriac, H.; Belleau, P.; Engle, D.D.; Plenker, D.; Deschenes, A.; Somerville, T.D.D.; Froeling, F.E.M.; Burkhart, R.A.; Denroche, R.E.; Jang, G.H.; et al. Organoid Profiling Identifies Common Responders to Chemotherapy in Pancreatic Cancer. Cancer Discov. 2018, 8, 1112–1129. [Google Scholar] [CrossRef]
- Chen, S.; Wang, M.; Liu, L.; Wang, G.; Wang, L.; Zhong, C.; Gao, C.; Wu, W.; Li, L. Ultrasound-guided fine-needle aspiration/biopsy-based pancreatic organoids establishment: an alternative model for basic and preclinical research. Gastroenterol. Rep. 2022, 11, goad019. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, H.; Ku, J.-L.; Chun, J.W.; Seo, H.Y.; Kim, S.C.; Paik, W.H.; Ryu, J.K.; Lee, S.K.; Lowy, A.M.; et al. Establishment of Patient-Derived Pancreatic Cancer Organoids from Endoscopic Ultrasound-Guided Fine-Needle Aspiration Biopsies. Gut Liver 2021, 16, 625–636. [Google Scholar] [CrossRef] [PubMed]
- Vilgelm, A.E.; Bergdorf, K.; Wolf, M.; Bharti, V.; Shattuck-Brandt, R.; Blevins, A.; Jones, C.; Phifer, C.; Lee, M.; Lowe, C.; et al. Fine-Needle Aspiration-Based Patient-Derived Cancer Organoids. iScience 2020, 23, 101408. [Google Scholar] [CrossRef] [PubMed]
- Collisson, E.A.; Sadanandam, A.; Olson, P.; Gibb, W.J.; Truitt, M.; Gu, S.; Cooc, J.; Weinkle, J.; Kim, G.E.; Jakkula, L.; et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat. Med. 2011, 17, 500–503. [Google Scholar] [CrossRef]
- Collisson, E.A.; Bailey, P.; Chang, D.K.; Biankin, A.V. Molecular subtypes of pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 207–220. [Google Scholar] [CrossRef]
- Brunton, H.; Caligiuri, G.; Cunningham, R.; Upstill-Goddard, R.; Bailey, U.-M.; Garner, I.M.; Nourse, C.; Dreyer, S.; Jones, M.; Moran-Jones, K.; et al. HNF4A and GATA6 Loss Reveals Therapeutically Actionable Subtypes in Pancreatic Cancer. Cell Rep. 2020, 31, 107625–107625. [Google Scholar] [CrossRef]
- O’Kane, G.M.; Grünwald, B.T.; Jang, G.-H.; Masoomian, M.; Picardo, S.; Grant, R.C.; Denroche, R.E.; Zhang, A.; Wang, Y.; Lam, B.; et al. GATA6 Expression Distinguishes Classical and Basal-like Subtypes in Advanced Pancreatic Cancer. Clin. Cancer Res. 2020, 26, 4901–4910. [Google Scholar] [CrossRef]
- García-Ortiz, M.V.; Cano-Ramírez, P.; Toledano-Fonseca, M.; Aranda, E.; Rodríguez-Ariza, A. Diagnosing and monitoring pancreatic cancer through cell-free DNA methylation: progress and prospects. Biomark. Res. 2023, 11, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Chen, X. , et al., Analysis of PD-L1 promoter methylation combined with immunogenic context in pancreatic ductal adenocarcinoma. Cancer Immunol Immunother, 2024. 73(8): p. 149.
- Hatziapostolou, M. , et al., Promoter Methylation Leads to Hepatocyte Nuclear Factor 4A Loss and Pancreatic Cancer Aggressiveness. Gastro Hep Adv, 2024. 3(5): p. 687-702.
- Yao, Y.; Lv, H.; Zhang, M.; Li, Y.; Herman, J.G.; Brock, M.V.; Gao, A.; Wang, Q.; Fuks, F.; Zhang, L.; et al. Epigenetic silencing of BEND4, a novel DNA damage repair gene, is a synthetic lethal marker for ATM inhibitor in pancreatic cancer. Front. Med. 2024, 18, 721–734. [Google Scholar] [CrossRef]
- Sakai, M.; Hibi, K.; Koshikawa, K.; Inoue, S.; Takeda, S.; Kaneko, T.; Nakao, A. Frequent promoter methylation and gene silencing of CDH13 in pancreatic cancer. Cancer Sci. 2004, 95, 588–591. [Google Scholar] [CrossRef]
- Matsubayashi, H.; Sato, N.; Fukushima, N.; Yeo, C.J.; Walter, K.M.; Brune, K.; Sahin, F.; Hruban, R.H.; Goggins, M. Methylation of cyclin D2 is observed frequently in pancreatic cancer but is also an age-related phenomenon in gastrointestinal tissues. Clin Cancer Res 2003, 9, 1446–52. [Google Scholar]
- Sato, N.; Fukushima, N.; Maehara, N.; Matsubayashi, H.; Koopmann, J.; Su, G.H.; Hruban, R.H.; Goggins, M. SPARC/osteonectin is a frequent target for aberrant methylation in pancreatic adenocarcinoma and a mediator of tumor–stromal interactions. Oncogene 2003, 22, 5021–5030. [Google Scholar] [CrossRef] [PubMed]
- Weng, Y.-C.; Ma, J.; Zhang, J.; Wang, J.-C. Long non-coding RNA LINC01133 silencing exerts antioncogenic effect in pancreatic cancer through the methylation of DKK1 promoter and the activation of Wnt signaling pathway. Cancer Biol. Ther. 2018, 20, 368–380. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, S.; Iwaya, H.; Akahane, T.; Hamada, T.; Higashi, M.; Hashimoto, S.; Tanoue, S.; Ohtsuka, T.; Ido, A.; Tanimoto, A. Sequential evaluation of MUC promoter methylation using next-generation sequencing-based custom-made panels in liquid-based cytology specimens of pancreatic cancer. Diagn. Cytopathol. 2022, 50, 499–507. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





