Submitted:
07 August 2024
Posted:
09 August 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Optimization of HEK Dual hTLR5 Sensor for Detection of Salmonella
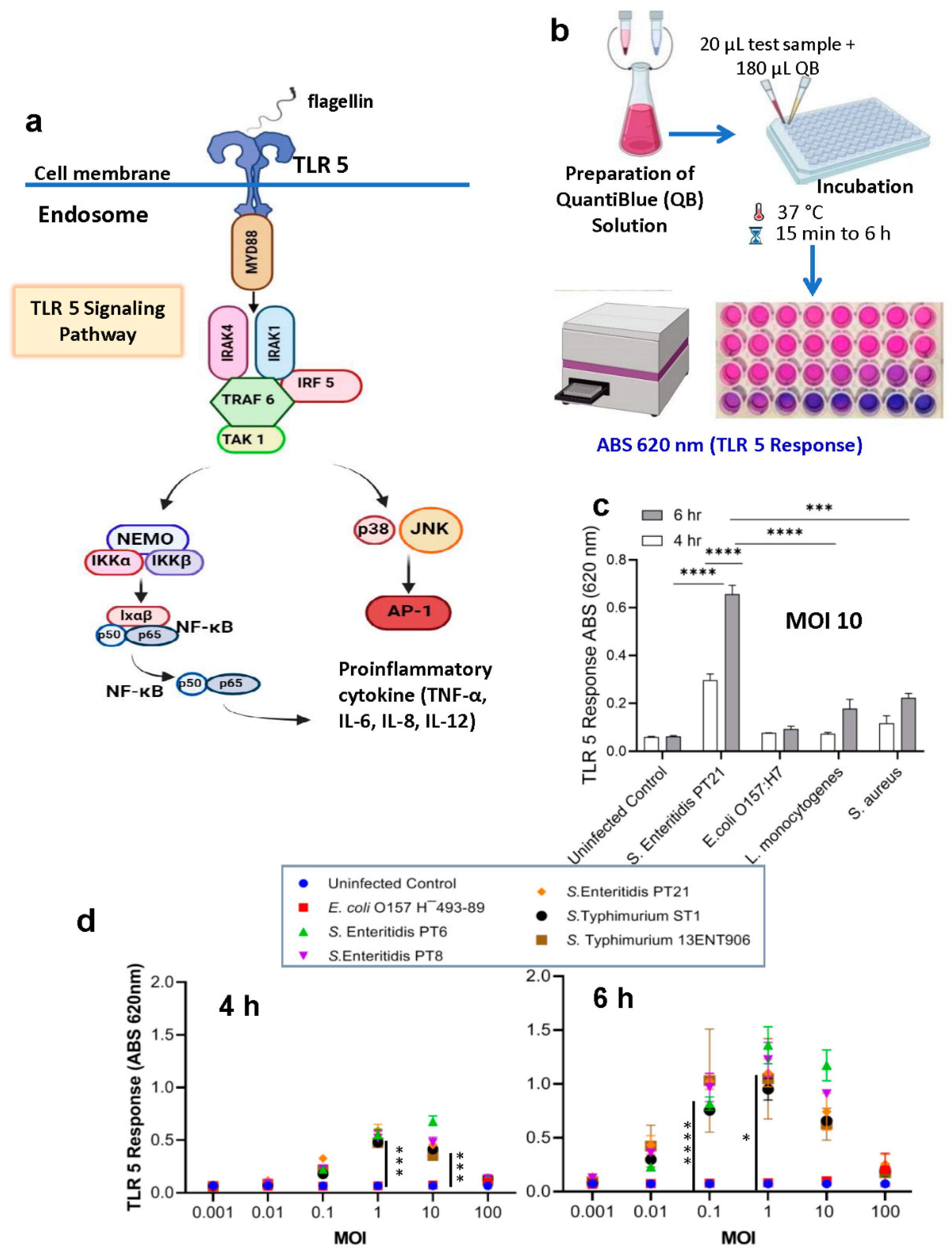
2.2. Specificity of HEK dual TLR5 sensor for detection of Salmonella serovars
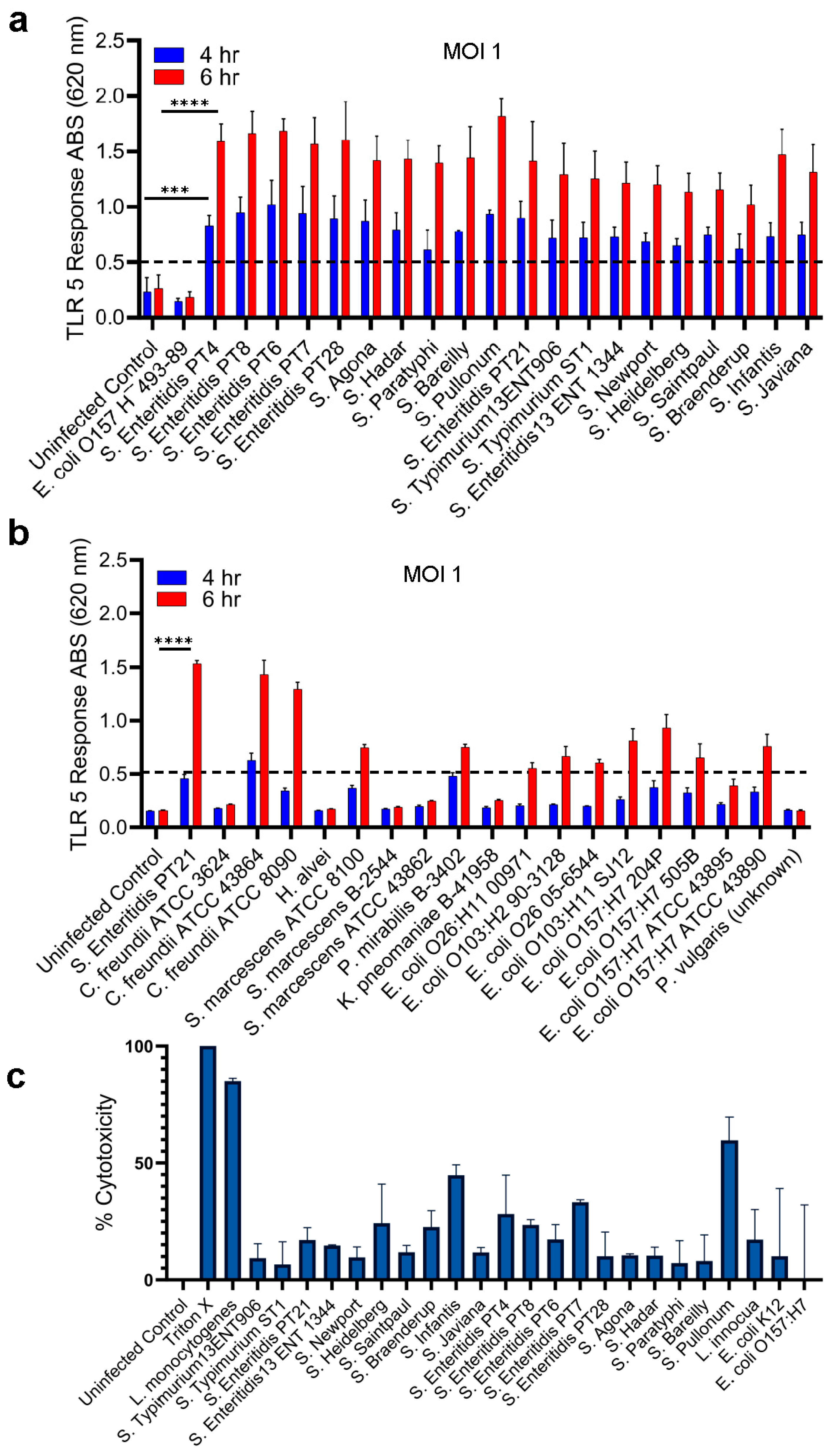
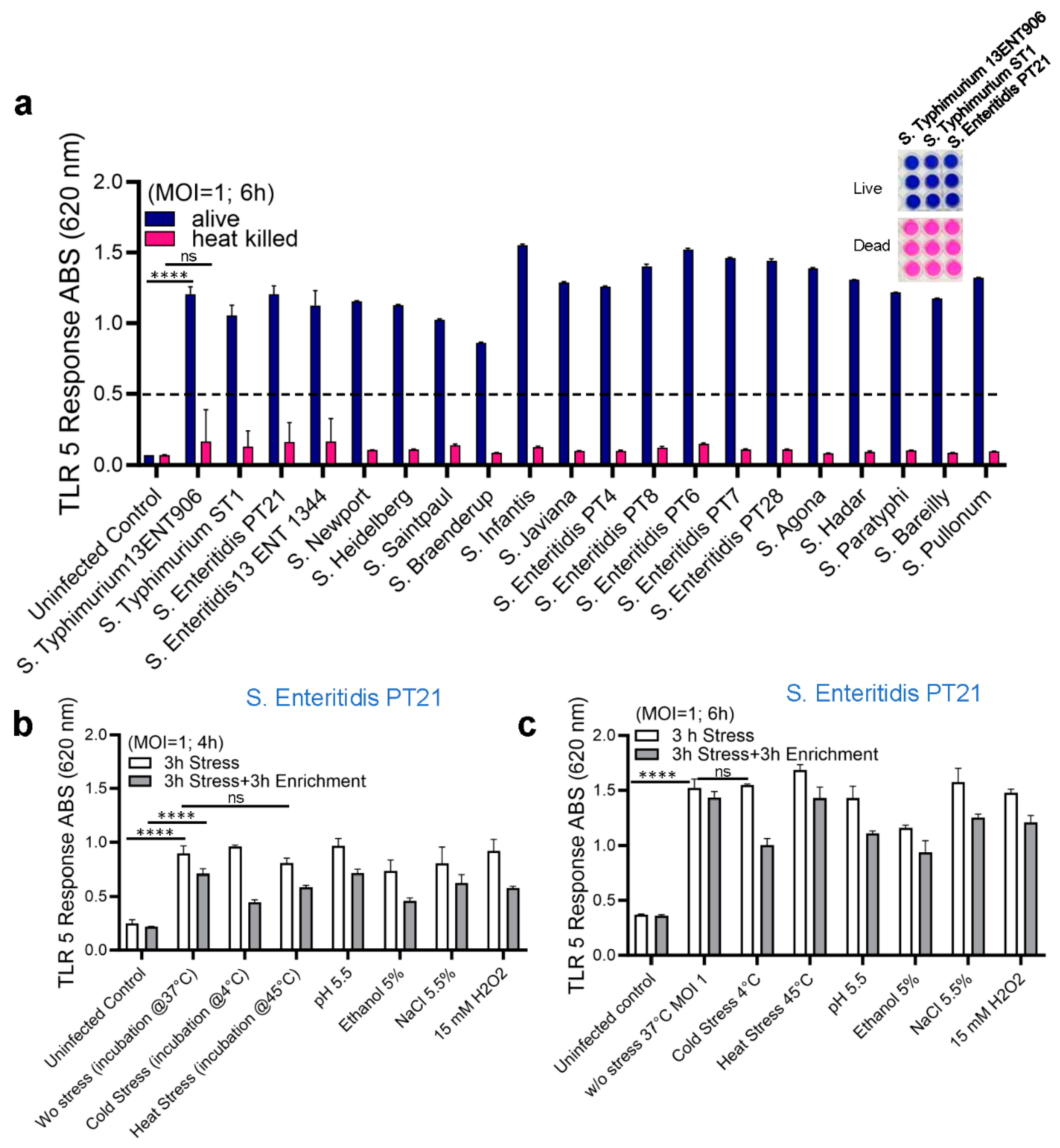
2.3. Detection of Live and Stress-Exposed Salmonella Using HEK dual TLR5 Sensor
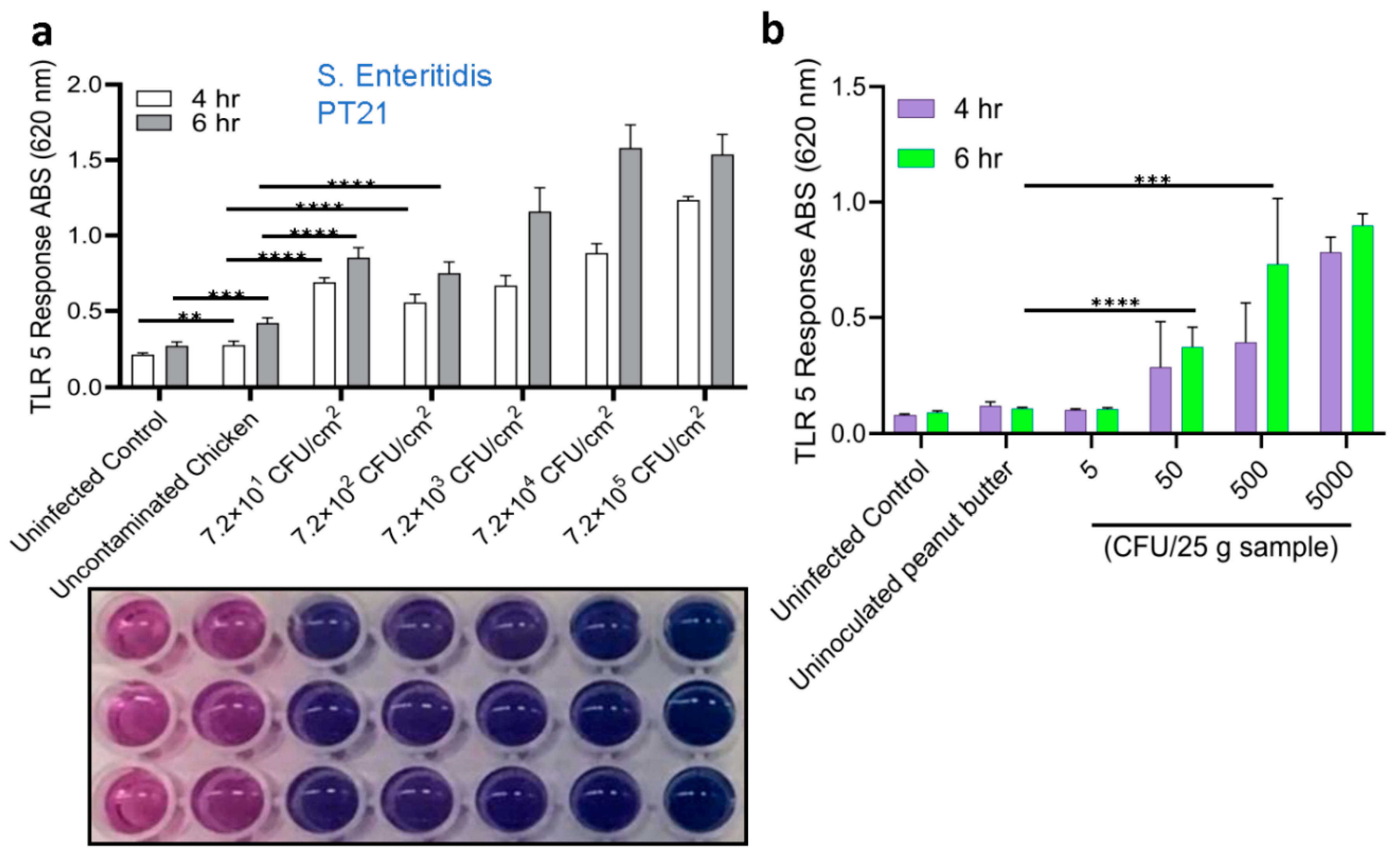
2.4. Detection of Salmonella from Spiked Food Samples Using HEK dual TLR5 Sensor
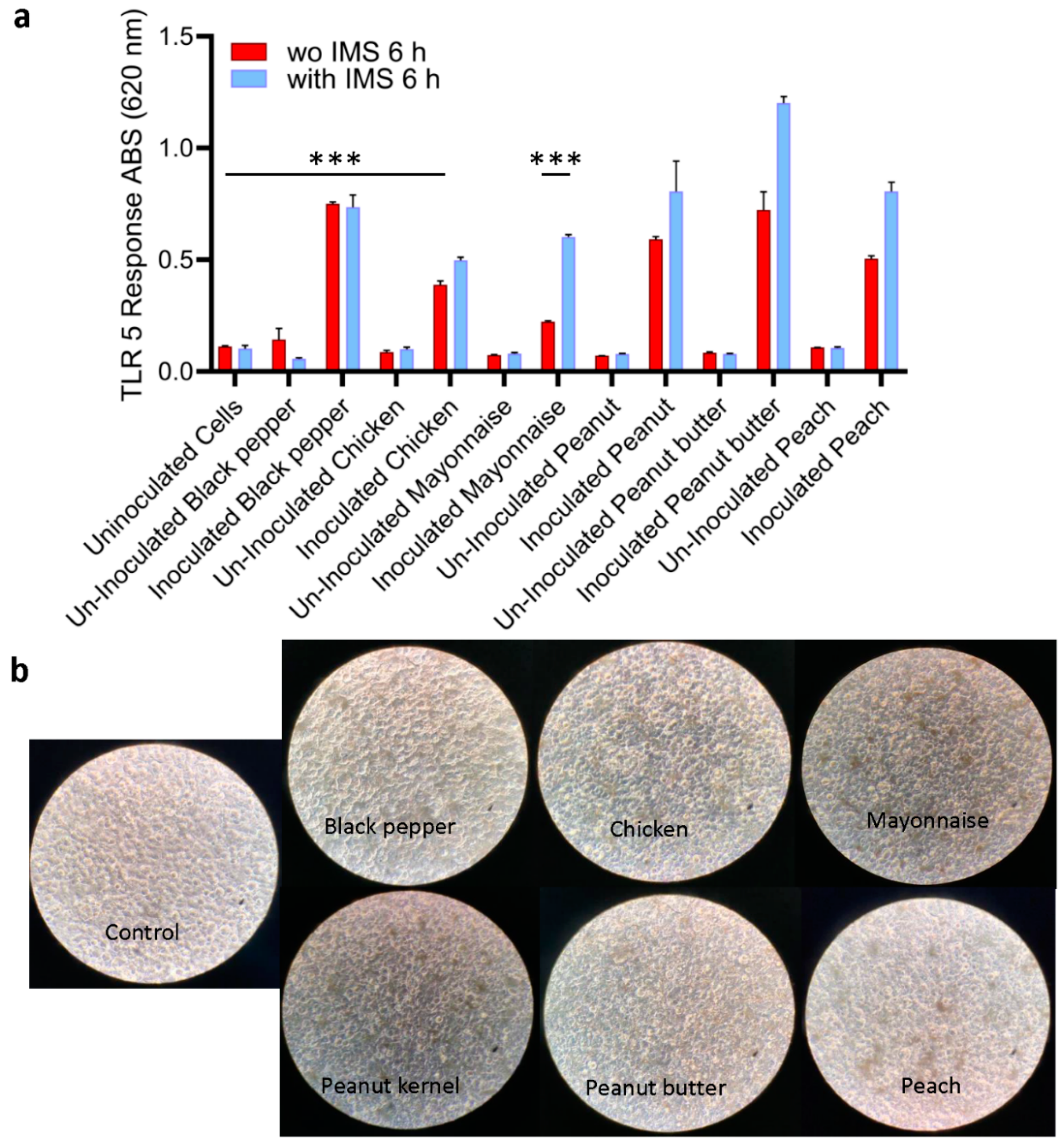
2.5. Validation of Sensor with Spiked Food Samples
3. Discussion
4. Materials and Methods
4.1. Bacterial Cultures and Motility Testing
4.2. Cell Culture
4.3. Cell-Based Detection of Bacterial Culture
4.4. Specificity/Selectivity of Cell-Based Sensor
4.5. Limit of Detection of Cell-Based Sensor
4.6. Salmonella Enteritidis Analysis in Spiked Food Samples
4.7. Detection of Stressed Cells
4.8. Cytotoxicity Assays
4.9. Polymerase Chain Reaction (PCR) confirmation
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgment
Conflict of Interest
References
- Bhunia, A. K., Salmonella enterica. In Foodborne Microbial Pathogens: Mechanisms and Pathogenesis, Bhunia, A. K., Ed. Springer New York: New York, NY, 2018; pp 271-287. 10.1007/978-1-4939-7349-1_15.
- Teklemariam, A. D.; Al-Hindi, R. R.; Albiheyri, R. S.; Alharbi, M. G.; Alghamdi, M. A.; Filimban, A. A.; Al Mutiri, A. S.; Al-Alyani, A. M.; Alseghayer, M. S.; Almaneea, A. M.; Albar, A. H.; Khormi, M. A.; Bhunia, A. K., Human Salmonellosis: A Continuous Global Threat in the Farm-to-Fork Food Safety Continuum. Foods 2023, 12, (9), 1756. [CrossRef]
- Popa, G. L.; Papa, M. I., Salmonella spp. infection-A continuous threat worldwide. Germs 2021, 11, (1), 88.
- Guard, J., Through the Looking Glass: Genome, Phenome, and Interactome of Salmonella enterica. Pathogens 2022, 11, (5), 581. 10.3390/pathogens11050581. [CrossRef]
- O'Bryan, C. A.; Ricke, S. C.; Marcy, J. A., Public health impact of Salmonella spp. on raw poultry: Current concepts and future prospects in the United States. Food Control 2022, 132, 108539. [CrossRef]
- Kowalska, B., Fresh vegetables and fruit as a source of Salmonella bacteria. Ann Agric Environ Med 2023, 30, (1), 9-14. [CrossRef]
- Peifer, M., beta-Catenin as Oncogene--The Smoking Gun. Science 1997, 275, (5307), 1752-0.
- CDC, Salmonella. In CDC; https://www.cdc.gov/salmonella/index.html: Atlanta, GA, 2020; Vol. https://www.cdc.gov/salmonella/index.html.
- Guillén, S.; Nadal, L.; Álvarez, I.; Mañas, P.; Cebrián, G., Impact of the Resistance Responses to Stress Conditions Encountered in Food and Food Processing Environments on the Virulence and Growth Fitness of Non-Typhoidal Salmonellae. Foods 2021, 10, (3), 617. [CrossRef]
- Horn, N.; Bhunia, A. K., Food-Associated Stress Primes Foodborne Pathogens for the Gastrointestinal Phase of Infection. Front. Microbiol. 2018, 9, (August), 1962. [CrossRef]
- Gruzdev, N.; Pinto, R.; Sela, S., Effect of desiccation on tolerance of Salmonella enterica to multiple stresses. Appl Environ Microbiol 2011, 77, (5), 1667-1673. [CrossRef]
- Gustavsen, S.; Breen, O., Investigation of an outbreak of Salmonella Oranienburg infections in Norway, caused by contaminated black pepper. Am. J. Epidemiol. 1984, 119, (5), 806-12. 10.1093/oxfordjournals.aje.a113801. [CrossRef]
- McCormic, Z. D.; Patel, K.; Higa, J.; Bancroft, J.; Donovan, D.; Edwards, L.; Cheng, J.; Adcock, B.; Bond, C.; Pereira, E., Bi-national outbreak of Salmonella Newport infections linked to onions: the United States experience. Epidemiol Infect 2022, 150, e199. [CrossRef]
- Mitchell Jr, M. R.; Kirchner, M.; Schneider, B.; McClure, M.; Neil, K. P.; Madad, A.; Jemaneh, T.; Tijerina, M.; Nolte, K.; Wellman, A., Multistate outbreak of Salmonella Oranienburg infections linked to bulb onions imported from Mexico–United States, 2021. Food Control 2024, 160, 110325.
- CDC, Multistate Outbreak of Human Salmonella Montevideo Infections https://www.cdc.gov/salmonella/2010/montevideo-5-4-2010.html.
- Kirk, M. D.; Little, C. L.; Lem, M.; Fyfe, M.; Genobile, D.; Tan, A.; Threlfall, J.; Paccagnella, A.; Lightfoot, D.; Lyi, H.; McIntyre, L.; Ward, L.; Brown, D. J.; Surnam, S.; Fisher, I. S., An outbreak due to peanuts in their shell caused by Salmonella enterica serotypes Stanley and Newport--sharing molecular information to solve international outbreaks. Epidemiol. Infect. 2004, 132, (4), 571-7. 10.1017/s095026880400216x. [CrossRef]
- Scheil, W.; Cameron, S.; Dalton, C.; Murray, C.; Wilson, D., A South Australian Salmonella Mbandaka outbreak investigation using a database to select controls. Aust N Z J Public Health 1998, 22, (5), 536-9. 10.1111/j.1467-842x.1998.tb01434.x. [CrossRef]
- Calhoun, S.; Post, L.; Warren, B.; Thompson, S.; Bontempo, A. R., Prevalence and concentration of Salmonella on raw shelled peanuts in the United States. J. Food Prot. 2013, 76, (4), 575-579.
- CDC, Outbreak of Salmonella Enteritidis Infections Linked to Peaches.
- CDC, Multistate Outbreak of Multidrug-Resistant Salmonella Heidelberg Infections Linked to Foster Farms Brand Chicken https://www.cdc.gov/salmonella/heidelberg-10-13/index.html.
- CDC, Outbreak of Salmonella Heidelberg Infections Linked to Tyson Brand Mechanically Separated Chicken at a Correctional Facility https://www.cdc.gov/salmonella/heidelberg-01-14/index.html.
- CDC, Outbreak of Salmonella Infections Linked to Chicken https://www.cdc.gov/salmonella/chicken-08-18/index.html.
- CDC, Multistate Outbreak of Multidrug-Resistant Salmonella Heidelberg Infections Linked to Foster Farms Brand Chicken https://www.cdc.gov/salmonella/heidelberg-10-13/index.html.
- Mitchell, E.; O'Mahony, M.; Lynch, D.; Ward, L. R.; Rowe, B.; Uttley, A.; Rogers, T.; Cunningham, D. G.; Watson, R., Large outbreak of food poisoning caused by Salmonella typhimurium definitive type 49 in mayonnaise. Brit. Med. J. 1989, 298, (6666), 99-101. 10.1136/bmj.298.6666.99. [CrossRef]
- Mason, B. W.; Williams, N.; Salmon, R. L.; Lewis, A.; Price, J.; Johnston, K. M.; Trott, R. M., Outbreak of Salmonella Indiana associated with egg mayonnaise sandwiches at an acute NHS hospital. Commun Dis Public Health 2001, 4, (4), 300-4.
- Carneiro, M. R.; Cabello, P. H.; Albuquerque-Junior, R. L.; Jain, S.; Candido, A. L., Characterization of a foodborne outbreak caused by Salmonella Enteritidis in Aracaju, State of Sergipe, Brazil. Revista da Sociedade Brasileira de Medicina Tropical 2015, 48, (3), 334-7. 10.1590/0037-8682-0260-2014. [CrossRef]
- von Wissmann, B.; Klinc, C.; Schulze, R.; Wolf, A.; Schreiner, H.; Rabsch, W.; Prager, R.; Hautmann, W., Outbreak of salmonellosis after a wedding party, Bavaria, Germany, summer 2010: the importance of implementing food safety concepts. Eurosurveillance 2012, 17, (6), 20076. doi:. [CrossRef]
- Bhunia, A. K., One day to one hour: how quickly can foodborne pathogens be detected? Fut. Microbiol. 2014, 9, (8), 935-946.
- Al-Hindi, R. R.; Teklemariam, A. D.; Alharbi, M. G.; Alotibi, I.; Azhari, S. A.; Qadri, I.; Alamri, T.; Harakeh, S.; Applegate, B. M.; Bhunia, A. K., Bacteriophage-Based Biosensors: A Platform for Detection of Foodborne Bacterial Pathogens from Food and Environment. Biosensors 2022, 12, (10), 905. 10.3390/bios12100905. [CrossRef]
- Foddai, A. C. G.; Grant, I. R., Methods for detection of viable foodborne pathogens: Current state-of-art and future prospects. Appl. Microbiol. Biotechnol. 2020, 104, 4281-4288. [CrossRef]
- Daquigan, N.; Grim, C. J.; White, J. R.; Hanes, D. E.; Jarvis, K. G., Early Recovery of Salmonella from Food Using a 6-Hour Non-selective Pre-enrichment and Reformulation of Tetrathionate Broth. Front. Microbiol. 2016, 7, (2103). 10.3389/fmicb.2016.02103. [CrossRef]
- Koyuncu, S.; Andersson, M. G.; Häggblom, P., Accuracy and Sensitivity of Commercial PCR-Based Methods for Detection of Salmonella enterica in Feed. Appl Environ Microbiol 2010, 76, (9), 2815. 10.1128/AEM.02714-09. [CrossRef]
- Moreirinha, C.; Trindade, J.; Saraiva, J. A.; Almeida, A.; Delgadillo, I., MIR spectroscopy as alternative method for further confirmation of foodborne pathogens Salmonella spp. and Listeria monocytogenes. J Food Sci Technol 2018, 55, (10), 3971-3978. 10.1007/s13197-018-3322-8. [CrossRef]
- Villamizar-Rodríguez, G.; Fernández, J.; Marín, L.; Muñiz, J.; González, I.; Lombó, F., Multiplex detection of nine food-borne pathogens by mPCR and capillary electrophoresis after using a universal pre-enrichment medium. Front. Microbiol. 2015, 6, (1194). 10.3389/fmicb.2015.01194. [CrossRef]
- Xu, L.; Bai, X.; Bhunia, A. K., Current State of Biosensors Development and their Application in Foodborne Pathogen Detection. J. Food Prot. 2021, 84, (7), 1213-1227. 10.4315/JFP-20-464. [CrossRef]
- Eijkelkamp, J. M.; Aarts, H. J. M.; van der Fels-Klerx, H. J., Suitability of Rapid Detection Methods for Salmonella in Poultry Slaughterhouses. Food Anal. Methods 2009, 2, (1), 1-13. 10.1007/s12161-008-9040-5. [CrossRef]
- Xu, L.; Bai, X.; Tenguria, S.; Liu, Y.; Drolia, R.; Bhunia, A. K., Mammalian cell-based immunoassay for detection of viable bacterial pathogens. Front. Microbiol. 2020, 11, 575615. 10.3389/fmicb.2020.575615. [CrossRef]
- Bhunia, A. K.; Singh, A. K.; Parker, K.; Applegate, B. M., Petri-plate, bacteria, and laser optical scattering sensor. Front. Cell. Infect. Microbiol. 2022, 12, 1087074. 10.3389/fcimb.2022.1087074. [CrossRef]
- To, C.; Banerjee, P.; Bhunia, A. K., Cell-Based Biosensor for Rapid Screening of Pathogens and Toxins. In Handbook of Cell Biosensors, Thouand, G., Ed. Springer International Publishing: Cham, 2020; pp 1-16. 10.1007/978-3-319-47405-2_102-1. [CrossRef]
- Lu, X.; Ye, Y.; Zhang, Y.; Sun, X., Current research progress of mammalian cell-based biosensors on the detection of foodborne pathogens and toxins. Crit. Rev. Food Sci. Nutr. 2021, 61, (22), 3819-3835. [CrossRef]
- Banerjee, P.; Bhunia, A. K., Mammalian cell-based biosensors for pathogens and toxins. Trends Biotechnol. 2009, 27, (3), 179-188. [CrossRef]
- Burkholder, K.; Bhunia, A., Salmonella enterica serovar Typhimurium adhesion and cytotoxicity during epithelial cell stress is reduced by Lactobacillus rhamnosus GG. Gut Pathog. 2009, 1, (1), 14. [CrossRef]
- Birhanu, B. T.; Park, N.-H.; Lee, S.-J.; Hossain, M. A.; Park, S.-C., Inhibition of Salmonella Typhimurium adhesion, invasion, and intracellular survival via treatment with methyl gallate alone and in combination with marbofloxacin. Vet. Res. 2018, 49, (1), 101. 10.1186/s13567-018-0597-8. [CrossRef]
- Chen, L. M.; Kaniga, K.; Galan, J. E., Salmonella spp. are cytotoxic for cultured macrophages. Mol. Microbiol. 1996, 21, (5), 1101-1115. [CrossRef]
- Yang, J.; Yan, H., TLR5: beyond the recognition of flagellin. Cell. Mol. Immunol. 2017, 14, (12), 1017-1019. [CrossRef]
- Song, W. S.; Jeon, Y. J.; Namgung, B.; Hong, M.; Yoon, S. I., A conserved TLR5 binding and activation hot spot on flagellin. Sci Rep 7: 40878. In 2017. [CrossRef]
- Gray, K. M.; Bhunia, A. K., Specific detection of cytopathogenic Listeria monocytogenes using a two-step method of immunoseparation and cytotoxicity analysis. J. Microbiol. Methods 2005, 60, (2), 259-268. [CrossRef]
- Mendonca, M.; Conrad, N.; Conceicao, F.; Moreira, A.; da Silva, W.; Aleixo, J.; Bhunia, A., Highly specific fiber optic immunosensor coupled with immunomagnetic separation for detection of low levels of Listeria monocytogenes and L. ivanovii. BMC Microbiol. 2012, 12, (1), 275. [CrossRef]
- de Cássia Dos Santos da Conceição, R.; Moreira, A. N.; Ramos, R. J.; Goularte, F. L.; Carvalhal, J. B.; Aleixo, J. A. G., Detection of Salmonella sp in chicken cuts using immunomagnetic separation. Braz J Microbiol 2008, 39, (1), 173-177. 10.1590/S1517-838220080001000034. [CrossRef]
- Mansfield, L. P.; Forsythe, S. J., The detection of Salmonella using a combined immunomagnetic separation and ELISA end-detection procedure. Lett Appl Microbiol 2000, 31, (4), 279-283. 10.1046/j.1472-765x.2000.00811.x. [CrossRef]
- Ha, D.-G.; Kuchma, S. L.; O’Toole, G. A., Plate-Based Assay for Swimming Motility in Pseudomonas aeruginosa. In Pseudomonas Methods and Protocols, Filloux, A.; Ramos, J.-L., Eds. Springer New York: New York, NY, 2014; pp 59-65. 10.1007/978-1-4939-0473-0_7. [CrossRef]
- Kearns, D. B.; Losick, R., Swarming motility in undomesticated Bacillus subtilis. Mol. Microbiol. 2003, 49, (3), 581-90. 10.1046/j.1365-2958.2003.03584.x. [CrossRef]
- Singh, A. K.; Bettasso, A. M.; Bae, E.; Rajwa, B.; Dundar, M. M.; Forster, M. D.; Liu, L.; Barrett, B.; Lovchik, J.; Robinson, J. P.; Hirleman, E. D.; Bhunia, A. K., Laser optical sensor, a label-free on-plate Salmonella enterica colony detection tool. mBio 2014, 5 (1), (1), e01019-13. 10.1128/mBio.01019-13. [CrossRef]
- Andersen-Nissen, E.; Smith, K. D.; Strobe, K. L.; Barrett, S. L. R.; Cookson, B. T.; Logan, S. M.; Aderem, A., Evasion of Toll-like receptor 5 by flagellated bacteria. Proc. Nat. Acad. Sci. USA 2005, 102, (26), 9247-9252. [CrossRef]
- Hahm, B. K.; Bhunia, A. K., Effect of environmental stresses on antibody-based detection of Escherichia coli O157:H7, Salmonella enterica serotype Enteritidis and Listeria monocytogenes. J. Appl. Microbiol. 2006, 100, (5), 1017-1027. [CrossRef]
- USDA-FSIS, Isolation and Identification of Salmonella from Meat, Poultry, Pasteurized Egg and Catfish Products. Method number MLG 4.06. In http://www.fsis.usda.gov/wps/wcm/connect/700c05fe-06a2-492a-a6e1-3357f7701f52/MLG-4.pdf?MOD=AJPERES: 2013.
- Bell, R. L.; Jarvis, K. G.; Ottesen, A. R.; McFarland, M. A.; Brown, E. W., Recent and emerging innovations in Salmonella detection: a food and environmental perspective. Microb.Biotechnol. 2016, 9, (3), 279-292. [CrossRef]
- FDA, Bacteriological Analytical Manual Online, 8th ed In AOAC International, Arlington, VA. [http://www.fda.gov/Food/ScienceResearch/LaboratoryMethods/BacteriologicalAnalyticalManualBAM/default.htm]: 2001. http://www.cfsan.fda.gov/~ebam/bam-toc.html.
- Rajapaksha, P.; Elbourne, A.; Gangadoo, S.; Brown, R.; Cozzolino, D.; Chapman, J., A review of methods for the detection of pathogenic microorganisms. Analyst 2019, 144, (2), 396-411. [CrossRef]
- Ricke, S. C.; Kim, S. A.; Shi, Z.; Park, S. H., Molecular-based identification and detection of Salmonella in food production systems: current perspectives. J. Appl. Microbiol. 2018, 125, (2), 313-327. 10.1111/jam.13888. [CrossRef]
- Lee, K.-M.; Runyon, M.; Herrman, T. J.; Phillips, R.; Hsieh, J., Review of Salmonella detection and identification methods: Aspects of rapid emergency response and food safety. Food Control 2015, 47, 264-276. [CrossRef]
- Buzby, J. C.; Farah-Wells, H.; Hyman, J., The estimated amount, value, and calories of postharvest food losses at the retail and consumer levels in the United States. USDA-ERS Economic Information Bulletin Number 121 2014, (121).
- Elkhishin, M. T.; Gooneratne, R.; Hussain, M. A., Microbial safety of foods in the supply chain and food security. Adv Food Technol Nutr Sci Open J 2017, 3, (1), 22-32. [CrossRef]
- Bhunia, A. K., Rapid pathogen screening tools for food safety. Food Technol. 2011, 65, (2), 38-43.
- Hayashi, F.; Smith, K. D.; Ozinsky, A.; Hawn, T. R.; Yi, E. C.; Goodlett, D. R.; Eng, J. K.; Akira, S.; Underhill, D. M.; Aderem, A., The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 2001, 410, (6832), 1099-1103. [CrossRef]
- Smith, K. D.; Andersen-Nissen, E.; Hayashi, F.; Strobe, K.; Bergman, M. A.; Barrett, S. L. R.; Cookson, B. T.; Aderem, A., Toll-like receptor 5 recognizes a conserved site on flagellin required for protofilament formation and bacterial motility. Nat. Immunol. 2003, 4, (12), 1247-1253.
- Yoon, S.-i.; Kurnasov, O.; Natarajan, V.; Hong, M.; Gudkov, A. V.; Osterman, A. L.; Wilson, I. A., Structural basis of TLR5-flagellin recognition and signaling. Science 2012, 335, (6070), 859-864. [CrossRef]
- Schrader, C.; Schielke, A.; Ellerbroek, L.; Johne, R., PCR inhibitors–occurrence, properties and removal. J. Appl. Microbiol. 2012, 113, (5), 1014-1026.
- Lin, A.; Singh, A.; Allred, A.; Allard, M.; Waltman, D.; Imanian, B.; Ng, J. H. J.; Sanahmadi, Y.; Khaksar, R., Targeted Next Generation Sequencing Assay for Direct Detection and Serotyping of Salmonella from Enrichment. J. Food Prot. 2024, 100256. [CrossRef]
- Vinayaka, A. C.; Ngo, T. A.; Kant, K.; Engelsmann, P.; Dave, V. P.; Shahbazi, M. A.; Wolff, A.; Bang, D. D., Rapid detection of Salmonella enterica in food samples by a novel approach with combination of sample concentration and direct PCR. Biosens Bioelectron 2019, 129, 224-230. 10.1016/j.bios.2018.09.078. [CrossRef]
- Kuang, H.; Cui, G.; Chen, X.; Yin, H.; Yong, Q.; Xu, L.; Peng, C.; Wang, L.; Xu, C., A one-step homogeneous sandwich immunosensor for Salmonella detection based on magnetic nanoparticles (MNPs) and quantum Dots (QDs). Int J Mol Sci 2013, 14, (4), 8603-8610. 10.3390/ijms14048603. [CrossRef]
- Cimaglia, F.; De Lorenzis, E.; Mezzolla, V.; Rossi, F.; Poltronieri, P., Detection of L. monocytogenes in Enrichment Cultures by Immunoseparation and Immunosensors. IEEE Sensors J. 2016, 16, (19), 7045-7052. 10.1109/JSEN.2016.2598700. [CrossRef]
| Log 10 (CFU/mL) ± SEM (N=3) | |||||||
| No stress | Cold Stress | Heat Stress | Low pH | Ethanol | NaCL | H2O2 | |
| 3 h stress | 9.86±0.19 | 9.38±0.21 | 9.35±0.11 | 9.67±0.14 | 9.60±0.22 | 9.24±0.28 | 9.33±0.21 |
| 3 h stress+3 h enrichment | 10.49±0.22 | 10.44±0.24 | 10.01±0.21 | 10.21±0.24 | 9.69±0.29 | 10.33±0.19 | 10.65±0.24 |
| Log 10 (CFU/ml) ± SEM (N=3) | ||||
| Inoculation Level (CFU/25g sample) | Pre-Enriched | Enriched | ||
| TSA | XLD | TSA | XLD | |
| 0 | No growth | No growth | No growth | No growth |
| ~5 | No growth | No growth | 2.86±0.21 | 3.09±0.18 |
| ~50 | 2.68±0.17 | 2.51±0.11 | 4.06±0.14 | 4.18±0.19 |
| ~500 | 3.94±0.21 | 3.85±0.19 | 6.32±0.22 | 6.39±0.24 |
| ~500 | 4.35±0.13 | 4.44±0.18 | 6.60±0.23 | 6.42±0.14 |
| Log 10 (CFU/ml)± SEM (N=3) | ||||||||
| Food Sample | After Selective Enrichment | Without IMS |
IMS |
Counts (CFU/100 µL) |
PCR (invA 796 bp) |
|||
| TSA | XLD | TSA | XLD | TSA | XLD | XLD | ||
| Black pepper (U) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | - |
| Black pepper (I) | 3.91±0.18 | 4.04±0.11 | 3.87±0.14 | 4.05±0.21 | 3.98±0.08 | 4.07±0.14 | 3.07±0.11 | - |
| Chicken (U) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | - |
| Chicken (I) | 3.85±0.15 | 3.92±0.08 | 3.94±0.19 | 4.10±0.21 | 3.91±0.15 | 4.10±0.11 | 3.1±0.09 | - |
| Mayonnaise (U) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | - |
| Mayonnaise (I) | 3.94±0.22 | 4.02±0.14 | 3.72±0.18 | 3.94±0.12 | 3.72±0.21 | 3.92±0.18 | 2.92±0.13 | - |
| Peanut kernel(U) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | - |
| Peanut kernel (I) | 3.92±0.26 | 4.09±0.19 | 4.26±0.22 | 4.29±0.18 | 4.07±0.14 | 4.13±0.19 | 3.13±0.18 | + |
| Peanut butter (U) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | - |
| Peanut butter (I) | 5.53±0.24 | 5.18±0.18 | 5.36±0.25 | 5.46±0.28 | 5.39±0.22 | 5.47±0.24 | 4.47±0.16 | - |
| Peach (U) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | - |
| Peach (I) | 3.71±0.14 | 3.88±0.16 | 3.72±0.12 | 4.08±0.19 | 3.91±0.16 | 4.33±0.14 | 3.33±0.11 | - |
| Salmonella enterica serovars | Non-Salmonella cultures |
| Enteritidis PT4 | Escherichia coli O157 H¯493-89 (flagella Negative control) |
| Enteritidis PT8 | E. coli O26:H11 00971 |
| Enteritidis PT6 | E. coli O103:H2 90-3128 |
| Enteritidis PT7 | E. coli O26 05-6544 |
| Enteritidis PT28 | E. coli O103:H11 SJ12 |
| Enteritidis PT21 | E. coli O157:H7 204P |
| Enteritidis 13ENT1344 | E. coli O157:H7 505B |
| Typhimurium 13ENT906 | E. coli O157:H7 ATCC 43895 |
| Typhimurium ST1 | E. coli O157:H7 ATCC 43890 |
| Agona 12ENT1356 | Citrobacter freundii ATCC 3624 |
| Hadar 13ENT979 | Citrobacter freundii ATCC43864 |
| Paratyphi 11J85 | Citrobacter freundii ATCC8090 |
| Bareilly 12ENT1164 | Hafnia alvei |
| Pullorum DUP-PVUII 1006 | Proteus vulgaris DUP-10086 |
| Newport 13ENT1060 | Proteus mirabilis B-3402 |
| Heidelberg 18ENT1418 | Serratia marcescens ATCC 8100 |
| Saintpaul 13ENT1045 | S. marcescens B-2544 |
| Bradenderup 12ENT1138 | S. marcescens ATCC 43862 |
| Infantis 13ENT866 | Klebsiella pneumoniae B-41958 |
| Javiana 13ENT86F | Listeria monocytogenes 104033S |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





