Submitted:
14 August 2025
Posted:
14 August 2025
You are already at the latest version
Abstract
Climate change increases microbial contamination risks in food, highlighting the need for real-time biosensors. However, food residues often interfere with detection signals, limiting direct application. An integrated system of filter-assisted sample preparation (FASP) and an immunoassay-based colorimetric biosensor offers rapid and simple on-site detection of foodborne pathogens in complex food matrices. The accuracy and stability of biosensor analysis were ensured by filter-assisted preprocessing, which separated food residues from bacteria. The system was applied to various food matrices, including vegetables, meats, and cheese brine, using samples spiked at contamination levels ranging from 10²–10³ CFU per 25 g, thereby demonstrating broad applicability. A detection limit of 10¹ CFU/mL was achieved for Escherichia coli O157:H7, Salmonella Typhimurium, and Listeria monocytogenes in the final preprocessed sample solutions. Sample preparation took under 3 minutes, and detection was completed within 2 hours under stationary conditions. This approach enables rapid pathogen detection in various food matrices without the need for special reading devices, contributing to food safety as a real-time, rapid-response food biosensor.
Keywords:
1. Introduction
| Preprocessing method | Detection method | Instrument | Preprocessing/ detection time |
LOD | Sample | Target | Reference | |
| Double filter method: GF/D and cellulose acetate filter with 0.45 μm-sized pore | Immunoassay-based colorimetric biosensor |
Stomacher Vacuum pump |
≥ 3 min/ ≥ 120 min |
101 CFU/mL |
Vegetables, meats, cheese brine etc. |
E. coli O157:H7 S. Typhimurium L. monocytogenes |
This work | |
| Immunomagnetic separation | Aptamer-based QCM sensor |
QCM crystal, Homogenizer, Frequency counter, Magnetic separator |
≥ 10 min/ ≥ 5 min |
102 CFU/mL |
Poultry, milk | L. monocytogenes | Beyazit et al., 2024 [25] |
|
| Filter method: Paper filter and Centrifugation method | Nanozyme-based colorimetric biosensor |
Centrifuge Paper-chip |
N.D.1) ≥ 120 min |
101 CFU/mL |
Milk | S. Typhimurium | Mirsadoughi et al., 2023 [26] |
|
| Pre-enrichment | Immunoassay-based optical biosensor |
Incubator, Photodetector Microfluidic system, Nanophotonic biosensor |
≥ Total 4 hours |
101–2 CFU/mL |
Hamburger patty | L. monocytogenes | Blanco et al., 2023 [27] |
|
| N.D. | DNA-based Ectrochemical biosensor |
Vibrator, Centrifuge, Heating source, Electrochemical instrument |
N.D. | 100 CFU/mL |
Human blood, Raw milk, egg, poultry |
S. Typhi | Bacchu et al., 2022 [28] |
|
| Filter method: glass wool, graphite electrode, and filter with 50 μm -sized pore, Continuous flow centrifuge |
Enzyme-linked immunoelectrochemical biosensor |
Stomacher Vacuum pump Continuous flow centrifuge Electrochemical instrument |
N.D. ≥ 3 hours |
102 CFU/mL |
Minced beef | E. coli O157:H7 | Capobianco et al., 2021 [29] |
|
| Centrifugation method, Pre-enrichment |
Immunoassay-based multistep lateral flow assay |
Stomacher, Centrifuge, Incubator, LFlIA strips |
≥ Total 7 hours |
100 CFU/g |
Lettuces |
E. coli O157:H7 S. Typhimurium S. aureus B. cereus |
Shin et al., 2018 [30] |
|
2. Materials and Methods
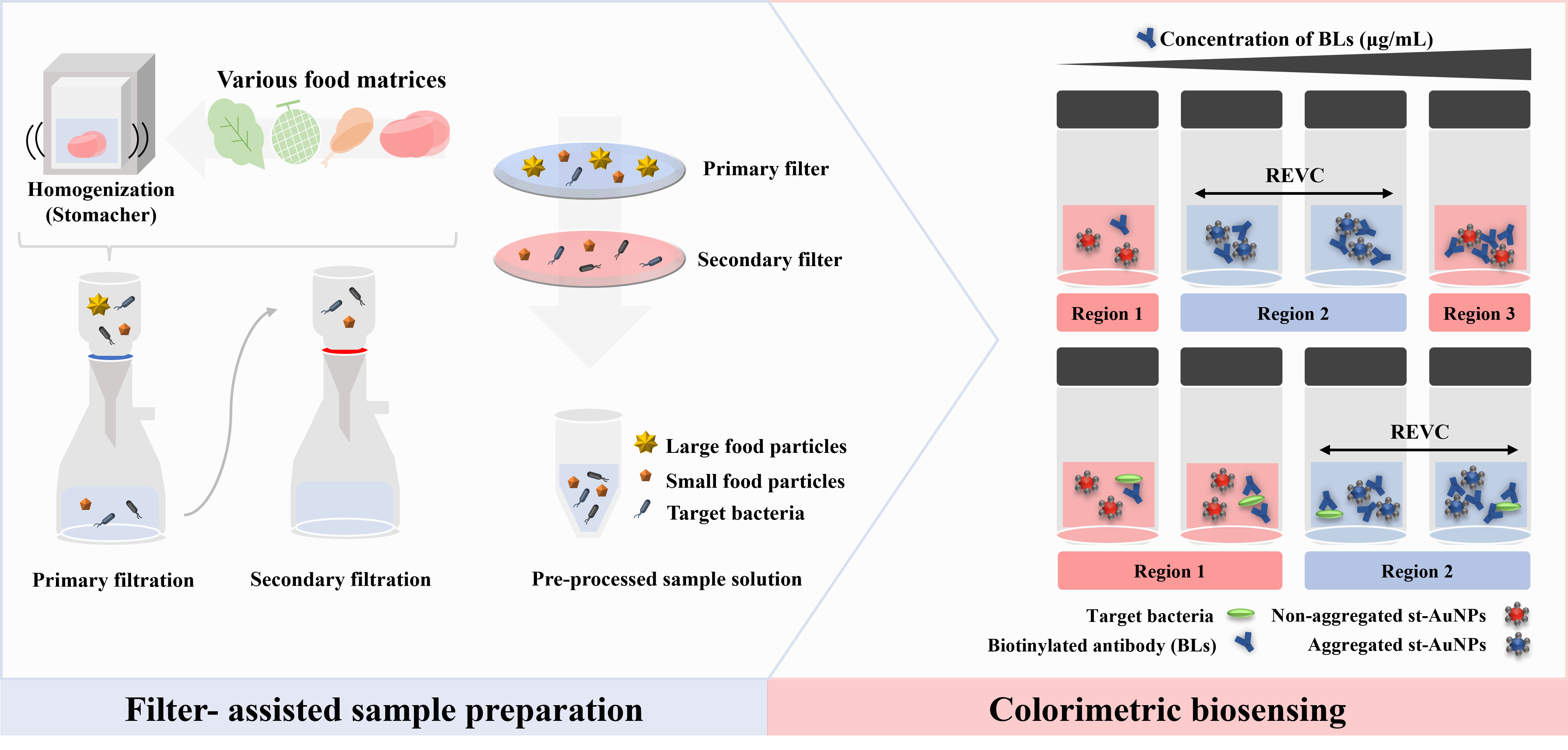
2.1. Filter-Assisted Sample Preparation
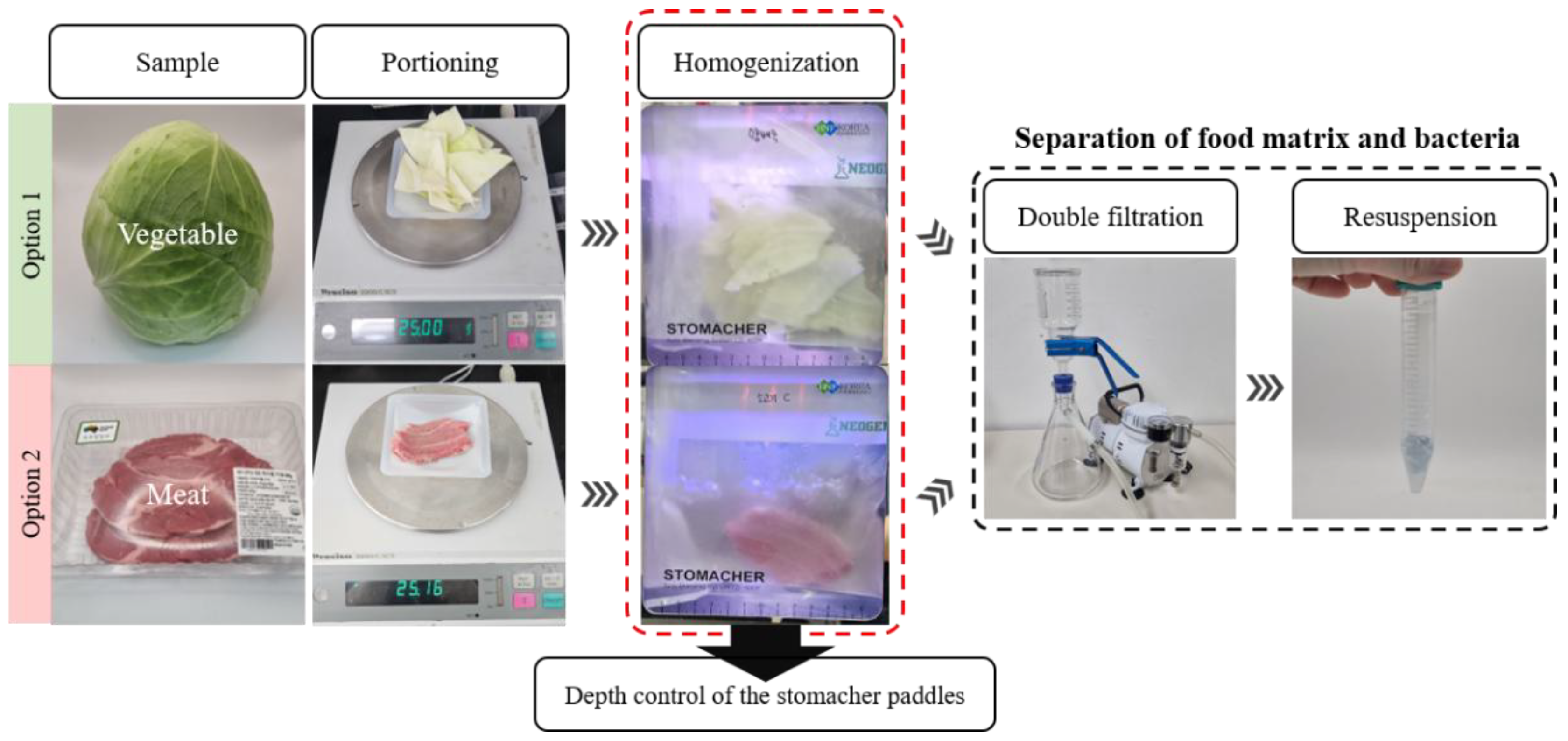
2.1.1. Food Samples
2.1.2. Filtration Process
2.1.3. Recovery of Bacteria from Various Food Matrices
2.2. Colorimetric Biosensing
2.2.1. Biosensor Materials
2.2.2. Synthesis of Streptavidin-Functionalized Gold Nanoparticles (AuNPs)
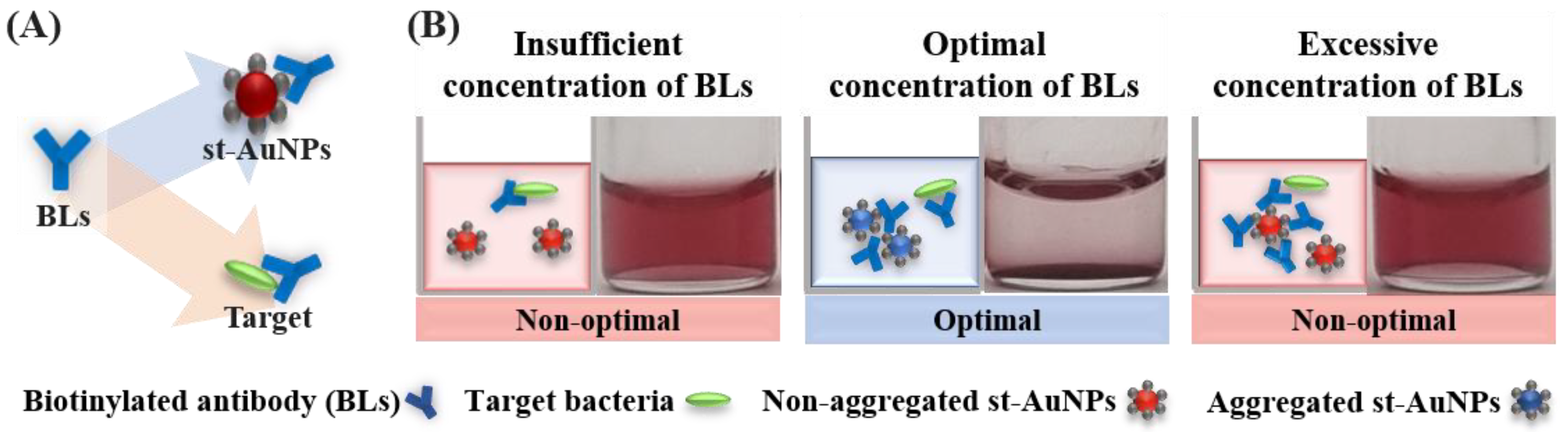
2.2.3. Immunoassay-Based Colorimetric Biosensor
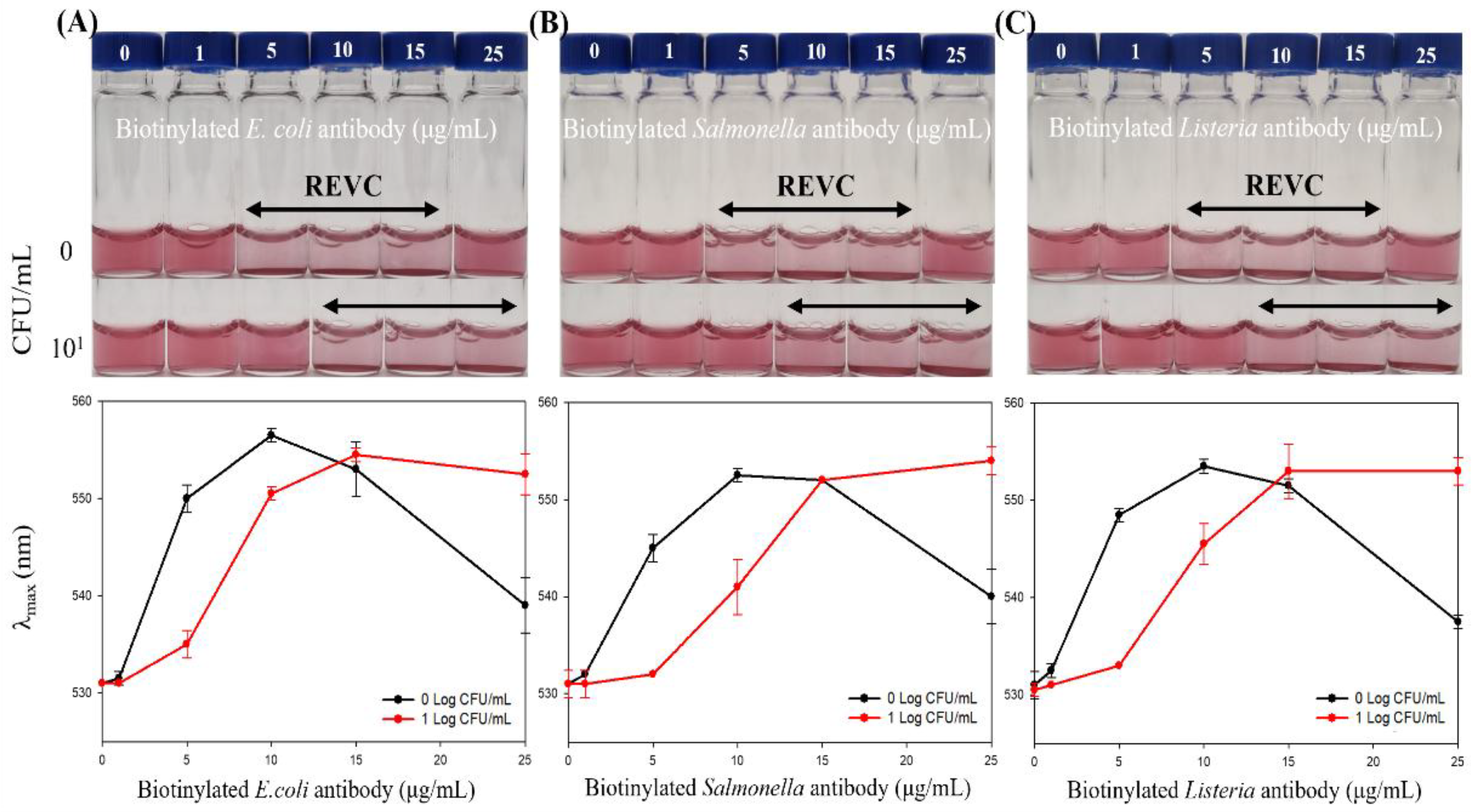
2.3. Impact of Sample Preparation on Biosensing Performance
2.4. Application of the Integrated Diagnostic System in Various Food Matrices
2.5. Bacteria Culture
3. Results and Discussion
3.1. Recovery of Bacteria from Various Food Matrices
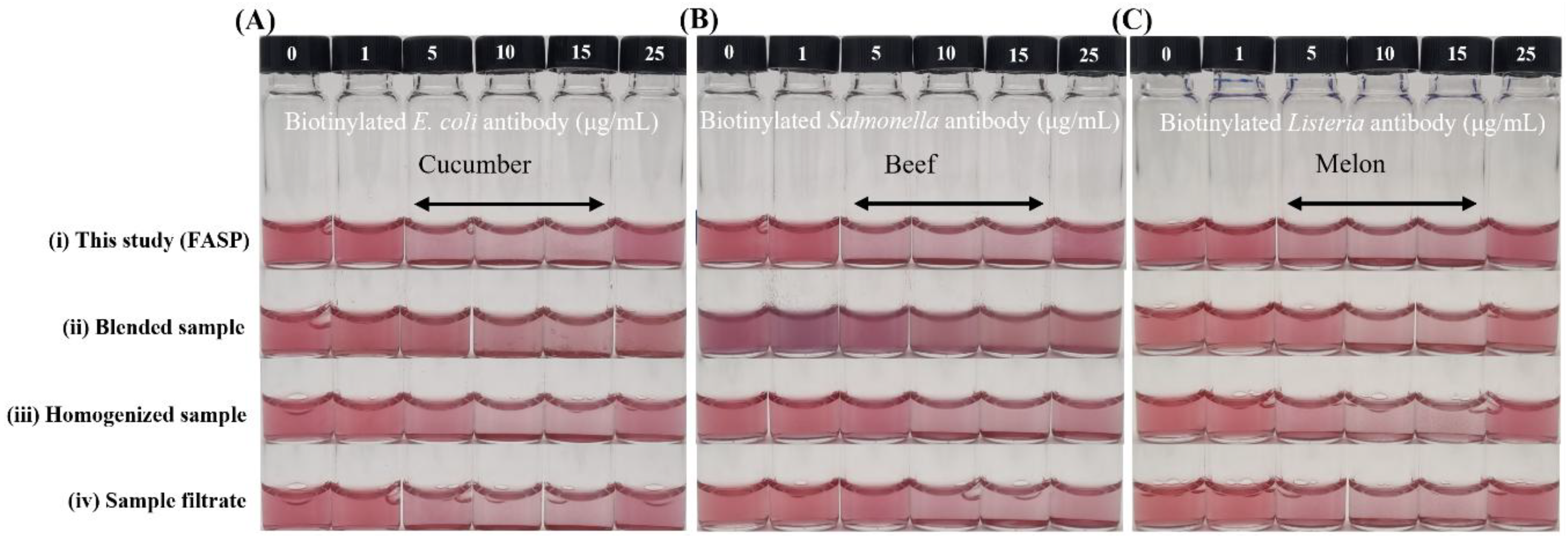
3.3. Impact of Sample Preparation on Biosensing Performance
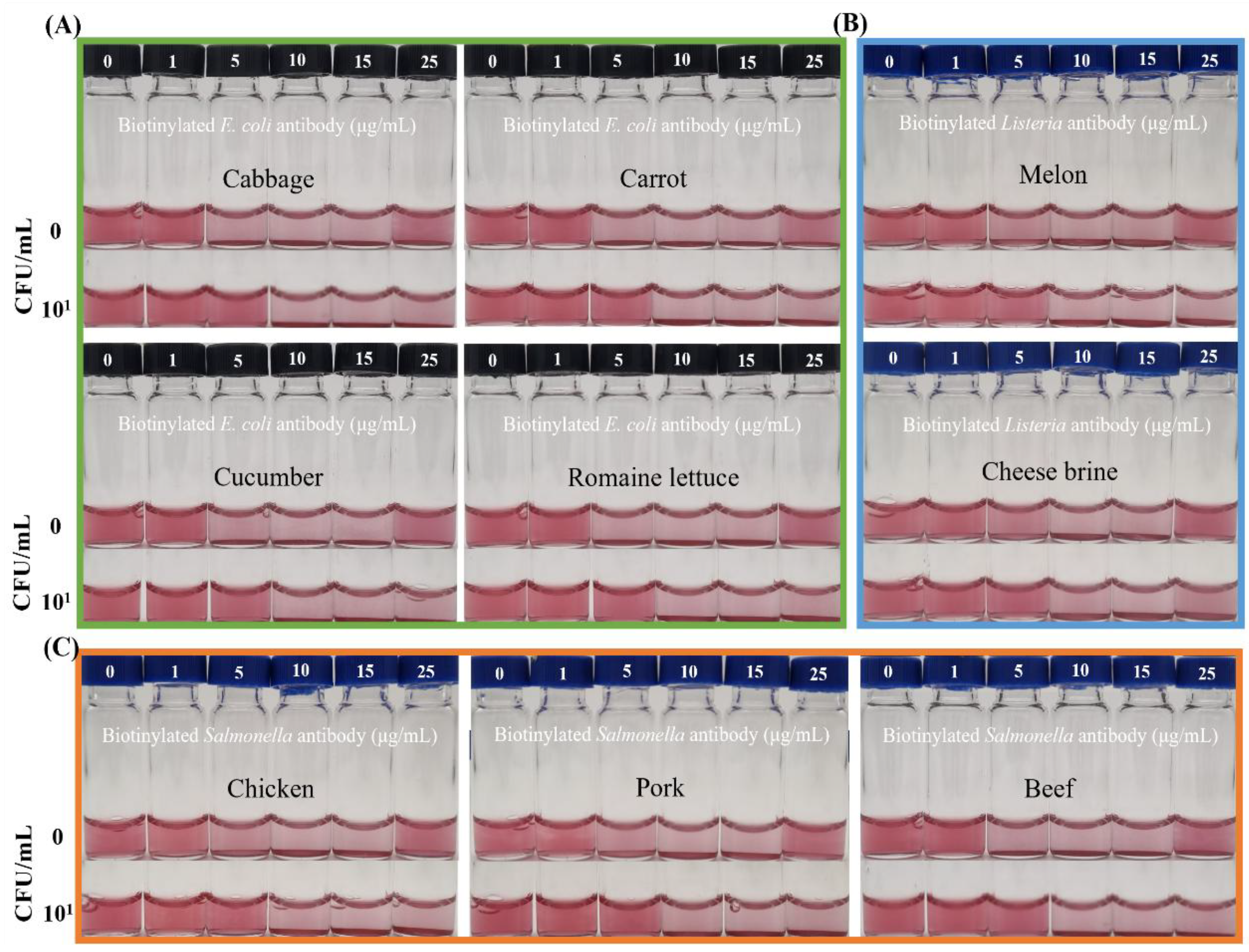
3.4. Application of the Integrated Diagnostic System in Various Food Matrices
4. Conclusions
Supplementary Materials
Author Contributions
Data Availability Statement
Acknowledgments
Conflicts of Interest
Declaration of generative AI and AI-assisted technologies in the writing process
References
- Chen, S.; Brahma, S.; Mackay, J.; Cao, C.; Aliakbarian, B. The Role of Smart Packaging System in Food Supply Chain. J. Food Sci. 2020, 85, 517–525. [Google Scholar] [CrossRef] [PubMed]
- Hua, Z.; Yu, T.; Liu, D.; Xianyu, Y. Recent Advances in Gold Nanoparticles-Based Biosensors for Food Safety Detection. Biosens. Bioelectron. 2021, 179, 113076. [Google Scholar] [CrossRef]
- Nnachi, R.C.; Sui, N.; Ke, B.; Luo, Z.; Bhalla, N.; He, D.; Yang, Z. Biosensors for Rapid Detection of Bacterial Pathogens in Water, Food and Environment. Environ. Int. 2022, 166, 107357. [Google Scholar] [CrossRef]
- Aladhadh, M. A Review of Modern Methods for the Detection of Foodborne Pathogens. Microorganisms 2023, 11, 1111. [Google Scholar] [CrossRef]
- Balbinot, S.; Srivastav, A.M.; Vidic, J.; Abdulhalim, I.; Manzano, M. Plasmonic Biosensors for Food Control. Trends Food Sci. Technol. 2021, 111, 128–140. [Google Scholar] [CrossRef]
- Duchenne-Moutien, R.A.; Neetoo, H. Climate Change and Emerging Food Safety Issues: A Review. J. Food Prot. 2021, 84, 1884–1897. [Google Scholar] [CrossRef]
- Xiao, F.; Li, W.; Xu, H. Advances in Magnetic Nanoparticles for the Separation of Foodborne Pathogens: Recognition, Separation Strategy, and Application. Compr. Rev. Food Sci. Food Saf. 2022, 21, 4478–4504. [Google Scholar] [CrossRef]
- Hu, R.; Wang, W.; Zheng, Q.; Lin, J.; Yang, H. Novel Microfluidic Chips Integrated with Smart Devices for In-Situ Detection of Foodborne Pathogenic Bacteria. Curr. Opin. Food Sci. 2024, 101269. [Google Scholar] [CrossRef]
- ISO 6887-1:2017. Microbiology of the Food Chain—Preparation of Test Samples, Initial Suspension and Decimal Dilutions for Microbiological Examination—Part 1: General Rules; ISO: Geneva, Switzerland. 2017. [Google Scholar]
- Rohde, A.; Hammerl, J.A.; Appel, B.; Dieckmann, R.; Al Dahouk, S. Sampling and Homogenization Strategies Significantly Influence the Detection of Foodborne Pathogens in Meat. Biomed Res. Int. 2015, 2015, 145437. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Salazar, J.K. Culture-independent Rapid Detection Methods for Bacterial Pathogens and Toxins in Food Matrices. Compr. Rev. Food Sci. Food Saf. 2016, 15, 183–205. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, C.M.; Gehring, A.G.; Paoli, G.C.; Chen, C.-Y.; He, Y.; Capobianco, J.A. Impacts of Clarification Techniques on Sample Constituents and Pathogen Retention. Foods. 2019, 8, 636. [Google Scholar] [CrossRef]
- Kim, J.H.; Oh, S.W. Development of a Filtration-Based LAMP–LFA Method as Sensitive and Rapid Detection of E. Coli O157: H7. J. Food Sci. Technol. 2019, 56, 2576–2583. [Google Scholar] [CrossRef]
- Dester, E.; Alocilja, E. Current Methods for Extraction and Concentration of Foodborne Bacteria with Glycan-Coated Magnetic Nanoparticles: A Review. Biosensors. 2022, 12, 112. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Jia, K.; Qu, X.; Li, Y.; Lin, J. Progress in Separation and Detection of Foodborne Bacteria for Food Safety. Curr. Opin. Food Sci. 2024, 101266. [Google Scholar] [CrossRef]
- Hosseinikebria, S.; Khazaei, M.; Dervisevic, M.; Judicpa, M.A.; Tian, J.; Razal, J.M.; Voelcker, N.H.; Nilghaz, A. Electrochemical Biosensors: The Beacon for Food Safety and Quality. Food Chem. 2025, 143284. [Google Scholar] [CrossRef]
- Yuan, H.; Yong, R.; Yuan, W.; Zhang, Q.; Lim, E.G.; Wang, Y.; Niu, F.; Song, P. Centrifugation-Assisted Lateral Flow Assay Platform: Enhancing Bioassay Sensitivity with Active Flow Control. Microsystems Nanoeng. 2025, 11, 101. [Google Scholar] [CrossRef]
- Yola, B.B.; Özdemir, N.; Yola, M.L. A Review Study on Molecularly Imprinting Surface Plasmon Resonance Sensors for Food Analysis. Biosensors. 2024, 14, 571. [Google Scholar]
- Bonyadi, F.; Kavruk, M.; Ucak, S.; Cetin, B.; Bayramoglu, G.; Dursun, A.D.; Arica, Y.; Ozalp, V.C. Real-Time Biosensing Bacteria and Virus with Quartz Crystal Microbalance: Recent Advances, Opportunities, and Challenges. Crit. Rev. Anal. Chem. 2024, 54, 2888–2899. [Google Scholar] [CrossRef] [PubMed]
- Suther, C.; Moore, M.D. Quantification and Discovery of PCR Inhibitors Found in Food Matrices Commonly Associated with Foodborne Viruses. Food Sci. Hum. Wellness 2019, 8, 351–355. [Google Scholar] [CrossRef]
- McMahon, T.; Abdelmesih, M.; Gill, A. Evaluation of DNA Extraction Methods for the Detection of Shiga Toxin Producing Escherichia Coli in Food by Polymerase Chain Reaction. Int. J. Food Microbiol. 2023, 404, 110317. [Google Scholar] [CrossRef]
- Su, W.; Liang, D.; Tan, M. Microfluidic Strategies for Sample Separation and Rapid Detection of Food Allergens. Trends Food Sci. Technol. 2021, 110, 213–225. [Google Scholar] [CrossRef]
- Kim, J.H.; Oh, S.W. Pretreatment Methods for Nucleic Acid-Based Rapid Detection of Pathogens in Food: A Review. Food Control. 2021, 121, 107575. [Google Scholar] [CrossRef]
- Han, H.B.; Ham, S.H.; You, Y.S.; Lee, J.H.; Hahn, J.W.; Choi, Y.J. Filter-Assisted Sample Preparation for on-Site Detection Using a Bi-Functional Linker-Based Biosensor Demonstrated with Escherichia Coli O157: H7. Food Chem. 2025, 471, 142714. [Google Scholar] [CrossRef]
- Beyazit, F.; Arica, M.Y.; Acikgoz-Erkaya, I.; Ozalp, C.; Bayramoglu, G. Quartz Crystal Microbalance–Based Aptasensor Integrated with Magnetic Pre-Concentration System for Detection of Listeria Monocytogenes in Food Samples. Microchim. Acta 2024, 191, 235. [Google Scholar] [CrossRef]
- Mirsadoughi, E.; Pebdeni, A.B.; Hosseini, M. Sensitive Colorimetric Aptasensor Based on Peroxidase-like Activity of ZrPr-MOF to Detect Salmonella Typhimurium in Water and Milk. Food Control. 2023, 146, 109500. [Google Scholar] [CrossRef]
- Fernández Blanco, A.; Hernández Pérez, M.; Moreno Trigos, Y.; García-Hernández, J. Development of Optical Label-Free Biosensor Method in Detection of Listeria Monocytogenes from Food. Sensors. 2023, 23, 5570. [Google Scholar] [CrossRef]
- Bacchu, M.S.; Ali, M.R.; Das, S.; Akter, S.; Sakamoto, H.; Suye, S.I.; Rahman, M.M.; Campbell, K.; Khan, M.Z.H. A DNA Functionalized Advanced Electrochemical Biosensor for Identification of the Foodborne Pathogen Salmonella Enterica Serovar Typhi in Real Samples. Anal. Chim. Acta 2022, 1192, 339332. [Google Scholar] [CrossRef]
- Capobianco, J.A.; Armstrong, C.M.; Lee, J.; Gehring, A.G. Detection of Pathogenic Bacteria in Large Volume Food Samples Using an Enzyme-Linked Immunoelectrochemical Biosensor. Food Control. 2021, 119, 107456. [Google Scholar] [CrossRef]
- Shin, J.H.; Hong, J.S.; Go, H.Y.; Park, J.H.; Kong, M.S; Ryu, S.Y.; Kim, K.P.; Roh, E.J.; Park, J.K. Multiplexed Detection of Foodborne Pathogens from Contaminated Lettuces Using a Handheld Multistep Lateral Flow Assay Device. J. Agric. Food Chem. 2018, 66, 290–297. [Google Scholar] [CrossRef] [PubMed]
- J, Turkevich. ; Stevenson, P.C.; J, Hillier. A Study of the Nucleation and Growth Processes in the Synthesis of Colloidal Gold. Discuss. faraday Soc. 1951, 11, 55–75. [Google Scholar] [CrossRef]
- Frens, G. Controlled Nucleation for the Regulation of the Particle Size in Monodisperse Gold Suspensions. Nat. Phys. Sci. 1973, 241, 20–22. [Google Scholar] [CrossRef]
- You, Y.S.; Lim, S.W.; Hahn, J.W.; Choi, Y.J.; Gunasekaran, S. Bifunctional Linker-Based Immunosensing for Rapid and Visible Detection of Bacteria in Real Matrices. Biosens. Bioelectron. 2018, 100, 389–395. [Google Scholar] [CrossRef]
- You, Y.S.; Lim, S.W.; Gunasekaran, S. Streptavidin-Coated Au Nanoparticles Coupled with Biotinylated Antibody-Based Bifunctional Linkers as Plasmon-Enhanced Immunobiosensors. ACS Appl. Nano Mater. 2020, 3, 1900–1909. [Google Scholar] [CrossRef]
- Popa, G.L.; Papa, M.I. Salmonella Spp. Infection-a Continuous Threat Worldwide. Germs. 2021, 11, 88. [Google Scholar] [CrossRef] [PubMed]
- Darabă, A. Pathogenic Escherichia Coli: An Overview on Pre-Harvest Factors That Impact the Microbial Safety of Leafy Greens; IntechOpen, 2021; ISBN 1839698705.
- Zhu, Q.; Gooneratne, R.; Hussain, M.A. Listeria Monocytogenes in Fresh Produce: Outbreaks, Prevalence and Contamination Levels. Foods. 2017, 6, 21. [Google Scholar] [CrossRef] [PubMed]
- Shamloo, E.; Hosseini, H.; Moghadam, Z.A.; Larsen, M.H.; Haslberger, A.; Alebouyeh, M. Importance of Listeria Monocytogenes in Food Safety: A Review of Its Prevalence, Detection, and Antibiotic Resistance. Iran. J. Vet. Res. 2019, 20, 241. [Google Scholar]
- Truchado, P.; Randazzo, W. New Challenges for Detection and Control of Foodborne Pathogens: From Tools to People. Foods. 2022, 11, 1788. [Google Scholar] [CrossRef]
- Meulenberg, E.P. Phenolics: Occurrence and Immunochemical Detection in Environment and Food. Molecules. 2009, 14, 439–473. [Google Scholar] [CrossRef]
- Sajali, N.; Wong, S.C.; Hanapi, U.K.; Abu Bakar@ Jamaluddin, S.; Tasrip, N.A.; Mohd Desa, M.N. The Challenges of DNA Extraction in Different Assorted Food Matrices: A Review. J. Food Sci. 2018, 83, 2409–2414. [Google Scholar] [CrossRef]
- Salehi, F. Physico-Chemical and Rheological Properties of Fruit and Vegetable Juices as Affected by High Pressure Homogenization: A Review. Int. J. Food Prop. 2020, 23, 1136–1149. [Google Scholar] [CrossRef]
- Martínez-Padilla, L.P. Rheology of Liquid Foods under Shear Flow Conditions: Recently Used Models. J. Texture Stud. 2024, 55, e12802. [Google Scholar] [CrossRef] [PubMed]
- Mammolenti, D.; Lupi, F.R.; Baldino, N.; Gabriele, D. Technological Advancements of Insoluble Dietary Fiber from Food By-Product Processing: A Review. Foods 2025, 14, 1822. [Google Scholar] [CrossRef]
- Esan, A.; Vanholsbeeck, F.; Swift, S.; McGoverin, C.M. Continuous Separation of Bacterial Cells from Large Debris Using a Spiral Microfluidic Device. Biomicrofluidics 2023, 17. [Google Scholar] [CrossRef] [PubMed]
- U.S. Food and Drug Administration (FDA). Elemental Analysis Manual (EAM) for Food and Related Products, Section 2.2 Food Homogenization; Version 3.0. U.S. Food and Drug Administration: Silver Spring, MD, USA, 2021.
- Lu, C.; Bao, Y.; Huang, J.Y. Fouling in Membrane Filtration for Juice Processing. Curr. Opin. Food Sci. 2021, 42, 76–85. [Google Scholar] [CrossRef]
- Li, X.; Ximenes, E.; Amalaradjou, M.A.R.; Vibbert, H.B.; Foster, K.; Jones, J.; Liu, X.; Bhunia, A.K.; Ladisch, M.R. Rapid Sample Processing for Detection of Food-Borne Pathogens via Cross-Flow Microfiltration. Appl. Environ. Microbiol. 2013, 79, 7048–7054. [Google Scholar] [CrossRef]
- Guron, G.K.P.; Cassidy, J.M.; Chen, C.Y.; Paoli, G.C. Transfer of Beef Bacterial Communities onto Food-Contact Surfaces. Front. Microbiol. 2024, 15, 1450682. [Google Scholar] [CrossRef]
- Hwang, S.H.; Lee, J.H.; Nam, T.G.; Koo, M.S.; Cho, Y.S. Changes in Physicochemical Properties and Bacterial Communities in Aged Korean Native Cattle Beef during Cold Storage. Food Sci. Nutr. 2022, 10, 2590–2600. [Google Scholar] [CrossRef]
- Jogdand, N.K.; Zende, R.J.; Vaidya, V.M.; Shirke, A.H.; Gaikwad, P.S. A Qualitative and Microbial Evaluation of Emu Meat Stored under Different Temperatures. LWT 2023, 188, 115412. [Google Scholar] [CrossRef]
- Long, M.; Yu, H.; Chen, L.; Wu, G.; Zhao, S.; Deng, W.; Chen, S.; Zhou, K.; Liu, S.; He, L. Recovery of Salmonella Isolated from Eggs and the Commercial Layer Farms. Gut Pathog. 2017, 9, 74. [Google Scholar] [CrossRef] [PubMed]
- Gantois, I.; Ducatelle, R.; Pasmans, F.; Haesebrouck, F.; Gast, R.; Humphrey, T.J.; Van Immerseel, F. Mechanisms of Egg Contamination by Salmonella Enteritidis. FEMS Microbiol. Rev. 2009, 33, 718–738. [Google Scholar] [CrossRef]
- Regmi, P.; Jones, D.R.; Gast, R.K.; Guard, J.Y.; Karcher, D.M. Egg Carton and Eggshell: Is There a Possibility of Salmonella Cross-Contamination? J. Appl. Poult. Res. 2021, 30, 100185. [Google Scholar] [CrossRef]
- Lay, H.T.; Yeow, R.J.E.; Ma, Y.; Zydney, A.L.; Wang, R.; Chew, J.W. Internal Membrane Fouling by Proteins during Microfiltration. J. Memb. Sci. 2021, 637, 119589. [Google Scholar] [CrossRef]
- D’Agata, R.; Palladino, P.; Spoto, G. Streptavidin-Coated Gold Nanoparticles: Critical Role of Oligonucleotides on Stability and Fractal Aggregation. Beilstein J. Nanotechnol. 2017, 8, 1–11. [Google Scholar] [CrossRef]
- Fuller, M.; Kӧper, I. Polyelectrolyte-Coated Gold Nanoparticles: The Effect of Salt and Polyelectrolyte Concentration on Colloidal Stability. Polymers. 2018, 10, 1336. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Wang, X.; Wang, L.; Hou, X.; Liu, W.; Chen, C. Interaction of Gold Nanoparticles with Proteins and Cells. Sci. Technol. Adv. Mater. 2015, 16, 34610. [Google Scholar] [CrossRef] [PubMed]
- Fratoddi, I. Hydrophobic and Hydrophilic Au and Ag Nanoparticles. Breakthroughs and Perspectives. Nanomaterials. 2017, 8, 11. [Google Scholar] [CrossRef]
- El Badawy, A.M.; Luxton, T.P.; Silva, R.G.; Scheckel, K.G.; Suidan, M.T.; Tolaymat, T.M. Impact of Environmental Conditions (PH, Ionic Strength, and Electrolyte Type) on the Surface Charge and Aggregation of Silver Nanoparticles Suspensions. Environ. Sci. Technol. 2010, 44, 1260–1266. [Google Scholar] [CrossRef] [PubMed]
- Masson, J.F. Consideration of Sample Matrix Effects and “Biological” Noise in Optimizing the Limit of Detection of Biosensors. ACS sensors 2020, 5, 3290–3292. [Google Scholar] [CrossRef]
- Kim, E.H.; Hahn, J.W.; You, Y.S.; Choi, Y.J. Design and Optimization of a Colorimetric Biosensor Using Functionalized Gold Nanoparticles and Bi-Functional Linker for Agriculture and Food Industry. Microchem. J. 2024, 206, 111600. [Google Scholar] [CrossRef]
| Target | Sample1) | Concentration of bacteria (CFU/total) | Concentration of bacteria (CFU/mL) |
||
| Resuspended second filter |
Control | Spiked sample | Resuspended second filter |
||
| Escherichia coli O157:H7 | Romaine lettuce | 163.7±8.2 | ND | 4.6±0.5 | 81.8±4.1 |
| Cabbage | 208.3±37.5 | ND | 5.6±1.2 | 104.1±18.8 | |
| Cucumber | 205.0±16.4 | ND | 6.6±1.2 | 102.5±8.2 | |
| Carrot | 181.0±19.1 | ND | 5.3±0.9 | 90.5±9.6 | |
| Salmonella Typhimurium | Chicken | 47.3 ± 8.22) | ND3) | 11.0±3.0 | 23.7±4.1 |
| Pork | 53.6±21.7 | ND | 12.0±3.3 | 26.8±10.8 | |
| Beef | 31.7±7.4 | ND | 11.0±2.2 | 15.8±3.7 | |
| Egg shell | N/A | ND | 6.6±0.9 | N/A | |
|
Listeria monocytogenes |
Melon | 122.7±38.0 | ND | 8.7±2.6 | 61.3±19.0 |
| Cheese brine | 132.7±11.0 | ND | 7.0±1.6 | 66.3±5.5 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





