Submitted:
29 July 2024
Posted:
30 July 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Mitochondrial Genome Composition
2.2. Protein-Coding Genes
2.4. Gene Order
2.5. Gene Collinearity
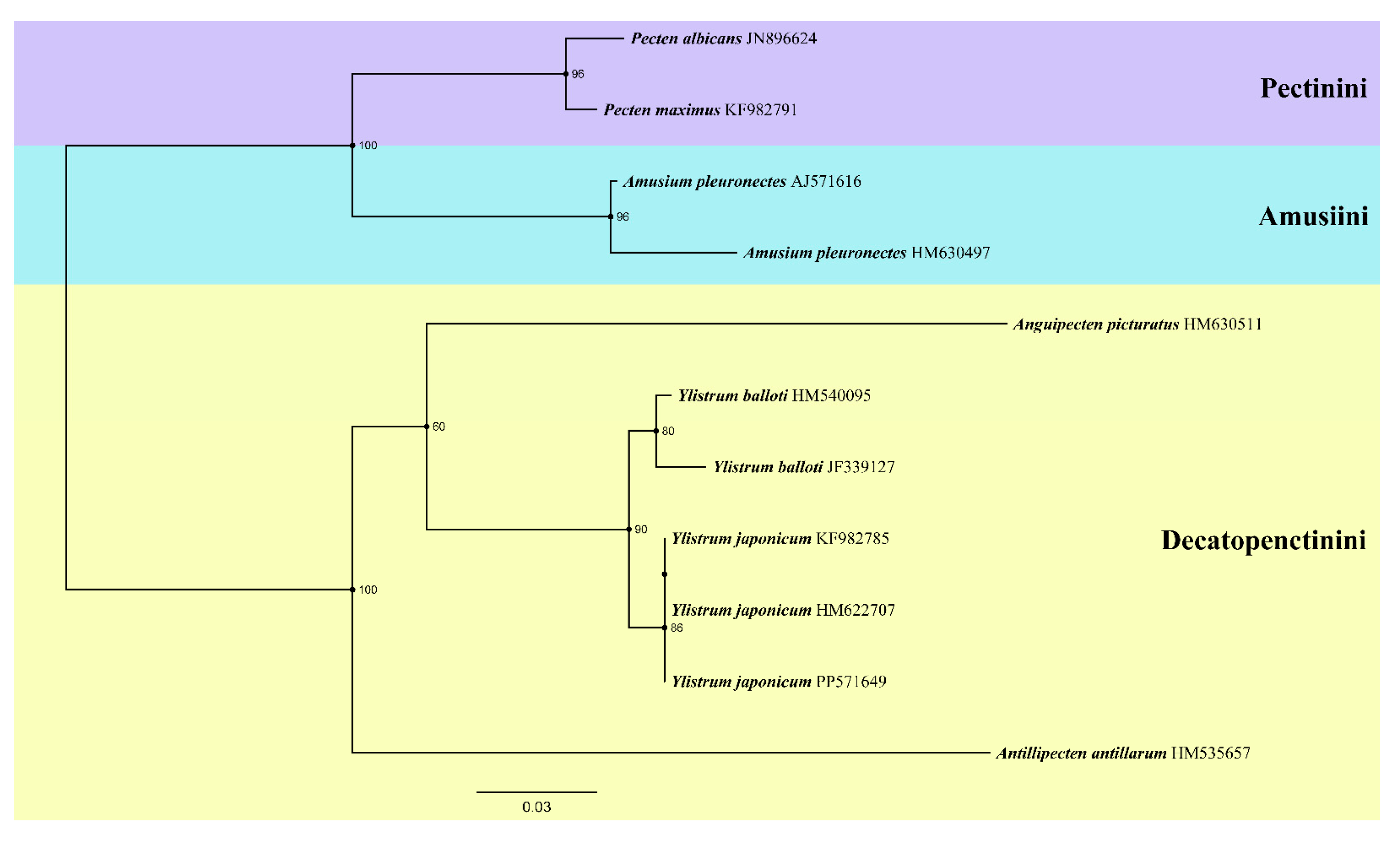
2.6. Phylogenetic Analysis
2.7. Systematic Descriptions
3. Discussion
4. Materials and Methods
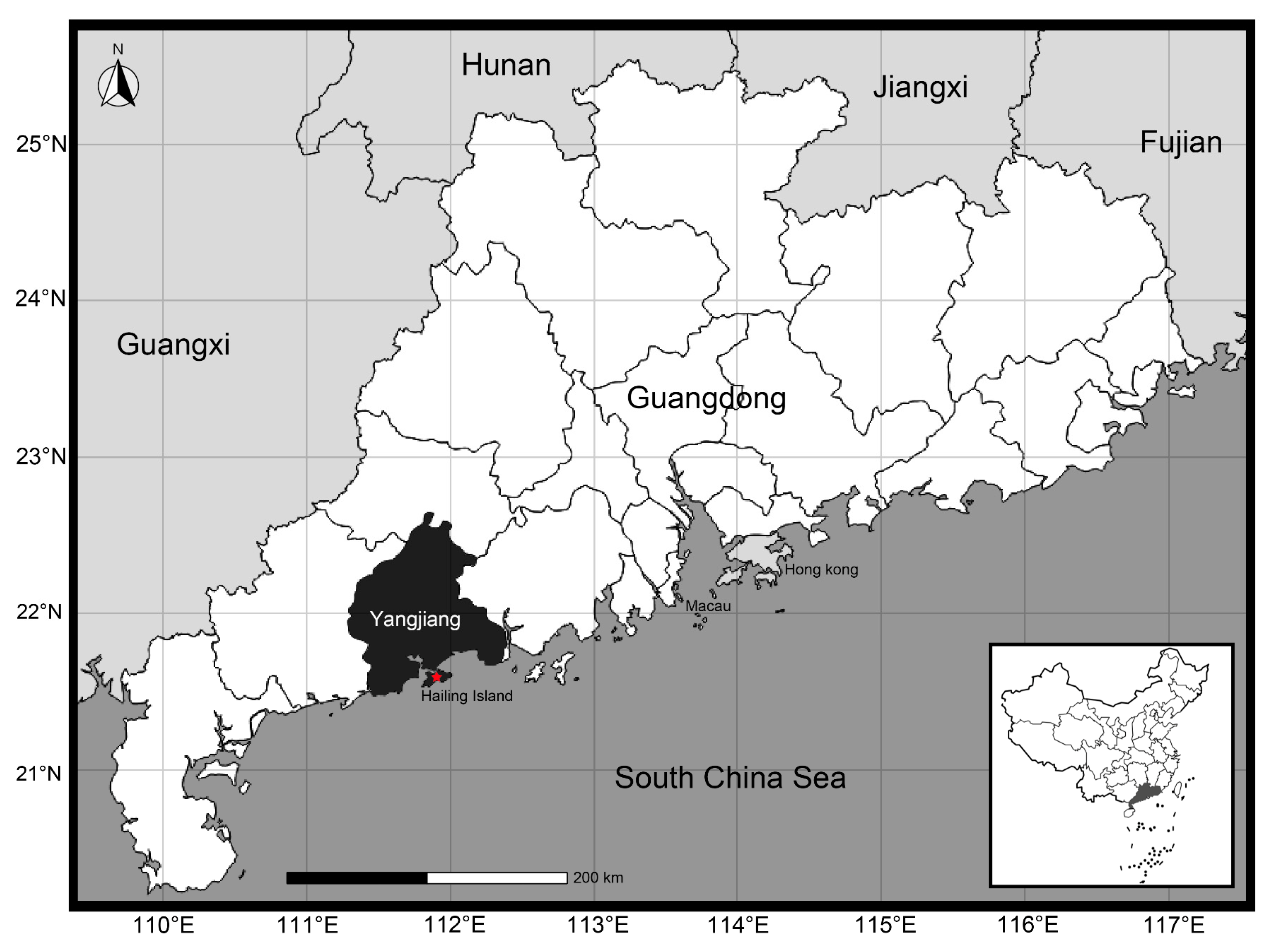
4.1. Sample Collection
4.2. DNA Extraction, Library Preparation and Next Generation Sequence
4.3. Sequence Assembly and Annotation
4.4. Gene Collinearity
4.5. Phylogenetic Analysis with Mitochondrial Genome
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mynhardt, G.; Alejandrino, A.; Puslednik, L.; Corrales, J.; Serb, J.M. Shell shape convergence masks biological diversity in gliding scallops: Description of Ylistrum n. gen. (Pectinidae) from the Indo-Pacifc Ocean. J. Mollus. Stud. 2014, 80, 400–411. [Google Scholar] [CrossRef]
- Dijkstra, H.H. Pectinoidea (Bivalvia: Propeamussiidae and Pectinidae) from the Panglao region, Philippine Islands. Vita Malacologica. 2013, 10, 1–108. [Google Scholar]
- Dijkstra, H.H.; Beu, A.G. Living scallops of Australia and adjacent waters (Mollusca: Bivalvia: Pectinoidea: Propeamussiidae, Cyclochlamydidae and Pectinidae). Rec. Aust. Mus. 2018, 70, 113–330. [Google Scholar] [CrossRef]
- Alejandrino, A.; Puslednik, L.; Serb, J.M. Convergent and parallel evolution in life habit of the scallops (Bivalvia: Pectinidae). BMC Evol Biol. 2011, 11, 1–9. [Google Scholar] [CrossRef]
- Sherratt, E.; Alejandrino, A.; Kraemer, A.C.; Serb, J.M.; Adams, D.C. Trends in the sand: directional evolution in the shell shape of recessing scallops (Bivalvia: Pectinidae). Evolution, 2016, 70, 2061–2073. [Google Scholar] [CrossRef] [PubMed]
- Serb, J.M. Reconciling morphological and molecular approaches in developing a phylogeny for the Pectinidae (Mollusca: Bivalvia). In Scallops Biology, Ecology, Aquaculture, and Fisheries, 3rd ed.; Shumway, S.E., Parsons, G.J., Eds.; Elsevier: Amsterdam, The Netherlands, 2016; pp. 1–29. [Google Scholar]
- Wang, Z.R. Fauna Sinica Invertebrate Vol. 31 Mollusca Bivalvia. Science Press: Beijing, BJ, CN, 2002; pp.1-374.
- Gmelin, J.F. In Gmelin J.F. (Ed.) Caroli a Linnaei Systema Naturae per Regna Tria Naturae, 13 ed, Tom. 1, Pars 6. Lipsiae: Georg. Emanuel Deer, 1791; pp. 3021-3910.
- Bernardi, M. Description d‘espéces nouvellas. Jour. de conchyl, 1861, 9, 46–49. [Google Scholar]
- Abbott, R.T.; Dance, S.P. Compendium of seashells: A colour guide to more than 4,200 of the world‘s marine shells. EP Dutton Inc.: New York, NY, USA, 1982; pp.1-411.
- Carpenter, K.E.; Niem, V.H. The living marine resources of the Western Central Pacifc. Volume 2: Cephalopods, crustaceans, holothurians and sharks. Food and Agriculture Organization of the United Nations, Rome, 1998; pp. 1–686.
- Beu, A.G.; Darragh, T.A. Revision of southern Australian Cenozoic fossil Pectinidae (Mollusca: Bivalvia). Proceedings of the Royal Society of Victoria, 2001, 113, 1–205. [Google Scholar]
- Okada, Y.; Uchida, S.; Uchida, T. New Illustrated Encyclopediaof the Fauna of Japan. Ⅱ. Hokuryukan, Tokyo, TKY, JPN, 1998; pp. 1-803.
- Kanmizutaru, T.; Anraku, K. Inducement of Shell Opening of Amusium japonicum by MgCl2 Injection into the Adductor Muscle. Nippon Suisan Gakk. 1999, 65, 856–859. [Google Scholar] [CrossRef]
- Kanmizutaru, T.; Anraku, K.; Toyoda, S. Light perception capability of pallial eyes in Japanese moon scallop Amusium japonicum as determined by electroretinogram. Nippon Suisan Gakk. 2005, 71, 928–934. [Google Scholar] [CrossRef]
- Son, P.W.; Chung, E.Y. 1994. Annual Reproductive Cycle and Size at First Sexual Maturity of the Sun and Moon Scallop Amusium Japonlcum Japonicum (Gmelin, 1791) (Bivalvia: Pectinidae) in the Coastal Waters of Jejudo, Korea. Malacologia. 1994, 51, 119–129. [Google Scholar] [CrossRef]
- Son, P.W.; Chung, E.Y. Gonadal Development, First Sexual Maturity and Sex Ratio of the Sun and Moon Scallop Amusium japonicum japonicum on the Coastal Waters of Jejudo, Korea. Developmant & Reproduction. 2005, 9, 95–103. [Google Scholar]
- Son, P.W.; Ha, D.S.; Rho, S.; Chang, D.S. Studies on the Age and Growth fo Sun and Moon Scallop, Amusium japonicum japonicum (GMELIN). Journal of Aquaculture. 1996, 9, 409–417 [In Korean]. [Google Scholar]
- Ye, W.J.; Liang, G.Y. A proliminary study on artificial breeding of Amussium japonica (Gmelin). Transactions of Oceanology and Limnology. 1989, 4, 86–93 [In Chinese]. [Google Scholar]
- Ye, W.J.; Liang, G.Y. A preliminary observation on the ecology of Amussium japonica (Gmelin). Chinese Journal of Zoology. 1990, 25, 5–7 [In Chinese]. [Google Scholar]
- Xu, K.; Kanno, M.; Yu, H.; Li, Q.; Kijima, A. Complete mitochondrial DNA sequence and phylogenetic analysis of Zhikong scallop Chlamys farreri (Bivalvia: Pectinidae). Mol Biol. Rep. 2011, 38, 3067–3074. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.T.; Li, Y.X.; Sun, Y.; Tao, J.; Qiu, J.W. A new species of the genus Catillopecten (Bivalvia: Pectinoidea: Propeamussiidae): morphology, mitochondrial genome, and phylogenetic relationship. Front. Mar. Sci. 2023, 10, 1168991. [Google Scholar] [CrossRef]
- Boore, J.L; Medina, M.; Rosenberg, L.A. Complete sequences of the highly rearranged molluscan mitochondrial genomes of the scaphopod Graptacme eborea and the bivalve Mytilus edulis. Mol. Biol. Evol. 2004, 21, 1492–1503. [Google Scholar] [CrossRef]
- Smith, D.R.; Snyder, M. Complete mitochondrial DNA sequence of the scallop Placopecten magellanicus: evidence of transposition leading to an uncharacteristically large mitochondrial genome. J. Mol. Evol. 2007, 65, 380–391. [Google Scholar] [CrossRef]
- Malkócs, T.; Viricel, A.; Becquet, V.; Evin, L.; Dubillot, E.; Pante, E. Complex mitogenomic rearrangements within the Pectinidae (Mollusca: Bivalvia). BMC Ecol Evol. 2022, 22, 29. [Google Scholar] [CrossRef]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–80. [Google Scholar] [CrossRef]
- Li, F.; Zhang, Y.; Zhong, T.; Heng, X.; Ao, T.; Gu, Z.; Wang, A.; Liu, C.; Yang, Y. The Complete Mitochondrial Genomes of Two Rock Scallops (Bivalvia: Spondylidae) Indicate Extensive Gene Rearrangements and Adaptive Evolution Compared with Pectinidae. Int. J. Mol. Sci. 2023, 24, 13844. [Google Scholar] [CrossRef] [PubMed]
- Smedley, G.D.; Audino, J.A.; Grula, C.; Porath-Krause, A.; Pairett, A.N.; Alejandrino, A.; Serb, J.M. Molecular phylogeny of the Pectinoidea (Bivalvia) indicates Propeamussiidae to be a non-monophyletic family with one clade sister to the scallops (Pectinidae). Mol. Phylogenet. Evol. 2019, 137, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.J.; Yu, H.W.; Liu, Y.R.; Zhang, Y.H.; Li, Y.l. The complete mitochondrial genome and phylogenetic analysis of Amusium pleuronectes. Mitochondrial DNA B. 2020, 5, 2318–2319. [Google Scholar] [CrossRef] [PubMed]
- Waller, T.R. Evolutionary relationships among commercial scallops (Mollusca: Bivalvia: Pectinidae). In Scallops: Biology, Ecology and Aquaculture; Shumway, S.E., Ed.; Elsevier: New York, NY, USA, 1991; pp. 1–73. [Google Scholar]
- Waller, T.R. New phylogenies of the Pectinidae (Mollusca: Bivalvia): Reconciling morphological and molecular approaches. In Scallops: Biology, Ecology and Aquaculture. Shumway S.E., Parsons G.J., Ed.; Elsevier B.V.: Amsterdam, The Netherlands, 2006; pp. 1–44. [Google Scholar]
- Waller, T.R. The ctenolium of scallop shells: functional morphology and evolution of a key family-level character in the Pectiniacea (Mollusca: Bivalvia). Malacologia. 1984, 25, 20. [Google Scholar]
- Saavedra, C.; Peña, J.B. Phylogenetics of American scallops (Bivalvia: Pectinidae) based on partial 16S and 12S ribosomal RNA gene sequences. Mar Biol. 2006, 150, 111–119. [Google Scholar] [CrossRef]
- Feng, Y.W.; Li, Q.; Kong, L.F.; Zheng, X.D. DNA barcoding and phylogenetic analysis of Pectinidae (Mollusca: Bivalvia) based on mitochondrial COI and 16S rRNA genes. Mol Biol. Rep. 2011, 38, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Barucca, M.; Olmo, E.; Schiaparelli, S.; Canapa, A. Molecular phylogeny of the family Pectinidae (Mollusca: Bivalvia) based on mitochondrial 16S and 12S rRNA genes. Mol. Phylogenet. Evol. 2004, 31, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Marin, A.; Fujimoto, T.; Arai, K. The variable 5′end of the 16S rRNA gene as a novel barcoding tool for scallops (Bivalvia, Pectinidae). Fisheries sci. 2015, 81, 73–81. [Google Scholar] [CrossRef]
- Habe, T. New name for Amusium japonicum formosum. Venus. 1992, 50, 235. [Google Scholar]
- Zhang, X.; Qi, Z.Y.; Li, J.M.; Ma, X.T.; Wang, Z.R.; Huang, X.M.; Zhuang, Q.Q. Bivalve mollusca of The South China sea. Science Press: Beijing, BJ, CHN. 1960; pp.1-274.
- Matsumoto, M.; Hayami, I. Phylogenetic analysis of the family Pectinidae (Bivalvia) based on mitochondrial cytochrome C oxidase subunit I. J. Mollus. Stud. 2000, 66, 477–488. [Google Scholar] [CrossRef]
- Malkowsky, Y.; Klussmann-Kolb, A. Phylogeny and spatio-temporal distribution of European Pectinidae (Mollusca: Bivalvia). Syst. Biodivers. 2012, 10, 233–242. [Google Scholar] [CrossRef]
- Puslednik, L.; Serb, J.M. Molecular phylogenetics of the Pectinidae (Mollusca: Bivalvia) and the effect of outgroup selection and increased taxon sampling on tree topology. Mol. Phylogenet. Evol. 2008, 48, 1178–1188. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.-X.; Li, Y.; Hittinger, C.T.; Chen, X.; Rokas, A. An investigation of irreproducibility in maximum likelihood phylogenetic inference. Nat. Commun. 2020, 11, 6096. [Google Scholar] [CrossRef]
- Haag, J.; Höhler, D.; Bettisworth, B.; Stamatakis, A. From Easy to Hopeless-Predicting the Difficulty of Phylogenetic Analyses. Mol. Biol. Evol. 2022, 39, msac254. [Google Scholar] [CrossRef]
- Zhang, S.P. Atlas of Marine Mollusks of China. China Ocean Press:Beijing, BJ, CHN. 2008, pp.1-383.
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics, 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A. S.; Lesin, V. M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Darling, A.C.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. http://wwwgenomeorg/cgi/doi/101101/gr2289704. 2004, 14, 1394–1403. [Google Scholar] [CrossRef]
- Xiang, C.; Gao, F.; Jakovlić, I.; Lei, H.; Hu, Y.; Zhang, H.; Zou, H.; Wang, G.; Zhang, D. Using PhyloSuite for Molecular Phylogeny and Tree-based Analyses. iMeta. 2023, 2, e87. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Ranwez, V.; Douzery, E.J.P.; Cambon, C.; Chantret, N.; Delsuc, F. MACSE v2: toolkit for the alignment of coding sequences accounting for frameshifts and stop codons. Mol. Biol. Evol. 2018, 35, 2582–2584. [Google Scholar] [CrossRef]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Nguyen, M.A.; von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
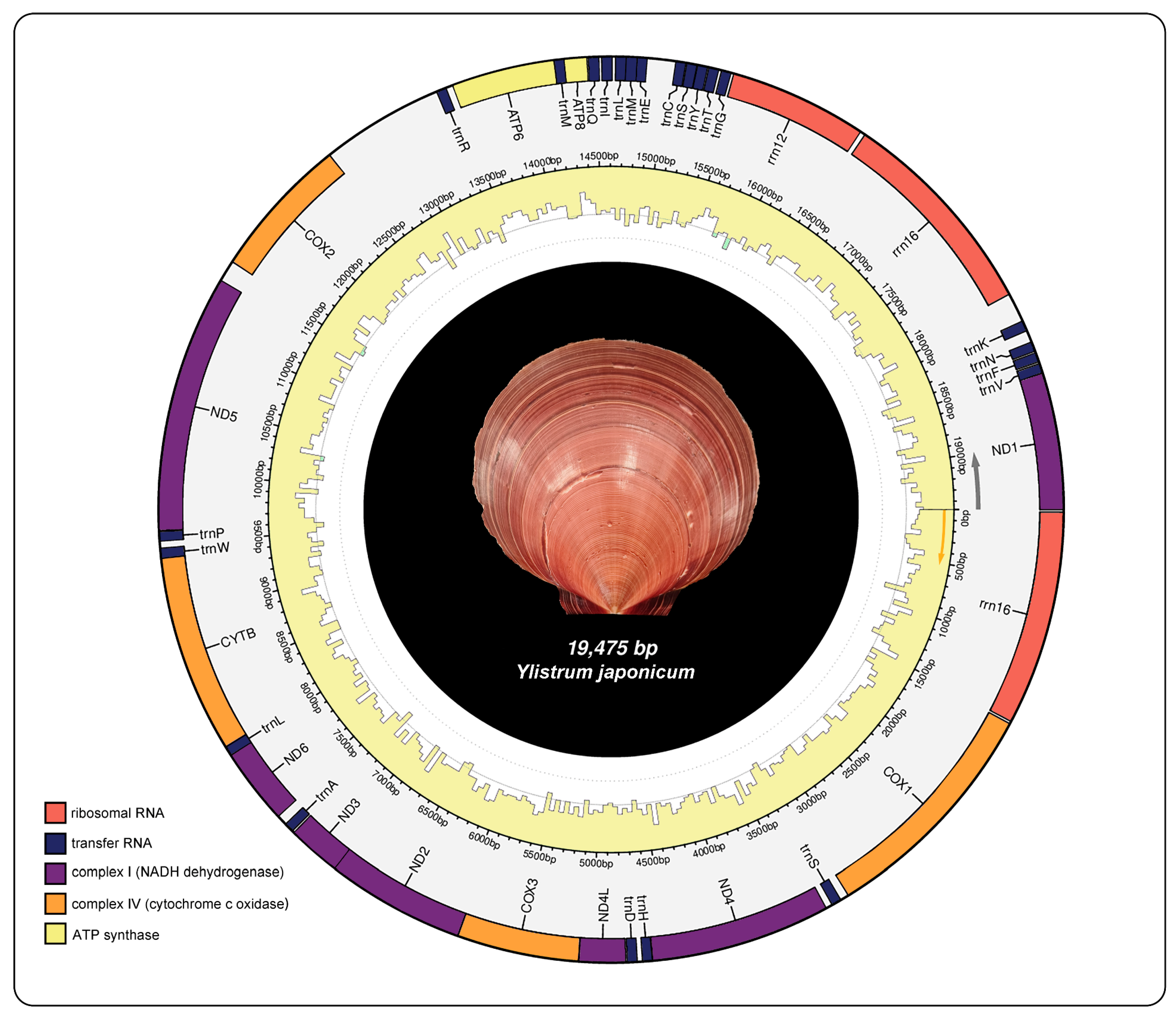
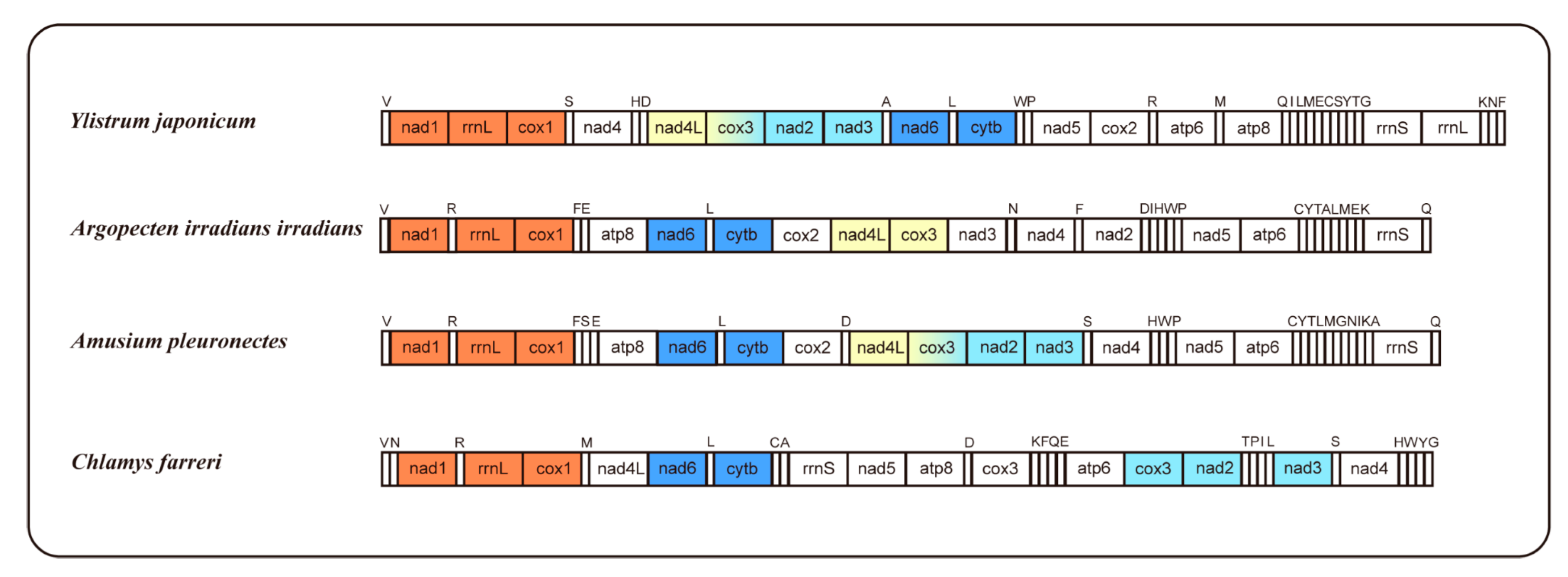
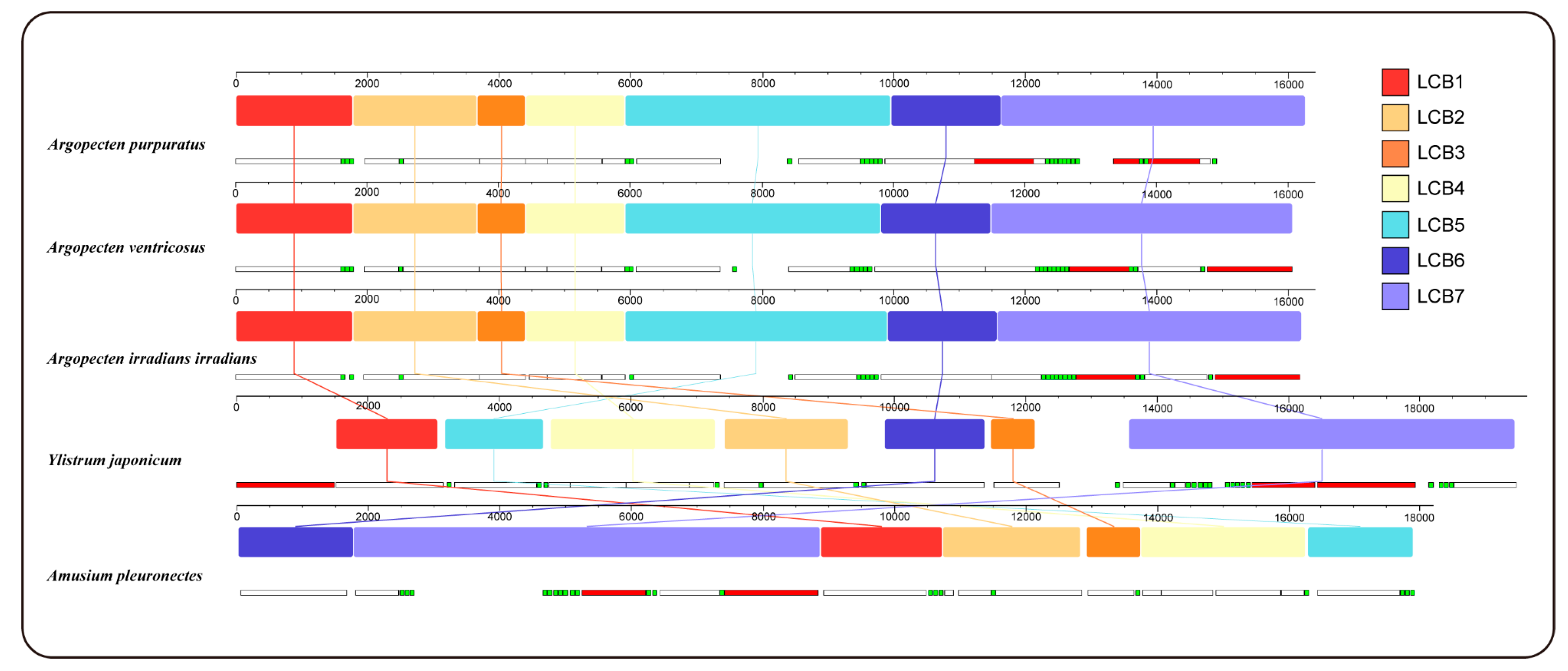
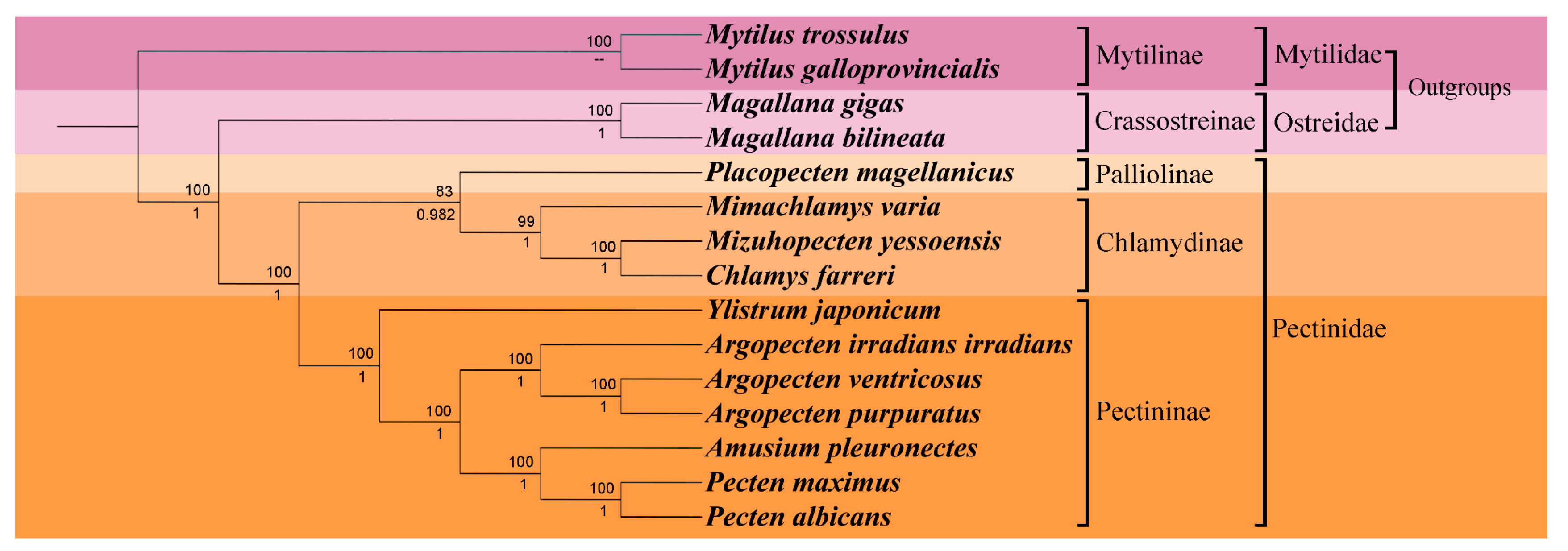
| Gene | Sequence location | Size (bp) | Start codon | Stop codon | Intergenic nucleotide (bp) |
|---|---|---|---|---|---|
| rrnL | 18-1509 | 1492 | 17 | ||
| cox1 | 1527-3158 | 1632 | ATG | TAA | 17 |
| trnS | 3219-3286 | 68 | 60 | ||
| nad4 | 3334-4581 | 1248 | GTG | TAG | 47 |
| trnH | 4588-4652 | 65 | 6 | ||
| trnD | 4690-4757 | 68 | 37 | ||
| nad4L | 4769-5089 | 321 | ATG | TAA | 11 |
| cox3 | 5094-5939 | 846 | GTG | TAG | 4 |
| nad2 | 5942-6904 | 963 | GTG | TAA | 2 |
| nad3 | 6904-7275 | 372 | ATC | TAA | -1 |
| trnA | 7291-7356 | 66 | 15 | ||
| nad6 | 7431-7958 | 528 | GTG | TAA | 74 |
| trnL | 7960-8024 | 65 | 1 | ||
| cytb | 8028-9402 | 1375 | TTG | T | 3 |
| trnW | 9403-9471 | 69 | 0 | ||
| trnP | 9522-9586 | 65 | 50 | ||
| nad5 | 9594-11381 | 1788 | TTG | TAG | 7 |
| cox2 | 11532-12524 | 993 | ATG | TAG | 150 |
| trnR | 13380-13447 | 68 | 855 | ||
| ato6 | 13498-14211 | 714 | ATG | TAG | 50 |
| trnM | 14216-14280 | 65 | 4 | ||
| atp8 | 14284-14439 | 156 | GTG | TAG | 3 |
| trnQ | 14451-14521 | 71 | 11 | ||
| trnI | 14542-14612 | 71 | 20 | ||
| trnL | 14638-14706 | 69 | 25 | ||
| trnM | 14713-14784 | 72 | 6 | ||
| trnE | 14788-14853 | 66 | 3 | ||
| trnC | 15053-15118 | 66 | 199 | ||
| trnS | 15129-15195 | 67 | 10 | ||
| trnY | 15205-15271 | 68 | 9 | ||
| trnT | 15283-15350 | 68 | 11 | ||
| trnG | 15372-15437 | 66 | 21 | ||
| rrnS | 15459-16415 | 957 | 21 | ||
| rrnL | 16453-17939 | 1486 | 37 | ||
| trnK | 18145-18216 | 72 | 205 | ||
| trnN | 18301-18367 | 67 | 84 | ||
| trnF | 18381-18445 | 65 | 13 | ||
| trnV | 18459-18525 | 67 | 13 | ||
| nad1 | 18528-19475 | 948 | GTG | TAG | 2 |
| Subfamily | Tribe in this research | Previous Tribe | Species | Genbank accession numbers | Genetic compartments |
| Ingroup | |||||
| Aequipectinini | Aequipectinini | Argopecten irradians irradians | DQ665851 | mitogenome | |
| Aequipectinini | Aequipectinini | Argopecten purpuratus | KT161260 | mitogenome | |
| Aequipectinini | Aequipectinini | Argopecten ventricosus | KT161261 | mitogenome | |
| Pectininae | Amusiini | Amusiini | Amusium pleuronectes | MT419374 | mitogenome |
| Amusiini | Amusiini | Amusium pleuronectes | AJ571616 | 16S rRNA | |
| Amusiini | Amusiini | Amusium pleuronectes | HM630497 | 16S rRNA | |
| Decatopectinini | Amusiini | Ylistrum japonicum | PP571649 | mitogenome | |
| Decatopectinini | Amusiini | Ylistrum japonicum | KF982785 | 16S rRNA | |
| Decatopectinini | Amusiini | Ylistrum japonicum | HM622707 | 16S rRNA | |
| Decatopectinini | Amusiini | Ylistrum balloti | HM540095 | 16S rRNA | |
| Decatopectinini | Amusiini | Ylistrum balloti | JF339127 | 16S rRNA | |
| Decatopectinini | Decatopectinini | Anguipecten picturatus | HM630511 | 16S rRNA | |
| Decatopectinini | Decatopectinini | Antillipecten antillarum | HMS35657 | 16S rRNA | |
| Pectinini | Pectinini | Pecten maximus | KP900975 | mitogenome | |
| Pectinini | Pectinini | Pecten maximus | KF982791 | 16S rRNA | |
| Pectinini | Pectinini | Pecten albicans | KP900974 | mitogenome | |
| Pectinini | Pectinini | Pecten maximus | JN896624 | 16S rRNA | |
| Palliolinae | Palliolini | Palliolini | Placopecten magellanicus | DQ088274 | mitogenome |
| Chlamydini | Chlamydini | Chlamy farreri | EF473269 | mitogenome | |
| Chlamydinae | Fortipectinini | Fortipectinini | Mizuhopecten yessoensis | FJ595959 | mitogenome |
| Mimachlamydini | Mimachlamydini | Mimachlamys varia | MZ520326 | mitogenome | |
| Outgroup | |||||
| Crassostreinae | Magallana bilineata | MT985154 | mitogenome | ||
| Magallana gigas | MZ497416 | mitogenome | |||
| Mytilinae | Mytilus galloprovincialis | DQ399833 | mitogenome | ||
| Mytilus trossulus | AY823625 | mitogenome |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




