Submitted:
02 August 2024
Posted:
02 August 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
3. Results
3.1. De Novo Assembly Overview
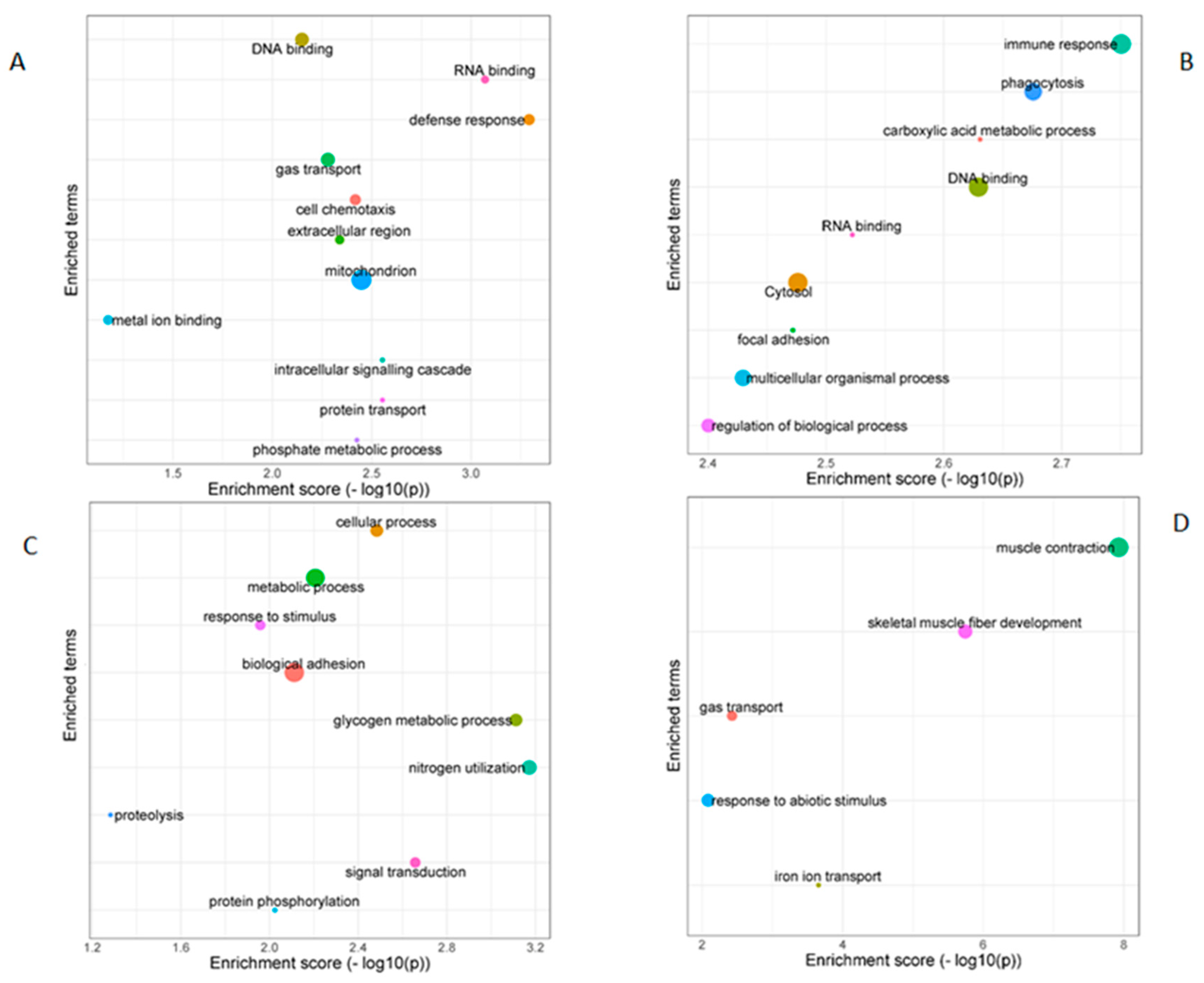
3.2. Identification and Characterization of lncRNAs in Blue Sharks
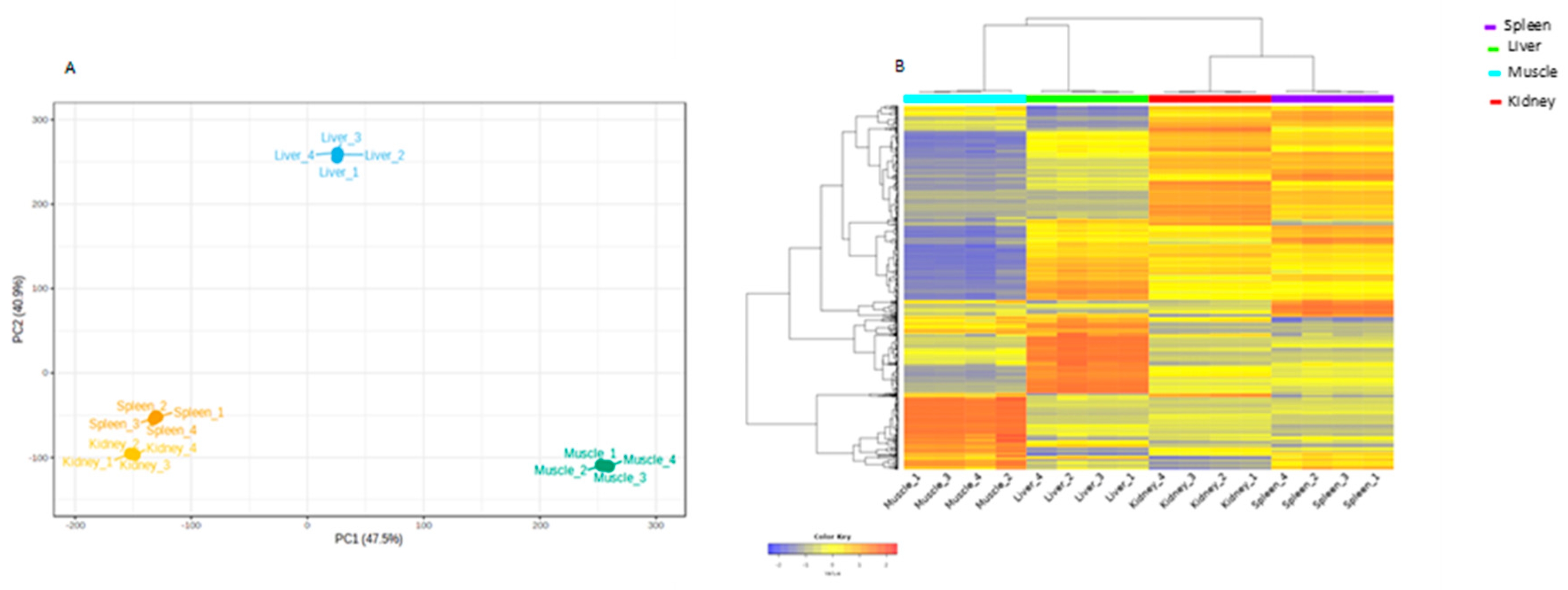
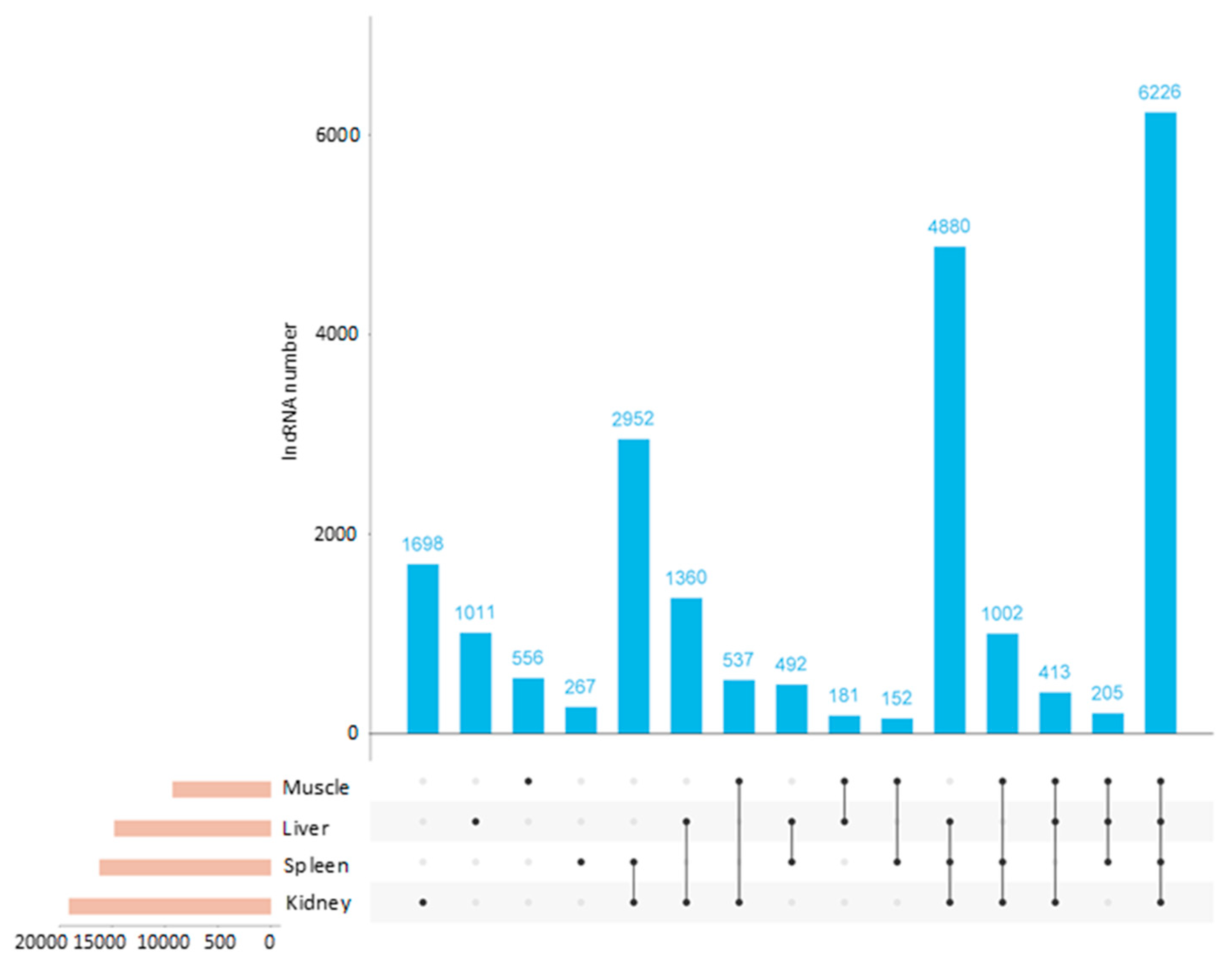
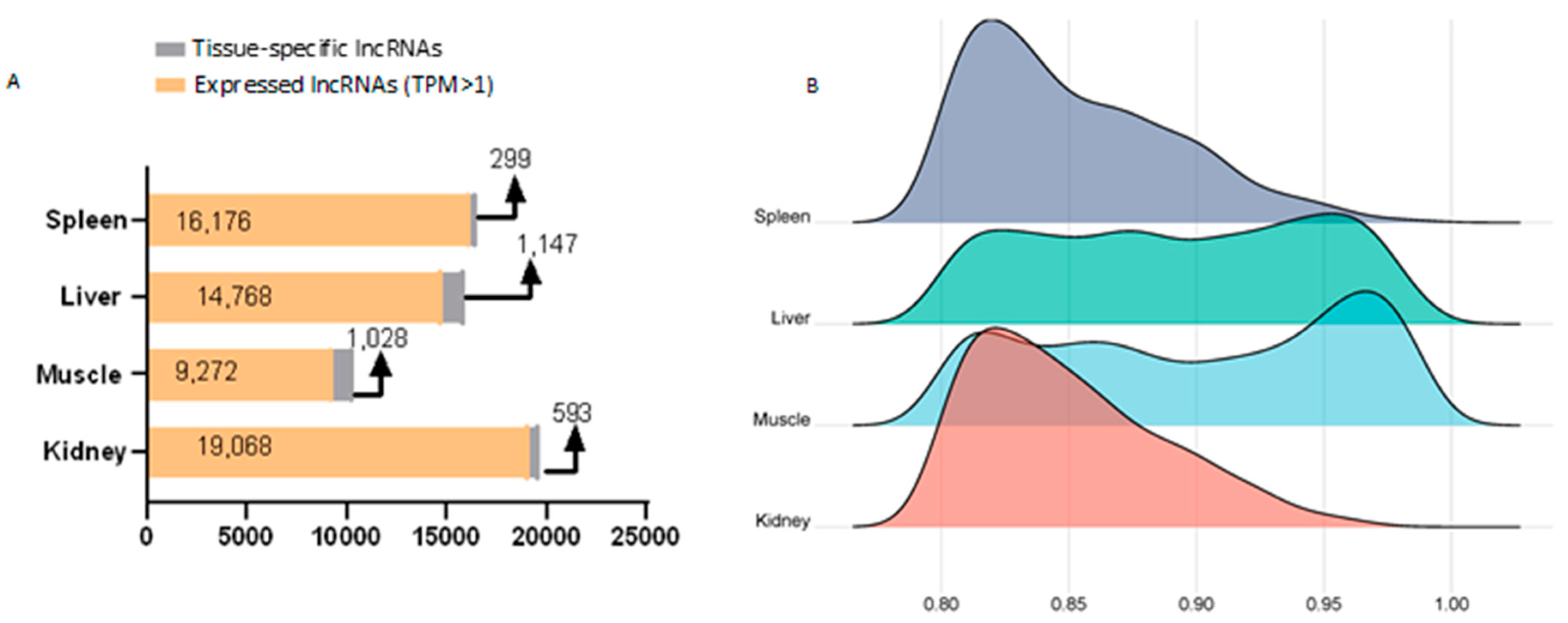
3.3. Identification of lncRNAs Among Tissues
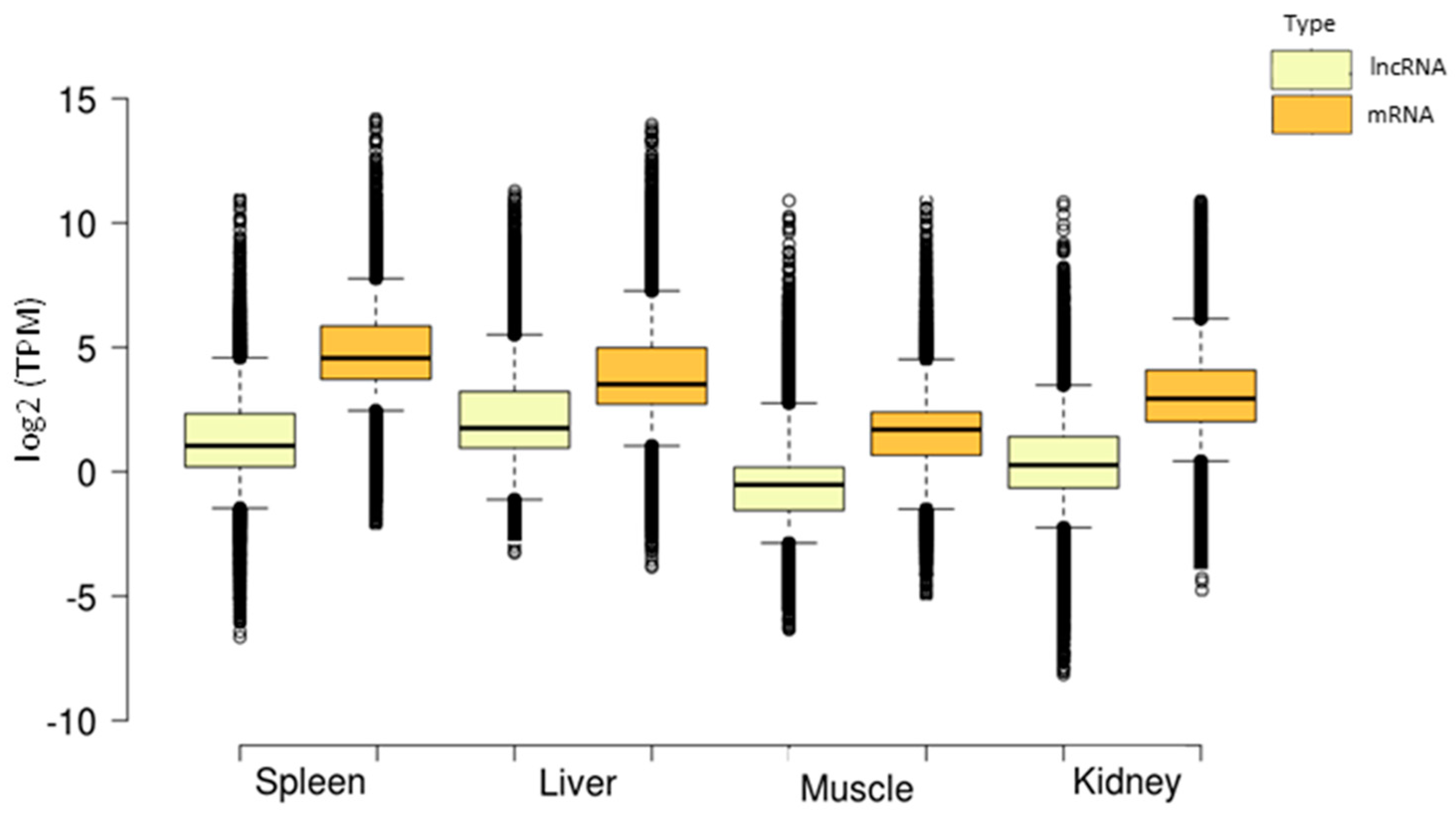
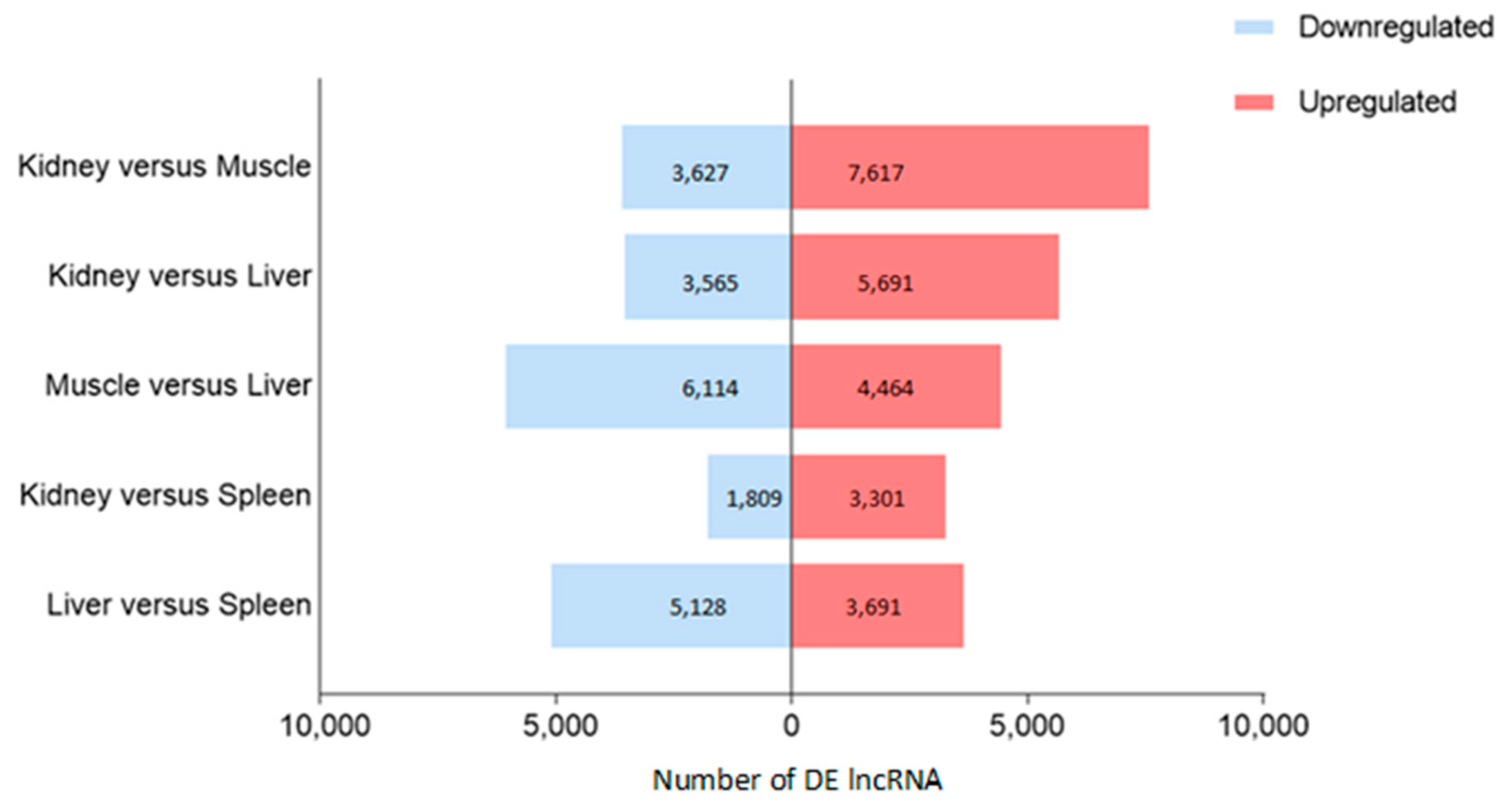
3.4. Tissue-Specific lncRNA Expression
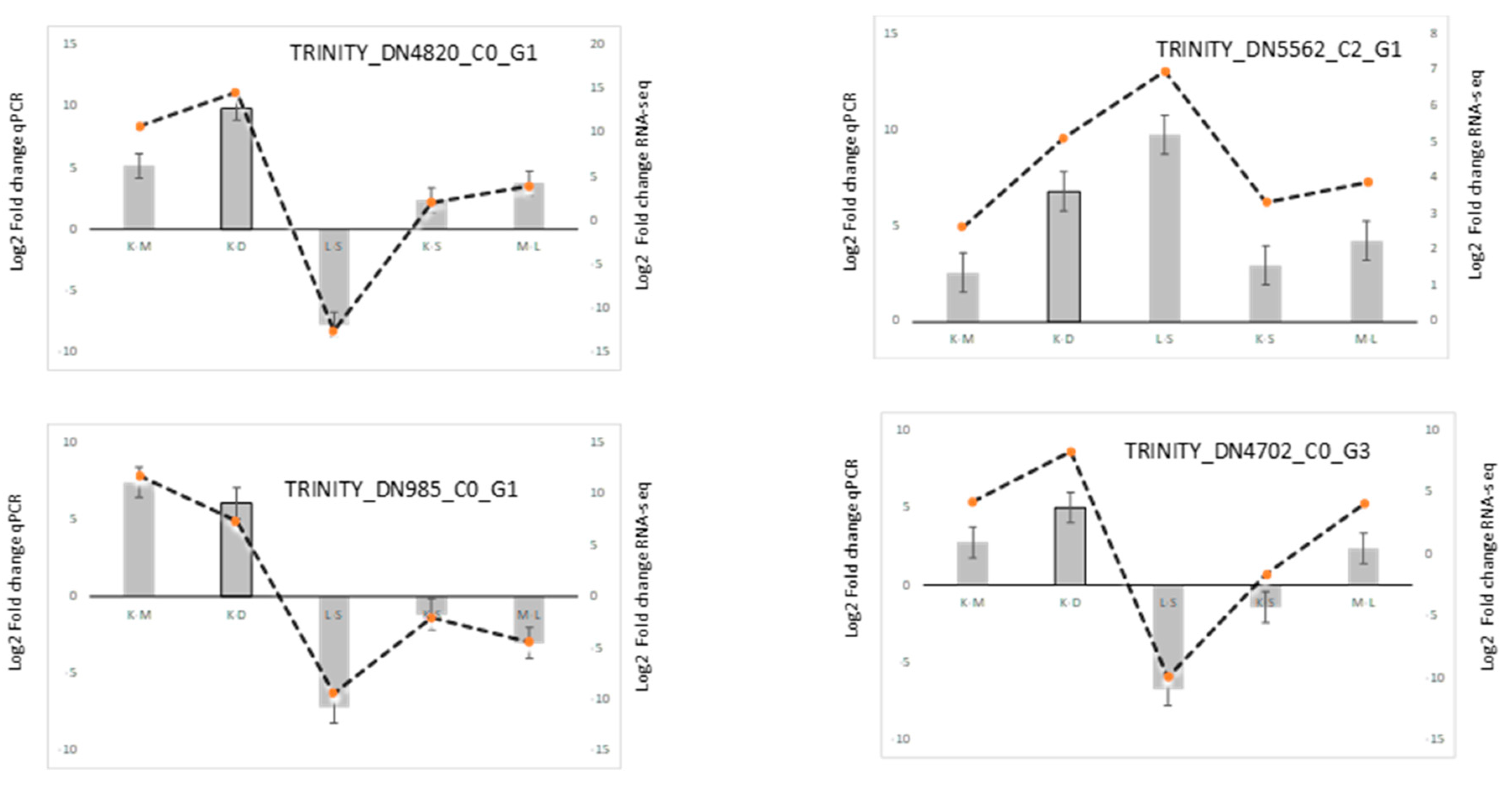
3.5. Experimental RT-qPCR Validation of lncRNAs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Turner.; S.; Miller.; R.M. New Ideas About Old Sharks. Am. Sci. 2005.; 93.
- Guinot, G.; Condamine, F.L. Global impact and selectivity of the Cretaceous-Paleogene mass extinction among sharks, skates, and rays. Science 2023, 379, 802–806. [CrossRef]
- Slater, T.S.; Ashbrook, K.; Kriwet, J. Evolutionary relationships among bullhead sharks (Chondrichthyes, Heterodontiformes). Pap. Palaeontol. 2020, 6, 425–437. [CrossRef]
- Ebert, D.A.; Dando, M.; Fowler, S. Sharks of the world. A Complete Guide.; Princeton University Press: Princeton, UK, 2021; pp 324-385.
- Rigby.; C.L.; Barreto.; R.; Carlson.; J.; Fernando.; D.; Fordham.; S.; Francis.; M.P.; Herman.; K.; Jabado.; R.W.; Liu.; K.M.; Marshall.; A.; Pacoureau.; N.; Romanov.; E.; Sherley.; R.B.; Winker.; H. 2019. Prionace glauca. The IUCN Red List of Threatened Species 2019: e.T39381A2915850.
- Camhi, M.D., Pikitch, E.K.; Babcock, E.A. Sharks of the Open Ocean: Biology, Fisheries and Conservation.; John Wiley & Sons, Chichester, 2009; 140-151.
- Martínez-Ortiz, J.; Aires-Da-Silva, A.M.; Lennert-Cody, C.E.; Maunder, M.N. The Ecuadorian Artisanal Fishery for Large Pelagics: Species Composition and Spatio-Temporal Dynamics. PLOS ONE 2015, 10, e0135136. [CrossRef]
- Vidal, A.; Cardador, L.; Garcia-Barcelona, S.; Macias, D.; Druon, J.-N.; Coll, M.; Navarro, J. The relative importance of biological and environmental factors on the trophodynamics of a pelagic marine predator, the blue shark (Prionace glauca). Mar. Environ. Res. 2023, 183, 105808. [CrossRef]
- Alves, L.M.; Lemos, M.F.; Moutinho, A.B.; Ceia, F.R.; Muñoz-Arnanz, J.; Jiménez, B.; Cabral, H.; Novais, S.C. Assessment of contaminants in blue sharks from the Northeast Atlantic: Profiles, accumulation dynamics, and risks for human consumers. Environ. Pollut. 2023, 316, 120467. [CrossRef]
- Page, T.M.; Lawley, J.W. The Next Generation Is Here: A Review of Transcriptomic Approaches in Marine Ecology. Front. Mar. Sci. 2022, 9. [CrossRef]
- Onimaru, K.; Tatsumi, K.; Shibagaki, K.; Kuraku, S. A de novo transcriptome assembly of the zebra bullhead shark, Heterodontus zebra. Sci. Data 2018, 5, 180197. [CrossRef]
- Goshima, M.; Sekiguchi, R.; Matsushita, M.; Nonaka, M. The complement system of elasmobranches revealed by liver transcriptome analysis of a hammerhead shark, Sphyrna zygaena. Dev. Comp. Immunol. 2016, 61, 13–24. [CrossRef]
- Seixas.; M.J.; Domingues.; R.R.; Antunes.; A. Decoding the Transcriptome of Sharks.; Rays.; and Chimaeras: Insights into Their Physiology.; Morphology.; Evolution.; and Biomedical Applications. Fishes. 2023, 8.; 271.
- Machado.; A.M.; Almeida.; T.; Mucientes.; G.; Esteves.; P.J.; Verissimo.; A.; Castro.; L.F.C. De novo assembly of the kidney and spleen transcriptomes of the cosmopolitan blue shark.; Prionace glauca. Mar. Genomics. 2018, 37.; 50-53.
- Li.; J.; Liu.; C. Coding or Noncoding.; the Converging Concepts of RNAs. Front. Genet. 2019, 10.; 496.
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.-L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long non-coding RNAs: definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [CrossRef]
- Ulitsky, I. Evolution to the rescue: using comparative genomics to understand long non-coding RNAs. Nat. Rev. Genet. 2016, 17, 601–614. [CrossRef]
- Zhang, X.; Wang, W.; Zhu, W.; Dong, J.; Cheng, Y.; Yin, Z.; Shen, F. Mechanisms and Functions of Long Non-Coding RNAs at Multiple Regulatory Levels. Int. J. Mol. Sci. 2019, 20, 5573. [CrossRef]
- Seifuddin, F.; Singh, K.; Suresh, A.; Judy, J.T.; Chen, Y.-C.; Chaitankar, V.; Tunc, I.; Ruan, X.; Li, P.; Chen, Y.; et al. lncRNAKB, a knowledgebase of tissue-specific functional annotation and trait association of long noncoding RNA. Sci. Data 2020, 7, 1–16. [CrossRef]
- Kosinska-Selbi, B.; Mielczarek, M.; Szyda, J. Review: Long non-coding RNA in livestock. Animal 2020, 14, 2003–2013. [CrossRef]
- Domínguez-Rosas, E.; Hernández-Oñate, M..; Fernandez-Valverde, S.-L.; Tiznado-Hernández, M.E. Plant long non-coding RNAs: identification and analysis to unveil their physiological functions. Front. Plant Sci. 2023, 14, 1275399. [CrossRef]
- Hezroni, H.; Koppstein, D.; Schwartz, M.G.; Avrutin, A.; Bartel, D.P.; Ulitsky, I. Principles of Long Noncoding RNA Evolution Derived from Direct Comparison of Transcriptomes in 17 Species. Cell Rep. 2015, 11, 1110–1122. [CrossRef]
- Hara, Y.; Yamaguchi, K.; Onimaru, K.; Kadota, M.; Koyanagi, M.; Keeley, S.D.; Tatsumi, K.; Tanaka, K.; Motone, F.; Kageyama, Y.; et al. Shark genomes provide insights into elasmobranch evolution and the origin of vertebrates. Nat. Ecol. Evol. 2018, 2, 1761–1771. [CrossRef]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [CrossRef]
- Ammunét, T.; Wang, N.; Khan, S.; Elo, L.L. Deep learning tools are top performers in long non-coding RNA prediction. Briefings Funct. Genom. 2022, 21, 230–241. [CrossRef]
- Breitwieser, F.P.; Baker, D.N.; Salzberg, S.L. KrakenUniq: confident and fast metagenomics classification using unique k-mer counts. Genome Biol. 2018, 19, 1–10. [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [CrossRef]
- Bushmanova, E.; Antipov, D.; Lapidus, A.; Suvorov, V.; Prjibelski, A.D. rnaQUAST: a quality assessment tool for de novo transcriptome assemblies. Bioinformatics 2016, 32, 2210–2212. [CrossRef]
- Kalvari, I.; Nawrocki, E.P.; Ontiveros-Palacios, N.; Argasinska, J.; Lamkiewicz, K.; Marz, M.; Griffiths-Jones, S.; Toffano-Nioche, C.; Gautheret, D.; Weinberg, Z.; et al. Rfam 14: expanded coverage of metagenomic, viral and microRNA families. Nucleic Acids Res. 2020, 49, D192–D200. [CrossRef]
- Nawrocki, E.P.; Eddy, S.R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 2013, 29, 2933–2935. [CrossRef]
- Yang.; C.; Yang.; L.; Zhou.; M.; Xie.; H.; Zhang.; C.; Wang.; M. D.; Zhu.; H. LncADeep: an ab initio lncRNA identification and functional annotation tool based on deep learning. Bioinformatics. 2018.; 34.; 3825–3834.
- Camargo, A.P.; Sourkov, V.; Pereira, G.A.G.; Carazzolle, M.F. RNAsamba: neural network-based assessment of the protein-coding potential of RNA sequences. NAR Genom. Bioinform. 2020, 2, lqz024. [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [CrossRef]
- Aubry, S.; Kelly, S.; Kümpers, B.M.C.; Smith-Unna, R.D.; Hibberd, J.M. Deep Evolutionary Comparison of Gene Expression Identifies Parallel Recruitment of Trans-Factors in Two Independent Origins of C4 Photosynthesis. PLOS Genet. 2014, 10, e1004365. [CrossRef]
- Kryuchkova-Mostacci, N.; Robinson-Rechavi, M. A benchmark of gene expression tissue-specificity metrics. Briefings Bioinform. 2017, 18, 205–214. [CrossRef]
- Love.; M. I.; Huber.; W.;Anders.; S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550.
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [CrossRef]
- Li, J.; Ma, W.; Zeng, P.; Wang, J.; Geng, B.; Yang, J.; Cui, Q. LncTar: a tool for predicting the RNA targets of long noncoding RNAs. Briefings Bioinform. 2015, 16, 806–812. [CrossRef]
- Rao X.; Huang X.; Zhou Z.; Lin X. An improvement of the 2ˆ(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat Bioinforma Biomath. 2013 Aug;3(3):71-85.
- Andersen.; C.L.; Jensen.; J.L.; Orntoft.; T.F. Normalization of real-time quantitative reverse transcription-PCR data: a model- based variance estimation approach to identify genes suited for normalization.; applied to bladder and colon cancer data sets. Cancer Res. 2004. 64.; 5245–5250.
- Pfaffl.; M.W.; Tichopad.; A.; Prgomet.; C.; Neuvians.; T.P. Determination of stable housekeeping genes.; differentially regulated target genes and sample integrity: bestKeeper–Excel- based tool using pair-wise correlations. Biotechnol. Lett. 2004. 26.; 509–515.
- Livak.; K. J.; Schmittgen.; T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. 2001.; 25, 402–408.
- Ruijter, J.M.; Ramakers, C.; Hoogaars, W.M.H.; Karlen, Y.; Bakker, O.; van den Hoff, M.J.B.; Moorman, A.F.M. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009, 37, e45. [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [CrossRef]
- Mallory, A.C.; Shkumatava, A. LncRNAs in vertebrates: Advances and challenges. Biochimie 2015, 117, 3–14. [CrossRef]
- Szcześniak, M.W.; Kubiak, M.R.; Wanowska, E.; Makałowska, I. Comparative genomics in the search for conserved long noncoding RNAs. Essays Biochem. 2021, 65, 741–749. [CrossRef]
- Press, C.; Evensen, . The morphology of the immune system in teleost fishes. Fish Shellfish. Immunol. 1999, 9, 309–318. [CrossRef]
- Criscitiello, M.F. What the shark immune system can and cannot provide for the expanding design landscape of immunotherapy. Expert Opin. Drug Discov. 2014, 9, 725–739. [CrossRef]
- Veríssimo, A.; Sampaio, Í.; McDowell, J.R.; Alexandrino. P.; Mucientes, G.; Queiroz N.; da Silva, C.; Jones, C.S.; Noble, L.R.; World without borders-genetic population structure of a highly migratory marine predator, the blue shark (Prionace glauca). Ecol Evol. 2017, 7:4768-4781.
- Ott, J.A.; Ohta, Y.; Flajnik, M.F.; Criscitiello, M.F. Lost structural and functional inter-relationships between Ig and TCR loci in mammals revealed in sharks. Immunogenetics 2021, 73, 17–33. [CrossRef]
- Zhao, T.; Zou, Y.; Yan, H.; Chang, Y.; Zhan, Y. (2023). Non-coding RNAs targeting NF-κB pathways in aquatic animals: A review. Front Immunol. 2023, 14, 1091607.
- de Melo, L.F.; Cabrera, M.L.; Rodrigues, A.C.B.; Turquetti, A.d.O.M.; Ruivo, L.P.; Bruno, C.E.M.; Rici, R.E.G. Morphological Description of Blue Shark Liver, Prionace glauca (Linnaeus, 1758), Elasmobranchii, Carcharhiniformes. Int. J. Adv. Eng. Res. Sci. 2019, 6, 286–290. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




