Submitted:
08 August 2024
Posted:
09 August 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
Materials and Methods
2.1. Plant Materials, Temperature and Relative Humidity
2.2. Experimental Design and Water Treatment
2.3. Crop Management
2.4. Measurements and Data Collection
2.5. Assessment of Phenotypic Variation to A. Flavus Colonization
2.6. Seed Coat Total Polyphenol Extraction and Quantification
2.7. Genotypes Aflatoxin Content Quantification
2.8. Analysis
Results
3.1. Yields and Yield Components
| Sources | PNP | IMPN | SNP | HY (gm-2) | PY (gm-2) | SY (gm-2) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| of variance | WW | WS | WW | WS | WW | WS | WW | WS | WW | WS | WW | WS |
| F value | 31.62 | 31.75 | 12 | 3.25 | 14.23 | 12 | 8.25 | 4.24 | 14 | 12 | 7.24 | 9.16 |
| F (prob) | <.001 | <.001 | <.001 | <.001 | <.001 | <.001 | <.001 | <.001 | <.001 | <.001 | <.001 | <.001 |
| WTrt | <.001 | <.001 | <.001 | <.001 | <.001 | <.001 | <.001 | <.001 | <.001 | <.001 | <.001 | <.001 |
| G×(Wtrt) | <.001 | <.001 | <.001 | 0.015 | 0.027 | 0.020 | ||||||
| Traits | Water Traitements | WS negative effect (%) | |
|---|---|---|---|
| WW | WS | ||
| Pods number per plant | 102±31 | 80±25 | 21.92 |
| Immature pods per plant (increasing) | 9±5 | 21±10 | 57.14 |
| Seeds number per plant | 123±31 | 89±27 | 27.36 |
| Pod Yield (gm-2) | 678.54±203.51 | 546.15±163.5 | 19.43 |
| Seed Yield (gm-2) | 316.67±90.26 | 230.37±69.3 | 27.24 |
| Haulm Yield (gm-2) | 828.53±85.30 | 645.57±150.13 | 22.07 |
| Well-Watered | Water Stressed | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotypes | PNP | SNP | IMPN | PY (gm-2) | SY (gm-2) | HY (gm-2) | PNP | SNP | IMPN | PY (gm-2) | SY (gm-2) | HY (gm-2) |
| 55-437 | 103±2 | 124±17 | 8±2 | 594.7±96 | 290.6±49 | 736.4±292 | 95±11 | 109±14 | 15±1 | 517±51 | 248.2±46 | 612.8±220 |
| Fleur 11 | 99±12 | 136±13 | 9±1 | 613.5±81 | 338.6±79 | 870.1±75 | 89±5 | 119±16 | 17±2 | 572±138 | 316.2±33 | 658±160 |
| ICG 10950 | 98±5 | 112±19 | 4±1 | 701.6±61 | 376.8±70 | 1022.4±154 | 79±8 | 70±14 | 16±2 | 638.8±30 | 284.3±10 | 740.3±97 |
| ICG 11322 | 76±22 | 72±12 | 4±1 | 567.8±33 | 272.8±39 | 691.6±118 | 62±14 | 65±9 | 26±2 | 529±49 | 242.3±31 | 583.9±127 |
| ICG 1142 | 112±6 | 112±8 | 12±2 | 1664.9±255 | 369.4±78 | 798.6±69 | 84±10 | 78±4 | 23±2 | 1247.9±283 | 328±82 | 666.6±151 |
| ICG 12235 | 207±16 | 116±18 | 34±3 | 766.6±154 | 188.5±32 | 930.1±82 | 141±6 | 56±13 | 76±3 | 677±128 | 183.4±17 | 756.6±148 |
| ICG 12697 | 112±7 | 137±5 | 9±2 | 560.6±76 | 395.4±70 | 637.2±245 | 75±6 | 100±9 | 17±3 | 363.4±50 | 245.3±19 | 513.6±85 |
| ICG 12879 | 101±2 | 129±11 | 8±2 | 472.2±68 | 306.9±27 | 584.7±184 | 86±6 | 64±20 | 16±2 | 351.7±33 | 211.7±48 | 536.9±58 |
| ICG 12988 | 101±17 | 131±21 | 10±2 | 649.7±148 | 280.9±92 | 562.2±80 | 76±13 | 102±11 | 17±2 | 388±38 | 206.5±35 | 533.6±86 |
| ICG 12991 | 115±8 | 153±21 | 4±1 | 525.3±42 | 243.8±36 | 657.5±203 | 79±10 | 86±4 | 17±3 | 443.9±33 | 199.5±25 | 629.5±169 |
| ICG 13099 | 111±3 | 133±9 | 8±1 | 889.1±235 | 370.8±120 | 925.1±86 | 88±10 | 85±20 | 21±2 | 653.8±98 | 286.3±51 | 741.1±133 |
| ICG 13603 | 177±12 | 205±26 | 10±1 | 780.9±83 | 356.2±111 | 775.4±58 | 134±9 | 163±20 | 25±1 | 662.5±131 | 256.6±17 | 589.9±102 |
| ICG 13858 | 76±8 | 112±14 | 8±3 | 475.7±80 | 196.0±57 | 910.5±48 | 48±4 | 67±14 | 21±4 | 397.5±20 | 134.8±32 | 552.6±159 |
| ICG 14106 | 92±8 | 142±17 | 8±2 | 833.5±47 | 321.4±35 | 724±162 | 53±8 | 74±13 | 19±2 | 776.9±145 | 252.1±50 | 513.4±50 |
| ICG 1415 | 100±5 | 98±11 | 13±2 | 655.3±78 | 282.7±28 | 767.3±76 | 86±10 | 71±12 | 22±5 | 624.3±67 | 209.6±34 | 589.7±135 |
| ICG 14523 | 159±13 | 174±12 | 10±2 | 906.0±154 | 378.6±31 | 954.3±58 | 121±12 | 111±3 | 21±3 | 746.5±27 | 298.8±25 | 780.2±125 |
| ICG 14630 | 53±7 | 84±10 | 9±2 | 725.4±107 | 206.9±61 | 893.5±224 | 50±4 | 67±8 | 15±3 | 654.8±68 | 150.6±12 | 636.2±262 |
| ICG 1519 | 86±5 | 79±15 | 8±2 | 590.5±96 | 214±55 | 761.5±185 | 63±11 | 70±16 | 20±3 | 485.6±16 | 180.9±36 | 695.4±208 |
| ICG 156 | 102±14 | 125±11 | 9±1 | 682±84 | 237.9±24 | 1602±468 | 84±2 | 83±9 | 21±3 | 432.6±58 | 193.2±38 | 1074.3±406 |
| ICG 163 | 104±7 | 113±10 | 10±3 | 733.9±204 | 433±83 | 1056.4±194 | 91±2 | 83±9 | 22±3 | 561.6±73 | 338±93 | 769.4±233 |
| ICG 2019 | 69±7 | 105±16 | 11±3 | 510.7±145 | 289.6±82 | 615.1±108 | 37±25 | 49±24 | 20±4 | 384.1±85 | 197.1±73 | 474.3±274 |
| ICG 2106 | 108±4 | 136±11 | 10±3 | 612.9±132 | 329.7±20 | 853.5±83 | 75±10 | 88±18 | 18±3 | 463.9±40 | 239.7±31 | 586.8±318 |
| ICG 3027 | 98±11 | 130±9 | 11±3 | 752.9±50 | 381.7±41 | 892.4±104 | 62±2 | 82±19 | 15±2 | 613.9±81 | 304.3±49 | 718.2±151 |
| ICG 311 | 106±9 | 135±10 | 7±2 | 638.5±106 | 393.6±71 | 676.2±124 | 86±8 | 101±11 | 14±1 | 562.8±102 | 269±20 | 543.1±54 |
| ICG 332 | 63±1 | 83±8 | 5±2 | 658.8±26 | 283.4±34 | 985±60 | 39±5 | 47±11 | 18±2 | 457.5±44 | 162.1±37 | 726.5±144 |
| ICG 334 | 105±9 | 159±20 | 8±1 | 884.7±22 | 444.3±29 | 813.5±172 | 98±7 | 143±15 | 24±2 | 692.3±89 | 286.5±32 | 553.5±138 |
| ICG 36 | 100±13 | 112±15 | 7±2 | 778.8±64 | 222.2±35 | 737.3±111 | 53±7 | 67±11 | 27±3 | 528.2±83 | 148.6±13 | 463.2±110 |
| ICG 3992 | 73±11 | 77±20 | 10±2 | 654.1±130 | 317.7±86 | 958.8±107 | 47±4 | 55±9 | 23±2 | 590.9±73 | 224.1±8 | 831.2±86 |
| ICG 4543 | 89±11 | 126±14 | 4±5 | 485.4±56 | 227.7±19 | 785.4±122 | 73±6 | 87±18 | 18±1 | 385.3±67 | 183.4±52 | 578±141 |
| ICG 4598 | 98±10 | 101±16 | 7±1 | 701.8±143 | 356.4±86 | 1050.8±62 | 76±7 | 89±18 | 19±1 | 497.8±41 | 197±9 | 803±66 |
| ICG 4684 | 87±8 | 121±25 | 10±1 | 633.8±95 | 347.9±41 | 765.3±97 | 71±5 | 98±18 | 19±2 | 445.7±53 | 246.8±49 | 652.1±87 |
| ICG 4729 | 80±2 | 94±6 | 10±2 | 393.3±64 | 229±17 | 711.7±115 | 66±7 | 80±13 | 23±1 | 329.7±47 | 155.4±28 | 530.2±91 |
| ICG 4750 | 89±4 | 116±5 | 9±1 | 580.1±27 | 338±28 | 764.5±86 | 75±9 | 85±10 | 27±4 | 439.8±46 | 186.7±32 | 635±109 |
| ICG 4764 | 84±7 | 125±7 | 8±2 | 727.3±157 | 369.3±68 | 839.8±200 | 79±3 | 108±7 | 25±3 | 549.6±104 | 231.5±24 | 643.7±165 |
| ICG 513 | 100±14 | 138±24 | 12±4 | 861.2±101 | 471.9±98 | 815.3±98 | 85±6 | 110±22 | 29±2 | 688.8±65 | 306±12 | 696.7±150 |
| ICG 5195 | 134±11 | 174±12 | 11±2 | 547.1±21 | 302.3±29 | 850.6±27 | 98±8 | 100±6 | 25±2 | 536±51 | 264.4±82 | 708.2±185 |
| ICG 532 | 90±3 | 114±7 | 10±1 | 729.9±123 | 298.2±55 | 824.7±145 | 71±8 | 104±2 | 18±2 | 532.4±96 | 213.1±60 | 677.5±110 |
| ICG 5609 | 60±6 | 98±21 | 6±2 | 524.9±81 | 278.9±14 | 750.7±171 | 49±10 | 58±9 | 15±3 | 411.5±34 | 146.8±17 | 556.8±147 |
| ICG 5663 | 144±9 | 135±6 | 17±2 | 667.1±78 | 313.9±64 | 735.7±2116 | 91±13 | 85±8 | 26±3 | 541.8±59 | 212.8±51 | 569.4±64 |
| ICG 6263 | 76±11 | 89±14 | 8±2 | 550.2±41 | 152.1±43 | 646.5±131 | 53±7 | 64±15 | 21±3 | 486.8±44 | 86.1±11 | 402±117 |
| ICG 6407 | 62±4 | 79±7 | 7±3 | 522.4±52 | 290.7±17 | 833.9±96 | 52±3 | 67±20 | 19±2 | 500.9±45 | 236.1±34 | 741±110 |
| ICG 6654 | 89±13 | 138±8 | 9±2 | 687.6±94 | 342.7±71 | 697.3±121 | 70±6 | 99±6 | 18±2 | 630.1±77 | 265.3±33 | 596±125 |
| ICG 6703 | 92±7 | 116±16 | 7±2 | 546.3±50 | 306±44 | 932.7±104 | 79±5 | 100±16 | 12±2 | 377.6±40 | 213±23 | 517.2±111 |
| ICG 6813 | 138±9 | 172±14 | 11±2 | 732.6±68 | 377.9±18 | 744.1±429 | 125±14 | 116±3 | 25±2 | 634.7±56 | 343.3±9 | 751.8±98 |
| ICG 6888 | 66±9 | 80±2 | 6±2 | 540.8±64 | 259.9±81 | 830.6±193 | 53±5 | 67±5 | 36±2 | 417.5±26 | 154.4±22 | 657±148 |
| ICG 7181 | 69±5 | 91±15 | 10±2 | 567.4±51 | 310.9±51 | 908.5±89 | 62±10 | 65±5 | 22±3 | 470.5±77 | 220.3±40 | 757±105 |
| ICG 721 | 106±8 | 132±21 | 11±2 | 804.5±75 | 409.4±33 | 1064.5±83 | 96±10 | 123±19 | 15±2 | 638.1±44 | 321.2±46 | 954.4±86 |
| ICG 76 | 149±22 | 162±10 | 9±2 | 933.6±51 | 467.7±69 | 807.4±106 | 132±9 | 131±18 | 19±3 | 727.2±72 | 342.7±41 | 618±155 |
| ICGIL 11102 | 141±31 | 159±29 | 8±2 | 603.7±62 | 282.9±62 | 974.6±139 | 119±8 | 127±21 | 23±2 | 568.6±56 | 248.3±62 | 761.1±177 |
| ICGIL 11110 | 132±13 | 151±8 | 12±2 | 833±102 | 418.1±55 | 848.4±120 | 89±9 | 103±18 | 25±2 | 578.8±132 | 270.8±29 | 567.8±224 |
| ICGIL 11114 | 70±9 | 109±22 | 5±1 | 712±110 | 423.5±85 | 825.2±158 | 65±5 | 91±19 | 21±3 | 442.2±135 | 264.8±32 | 627.4±182 |
| ICGIL 11125 | 121±4 | 132±12 | 11±1 | 724.4±119 | 398.2±56 | 728.5±116 | 105±10 | 108±5 | 19±2 | 674.3±194 | 207.8±61 | 594.2±184 |
| ICGIL 17108 | 135±7 | 131±5 | 15±1 | 765.7±98 | 180.7±35 | 1022.2±111 | 115±6 | 89±9 | 21±2 | 582.3±52 | 104.3±23 | 808±181 |
| J 11 | 116±8 | 127±9 | 8±1 | 497.4±39 | 259.1±39 | 697.3±61 | 79±8 | 87±8 | 17±2 | 443.7±70 | 205.3±13 | 493.5±183 |
| JL24 | 105±9 | 140±19 | 13±2 | 565.8±69 | 280.1±37 | 720.6±33 | 88±3 | 126±26 | 28±2 | 488.5±30 | 245±52 | 539.2±214 |
| Means | 102±31 | 123±31 | 9±4 | 678.5±204 | 316.7±90 | 828.5±279 | 80±25 | 89±27 | 22±8 | 546.2±164 | 230.4±69 | 645.6±215 |
| LSD | 15 | 21 | 5 | 131 | 79 | 145 | 12 | 20 | 9 | 107 | 56 | 140 |
3.2. Intermittent Water Deficit (WS) Effect on Pre-Harvest Aflatoxin Contamination (AC)
| Aflatoxin B1 | ||
|---|---|---|
| Genotypes | WW | WS |
| Means | 1.81 | 5.51 |
| LSD | 1.14 | 1.49 |
| F value | 9.55 | 56 |
| F (Prob) | <.001 | <.001 |
| G×W(Trt) | <.001 | |
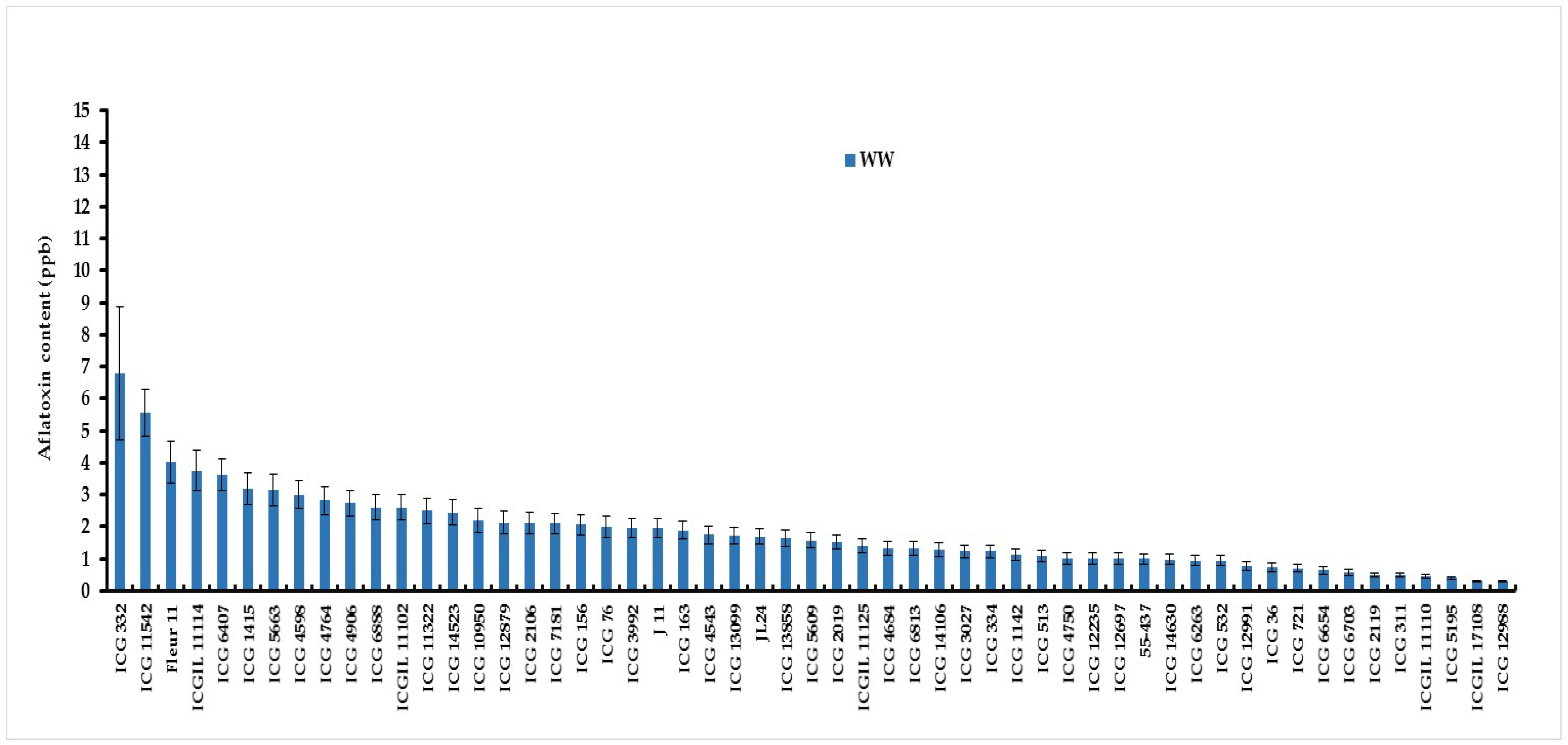
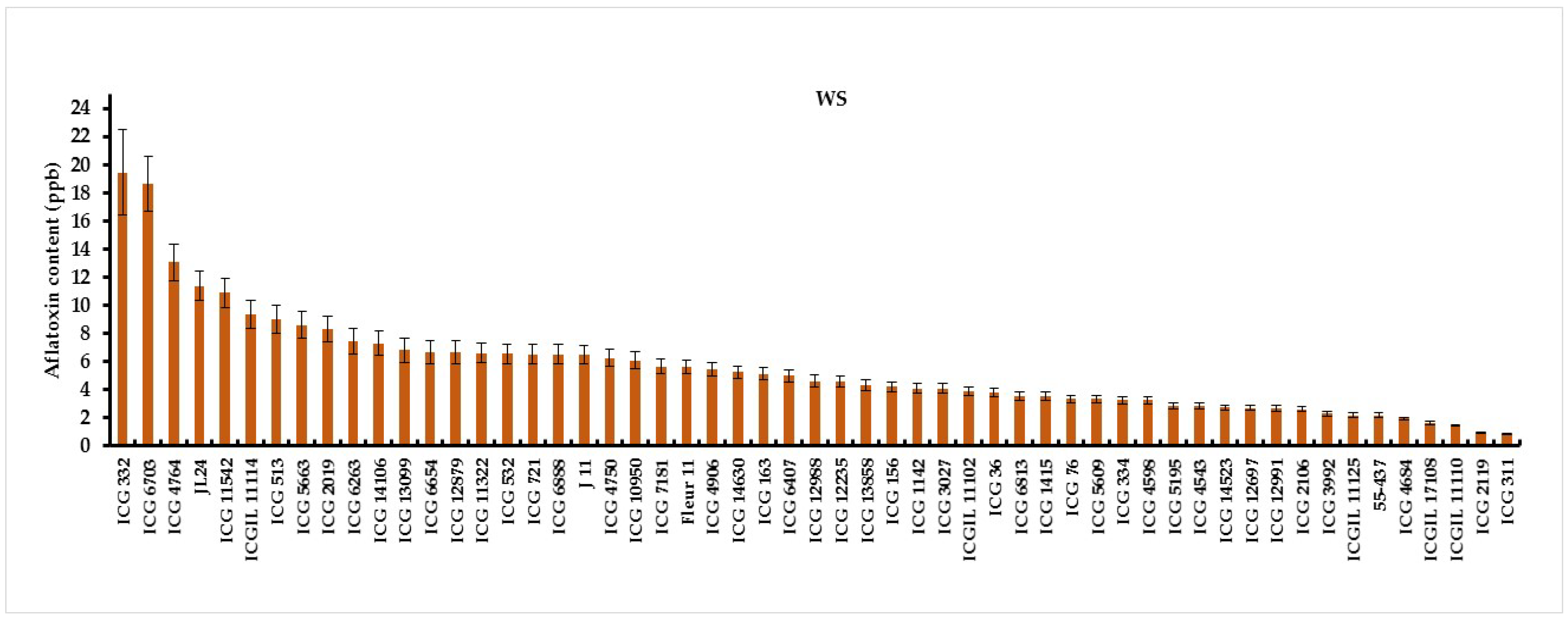
3.3. Seeds Colonisation and Total Polyphenol Content of Seed Coat
| Genotypes | %TPP | % AFC |
|---|---|---|
| Means | 50.50 | 85 |
| LSD | 2.54 | 16.63 |
| F value | 19.38 | 4.12 |
| F (Prob) | <.001 | <.001 |
| Genotypes | % AFC | %TPP | Genotypes | % AFC | %TPP | Genotypes | % AFC | %TPP | |
| CI ≤25% | ICG 311 | 19 | 53.08 | 55-437 | 21 | 49 | ICG 12988 | 18 | 52.1 |
| CI˃25%≤50% | ICG 11322 | 50 | 50.43 | ICG 14523 | 43 | 51.61 | ICG 14523 | 43 | 51.61 |
| ICG 11542 | 28 | 43.90 | ICG 163 | 45 | 47.38 | ICG 3027 | 83 | 57.50 | |
| ICG 4543 | 28 | 47.55 | ICG 76 | 42 | 52.57 | J 11 | 40 | 55.16 | |
| CI ˃50% | Fleur 11 | 86 | 47.90 | ICG 10950 | 91 | 56.05 | ICG 1142 | 100 | 50.32 |
| ICG 12235 | 85 | 56.67 | ICG 12697 | 91 | 51.47 | ICG 12879 | 100 | 45.50 | |
| ICG 12991 | 81 | 48.75 | ICG 13099 | 66 | 50.35 | ICG 4906 | 83 | 46.86 | |
| ICG 13858 | 91 | 53.27 | ICG 14106 | 100 | 48.76 | ICG 1415 | 100 | 43.68 | |
| ICG 14630 | 81 | 56.95 | ICG 14630 | 81 | 56.95 | ICG 156 | 90 | 51.86 | |
| ICG 2019 | 100 | 52.34 | ICG 2106 | 83 | 48.63 | ICG 332 | 100 | 53.22 | |
| ICG 334 | 66 | 50.76 | ICG 36 | 66 | 49.14 | ICG 3992 | 83 | 53.17 | |
| ICG 4598 | 100 | 48.16 | ICG 4684 | 75 | 45.97 | ICG 2119 | 100 | 52.41 | |
| ICG 4750 | 65 | 47.38 | ICG 4764 | 70 | 50.23 | ICG 513 | 100 | 53.69 | |
| ICG 5195 | 100 | 52.36 | ICG 532 | 100 | 42.74 | ICG 5609 | 83 | 43.74 | |
| ICG 5663 | 81 | 49.36 | ICG 6263 | 91 | 54.01 | ICG 6407 | 61 | 52.14 | |
| ICG 6654 | 91 | 48.31 | ICG 6703 | 91 | 46.32 | ICG 6813 | 46 | 46.53 | |
| ICG 6888 | 83 | 46.09 | ICG 7181 | 100 | 45.92 | ICG 721 | 91 | 50.20 | |
| ICGIL 11102 | 100 | 53.09 | ICGIL 11110 | 75 | 51.29 | ICGIL 11114 | 75 | 54.34 | |
| ICGIL 11125 | 61 | 56.30 | ICGIL 17108 | 91 | 62.96 | JL24 | 100 | 47.93 | |
3.4. Relationship between Aflatoxin Contaminations with Total Polyphenol, Pod Yield, Seed Yield, and Haulm Yield
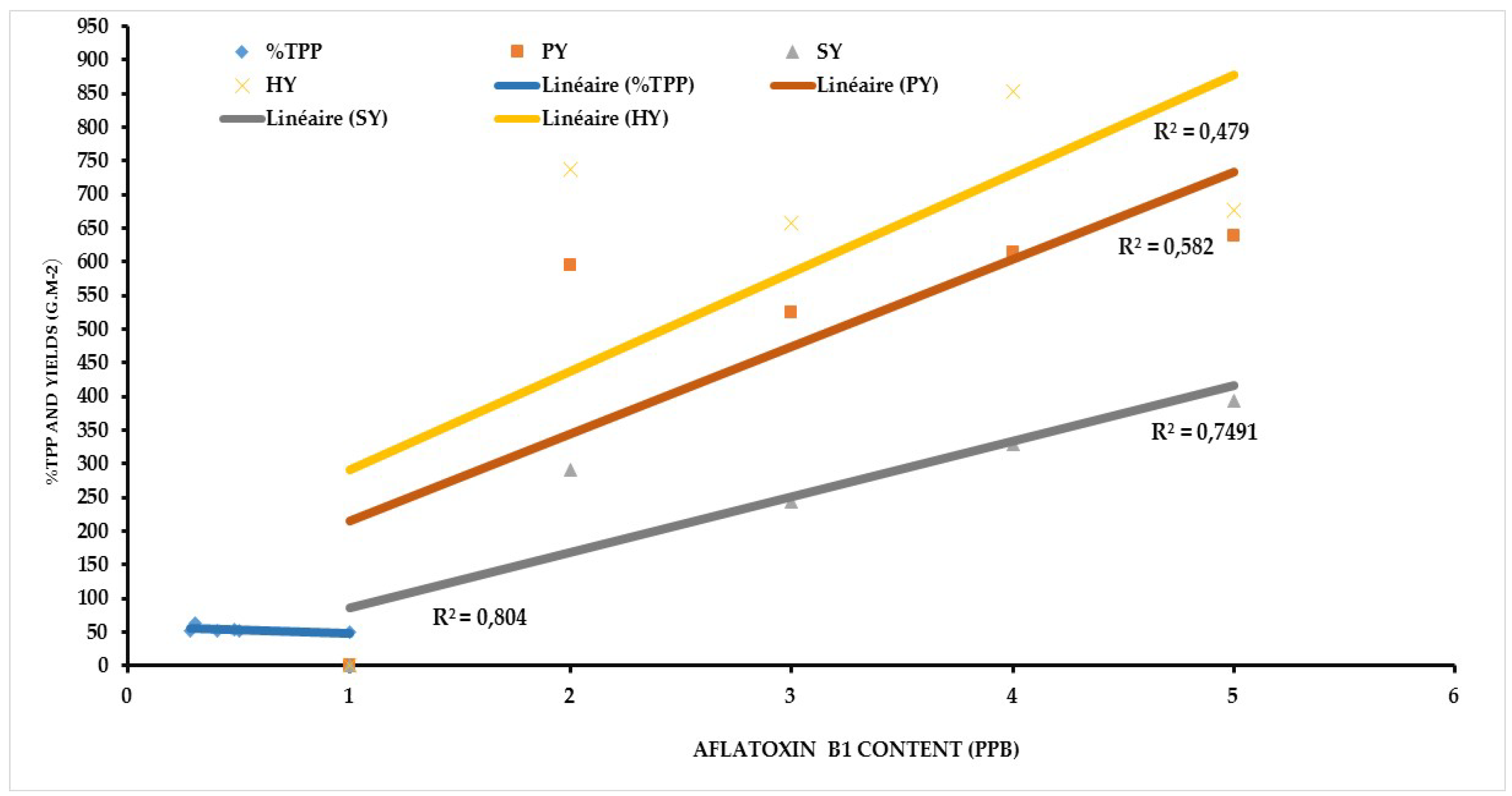
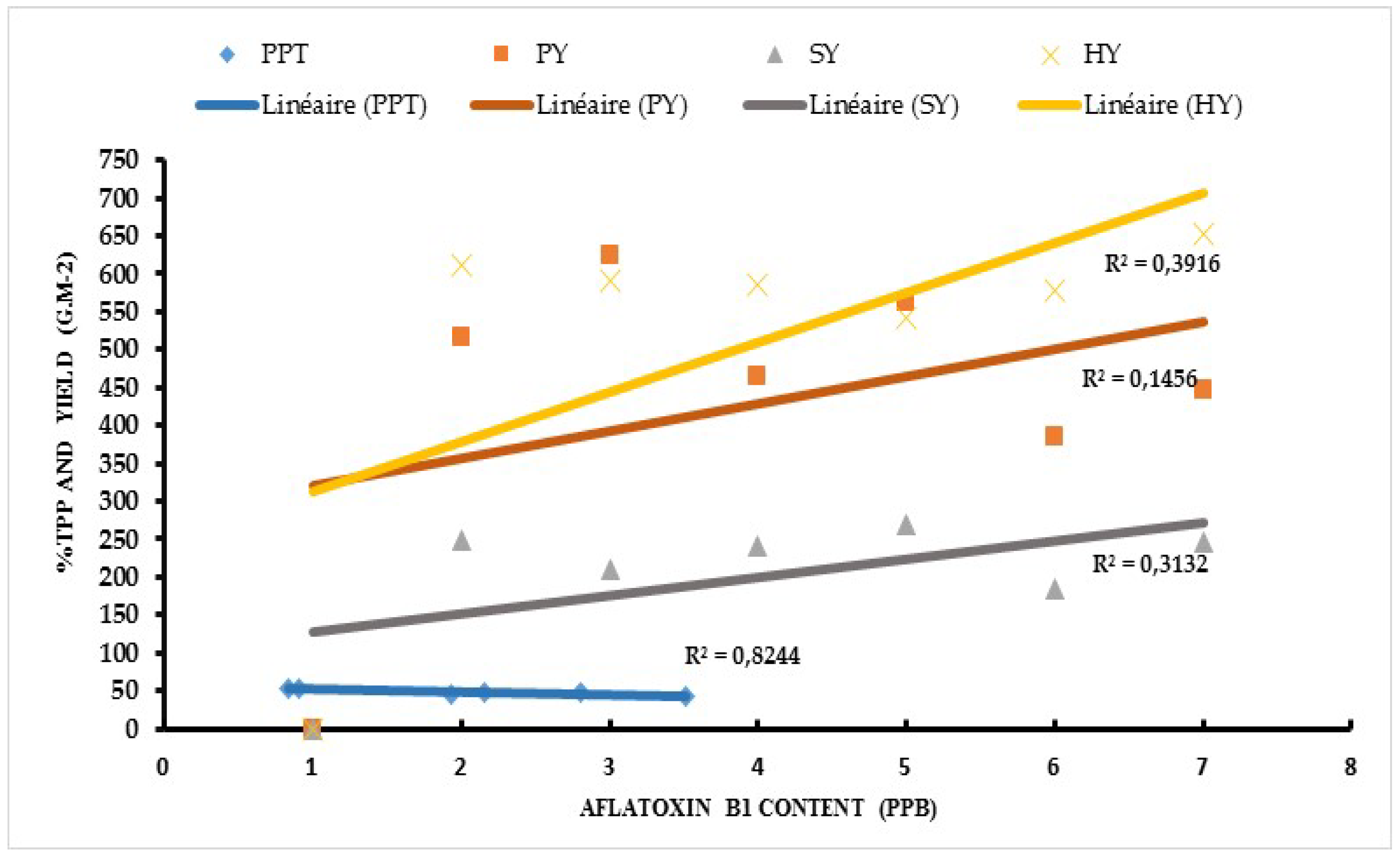
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Underwood C, Taylor H, Hoveland C. Soil Physical Factors Affecting Peanut Pod Development Agronomy Journal. 1971;63(6):953-4. [CrossRef]
- Horn B. Colonization of wounded peanut seeds by soil fungi: Selectivity for species from Aspergillus section Flavi. Mycologia. . 97 202-17 103852/mycologia971202. (2005). [CrossRef]
- Arunyanark A, Pimratch S, Jogloy S, Wongkaew S, Vorasoot N, Akkasaeng C, et al. Association between aflatoxin contamination and N2 fixationin peanut under drought conditions. 2012.
- Ndunguru B, Ntare B, Williams J, Greenberg D. Assessment of groundnut cultivars for end-of-season drought tolerance in a Sahelian environment. The Journal of Agricultural Science. 1995;125(1):79-85. [CrossRef]
- Hamidou F, Halilou O, Vadez V. Assessment of groundnut under combined heat and drought stress. Journal of Agronomy and Crop Science. 2013;199(1):1-11. [CrossRef]
- Waliyar F, Traore A, Fatondji D, Ntare B. Effect of irrigation interval, planting date, and cultivar on Aspergillus flavus and aflatoxin contamination of peanut in a sandy soil of Niger. Peanut Science. 2003;30(2):79-84. [CrossRef]
- Craufurd P, Prasad P, Waliyar F, Taheri A. Drought, pod yield, pre-harvest Aspergillus infection and aflatoxin contamination on peanut in Niger. Field Crops Research. 2006;98(1):20-9. [CrossRef]
- Dorner JW, Cole RJ, Sanders TH, Blankenship PD. Interrelationship of kernel water activity, soil temperature, maturity, and phytoalexin production in preharvest aflatoxin contamination of drought-stressed peanuts. Mycopathologia. 1989;105(2):117-28. [CrossRef]
- Girdthai T, Jogloy S, Vorasoot N, Akkasaeng C, Wongkaew S, Holbrook CC, et al. Heritability of, and genotypic correlations between, aflatoxin traits and physiological traits for drought tolerance under end of season drought in peanut (Arachis hypogaea L.). Field Crops Research. 2010;118(2):169-76. [CrossRef]
- Waliyar F, Hassan H, Bonkoungou S. Sources of resistance to Aspergillus flavus and aflatoxin contamination in groundnut genotypes in West Africa. Plant Disease. 1994;78(7):704-8. [CrossRef]
- Keller NP, Butchko RA, Sarr B, Phillips TD. A visual pattern of mycotoxin production in maize kernels by Aspergillus spp. Phytopathology. 1994;84(5):483-8.
- Upadhyaya H, Nigam S, Mehan V, Lenne J. Aflatoxin contamination of groundnut: prospects for a genetic solution through conventional breeding. 1997.
- SOUZA FH, Marcos-Filho J. The seed coat as a modulator of seed-environment relationships in Fabaceae. Brazilian Journal of Botany. 2001;24:365-75. [CrossRef]
- Pandey MK, Kumar R, Pandey AK, Soni P, Gangurde SS, Sudini HK, et al. Mitigating aflatoxin contamination in groundnut through a combination of genetic resistance and post-harvest management practices. Toxins. 2019;11(6):315. [CrossRef]
- Arunyanark A, Jogloy S, Vorasoot N, Akkasaeng C, Kesmala T, Patanothai A. Stability of Relationship Between Chlorophyll Density and Soil Plant Analysis Development Chlorophyll Meter Readings in Peanut Across Different Drought Stress Conditions. Asian Journal of Plant Sciences. 2009;8(2):102-10. [CrossRef]
- Dieme RMA, Faye I, Zoclanclounon YAB, Fonceka D, Ndoye O, Diedhiou PM. Identification of Sources of Resistance for Peanut Aspergillus flavus Colonization and Aflatoxin Contamination. International Journal of Agronomy. 2018;2018:1-7. [CrossRef]
- Jenipher Bisikwa FO. Tolerance Levels of Peanut Varieties against Aspergillus flavus Infection. Journal of Plant Pathology & Microbiology. 2013;04(08). [CrossRef]
- Mendu L, Cobos CJ, Tengey TK, Commey L, Balasubramanian VK, Williams LD, et al. Seed coat mediated resistance against Aspergillus flavus infection in peanut. Plant Gene. 2022;32:100381. [CrossRef]
- Commey L, Tengey TK, Cobos CJ, Dampanaboina L, Dhillon KK, Pandey MK, et al. Peanut Seed Coat Acts as a Physical and Biochemical Barrier against Aspergillus flavus Infection. Journal of Fungi. 2021;7(12):1000. [CrossRef]
- Mixon A. Reducing Aspergillus species infection of peanut seed using resistant genotypes. Wiley Online Library, 1986 0047-2425. [CrossRef]
- Waliyar F, Kumar K, Diallo M, Traore A, Mangala U, Upadhyaya H, et al. Resistance to pre-harvest aflatoxin contamination in ICRISAT’s groundnut mini core collection. European Journal of Plant Pathology. 2016;145(4):901-13. [CrossRef]
- Waliyar F, Bockelee-Morvan A. Resistance of groundnut varieties to Aspergillus flavus in Senegal. 1989.
- Thakur R, Rao V, Reddy S, Ferguson M. Evaluation of wild Arachis germplasm accessions for in vitro seed colonization and anatoxin production by Aspergillus flavus. International Arachis Newsletter. 2000;20:44-6.
- Li Y, Kong W, Li M, Liu H, Zhao X, Yang S, et al. Litsea cubeba essential oil as the potential natural fumigant: Inhibition of Aspergillus flavus and AFB1 production in licorice. Industrial Crops and Products. 2016;80:186-93. [CrossRef]
- Singleton VL, Rossi JA. Colorimetry of total phenolics with phosphomolybdic-phosphotungstic acid reagents. American journal of Enology and Viticulture. 1965;16(3):144-58. [CrossRef]
- Reddy S, Kiran Mayi D, Uma Reddy M, Thirumala-Devi K, Reddy D. Aflatoxins B1 in different grades of chillies (Capsicum annum L.) in India as determined by indirect competitive-ELISA. Food additives & contaminants. 2001;18(6):553-8. [CrossRef]
- Kalariya KA, Singh AL, Goswami N, Mehta D, Mahatma MK, Ajay B, et al. Photosynthetic characteristics of peanut genotypes under excess and deficit irrigation during summer. Physiology and Molecular Biology of Plants. 2015;21(3):317-27. [CrossRef]
- Boontang S, Girdthai T, Jogloy S, Akkasaeng C, Vorasoot N, Patanothai A, et al. Responses of released cultivars of peanut to terminal drought for traits related to drought tolerance. Asian Journal of Plant Sciences. 2010;9(7):423-31. [CrossRef]
- Songsri P, Jogloy S, Vorasoot N, Akkasaeng C, Patanothai A, Holbrook C. Root distribution of drought-resistant peanut genotypes in response to drought. Journal of Agronomy and Crop Science. 2008;194(2):92-103. [CrossRef]
- Sudhakar P, Latha P, Babitha M, Reddy P, Naidu P. Relationship of drought tolerance traits with aflatoxin contamination in groundnut. Indian Journal of Plant Physiology. 2007;12(3):261.
- Holbrook C, Kvien C, Rucker K, Wilson D, Hook J, Matheron M. Preharvest aflatoxin contamination in drought-tolerant and drought-intolerant peanut genotypes. Peanut Science. 2000;27(2):45-8. [CrossRef]
- Arunyanark A, Jogloy S, Wongkaew S, Akkasaeng C, Vorasoot N, Kesmala T, et al. Heritability of aflatoxin resistance traits and correlation with drought tolerance traits in peanut. Field Crops Research. 2010;117(2-3):258-64. [CrossRef]
- Amos M, Sy AT, Blaise K, Sondé ALMK. Assessment of sixteen varieties of groundnut in two agro ecological zones in Burkina Faso for yield and tolerance to aflatoxin. African Journal of Agricultural Research. 2021;17(1):66-78. [CrossRef]
- Okello DK, Kaaya AN, Bisikwa J, Were M, Oloka HK. Management of aflatoxins in groundnuts: A manual for farmers, processors, traders and consumers in Uganda. National Agricultural Research Organization; 2010.
- Hamidou F, Rathore A, Waliyar F, Vadez V. Although drought intensity increases aflatoxin contamination, drought tolerance does not lead to less aflatoxin contamination. Field crops research. 2014;156:103-10. [CrossRef]
- Wilson JS, Otsuki T. Global trade and food safety: winners and losers in a fragmented system: World Bank Publications; 2001.
- Holbrook CC, Stalker HT, Janick J. Peanut breeding and genetic resources. Plant breeding reviews. 2002;22:297-356. [CrossRef]
- LaPrade J, Bartz J, Norden A, Demuynk T. Correlation of peanut seed-coat surface wax accumulations with tolerance to colonization by Aspergillus flavus. J Am Peanut Res Educ Soc. 1973;5:89-94.
- Liang X, Zhou G, Pan R. Study on the relationship of wax and cutin layers in peanut seeds and resistance to invasion and aflatoxin production by Aspergillus flavus. J Trop Subtrop Bot. 2003;11:11-4.
- Dieme RMA, Faye I, Zoclanclounon YAB, Fonceka D, Ndoye O, Diedhiou PM. Identification of Sources of Resistance for Peanut Aspergillus flavus Colonization and Aflatoxin Contamination. International Journal of Agronomy. 2018. [CrossRef]
- Lindsey D, Turner RB. Inhibition of growth of Aspergillus flavus and Trichoderma viride by peanut embryos. Mycopathologia. 1975;55(3):149-52. [CrossRef]
- Mehan V. Screening groundnuts for resistance to seed invasion by Aspergillus flavus and to aflatoxin production. 1989.
- Kasno A, Trustinah T, Purnomo J, Sumartini S. Seed Coat Resistance of Groudnut to Aspergillus Flavus and Their Stability Performance in The Field. AGRIVITA, Journal of Agricultural Science. 2011;33(1):53-62. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




