Submitted:
27 June 2024
Posted:
01 July 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Genetic Material
2.2. Plant Material and DNA Extraction
2.3. Selection of Primers and Polymerase Chain Reaction Amplification
2.4. SSR Data Analysis
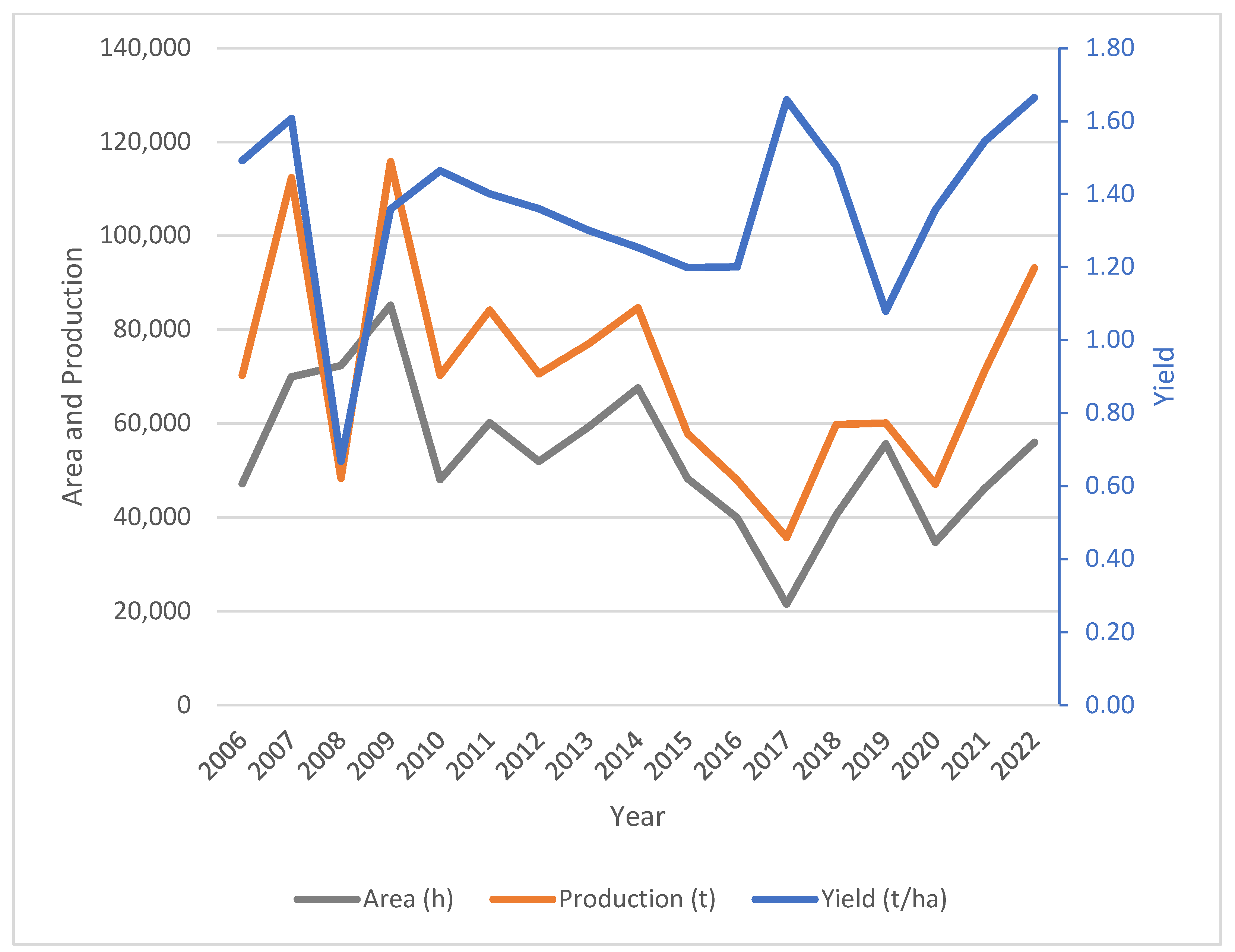
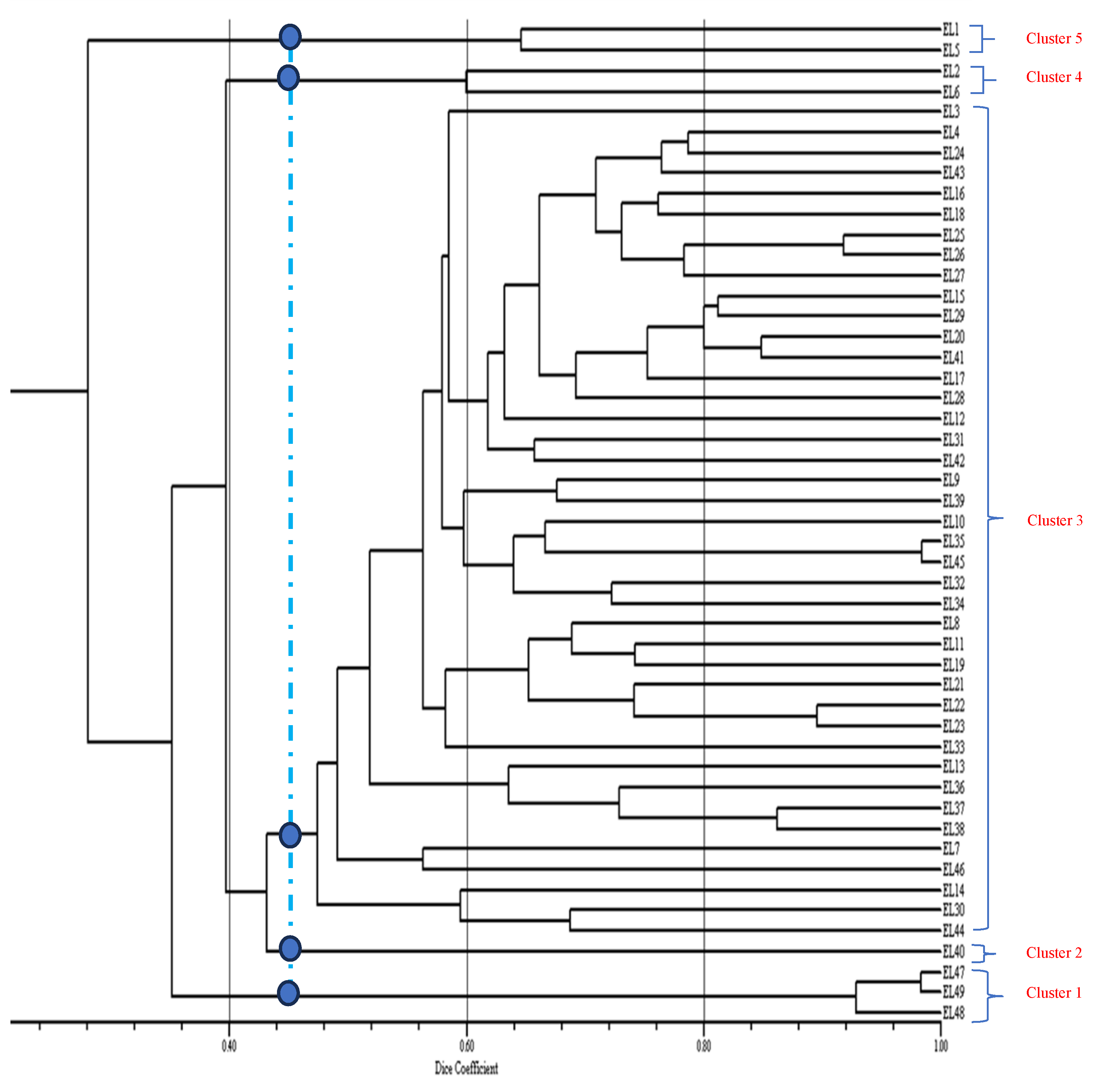
3. Results and Discussion
| Marker | Major allele frequency | Number of alleles | Gene diversity | Observed heterozygosity (Ho) | Polymorphic information content |
|---|---|---|---|---|---|
| Satt394 | 0.85 | 5.00 | 0.27 | 0.00 | 0.26 |
| Satt577 | 0.68 | 3.00 | 0.47 | 0.15 | 0.40 |
| Satt180 | 0.74 | 5.00 | 0.43 | 0.08 | 0.40 |
| Satt459 | 0.76 | 3.00 | 0.38 | 0.06 | 0.35 |
| Satt522 | 0.44 | 4.00 | 0.65 | 0.19 | 0.59 |
| Satt598 | 0.67 | 2.00 | 0.44 | 0.13 | 0.34 |
| Satt242 | 0.56 | 5.00 | 0.57 | 0.17 | 0.49 |
| Satt429 | 0.40 | 6.00 | 0.69 | 0.13 | 0.64 |
| Satt182 | 0.91 | 3.00 | 0.17 | 0.08 | 0.17 |
| Satt397 | 0.50 | 2.00 | 0.50 | 0.06 | 0.38 |
| Satt148 | 0.62 | 4.00 | 0.55 | 0.19 | 0.50 |
| Satt477 | 0.84 | 2.00 | 0.27 | 0.09 | 0.23 |
| Satt530 | 0.75 | 7.00 | 0.43 | 0.09 | 0.41 |
| Satt590 | 0.58 | 8.00 | 0.64 | 0.15 | 0.61 |
| Satt012 | 0.34 | 8.00 | 0.79 | 0.11 | 0.77 |
| Satt172 | 0.61 | 3.00 | 0.53 | 0.17 | 0.46 |
| Satt414 | 0.41 | 7.00 | 0.74 | 0.09 | 0.71 |
| Satt215 | 0.78 | 4.00 | 0.37 | 0.06 | 0.35 |
| Satt387 | 0.50 | 3.00 | 0.55 | 0.13 | 0.45 |
| Satt441 | 0.49 | 9.00 | 0.69 | 0.19 | 0.66 |
| Satt156 | 0.65 | 5.00 | 0.52 | 0.11 | 0.46 |
| Satt184 | 0.67 | 4.00 | 0.51 | 0.19 | 0.47 |
| Satt294 | 0.43 | 6.00 | 0.68 | 0.13 | 0.63 |
| Satt434 | 0.75 | 6.00 | 0.43 | 0.09 | 0.41 |
| Satt511 | 0.49 | 4.00 | 0.67 | 0.17 | 0.63 |
| Satt490 | 0.50 | 3.00 | 0.58 | 0.09 | 0.40 |
| Satt509 | 0.65 | 5.00 | 0.54 | 0.06 | 0.51 |
| Sct_034 | 0.95 | 3.00 | 0.09 | 0.02 | 0.10 |
| Satt372 | 0.35 | 6.00 | 0.73 | 0.21 | 0.68 |
| Sct_067 | 0.91 | 2.00 | 0.17 | 0.00 | 0.16 |
| Total | - | 135 | - | - | - |
| Mean | 0.63 | 4.57 | 0.50 | 0.11 | 0.45 |
| EL1 | EL2 | EL3 | EL4 | EL5 | EL6 | EL7 | EL8 | EL9 | EL10 | EL11 | EL12 | EL13 | EL14 | EL15 | EL16 | EL17 | EL18 | EL19 | EL20 | EL21 | EL22 | EL23 | EL24 | ||
| EL1 | 1.00 | ||||||||||||||||||||||||
| EL2 | 0.19 | 1.00 | |||||||||||||||||||||||
| EL3 | 0.29 | 0.30 | 1.00 | ||||||||||||||||||||||
| EL4 | 0.22 | 0.27 | 0.69 | 1.00 | |||||||||||||||||||||
| EL5 | 0.65 | 0.36 | 0.26 | 0.19 | 1.00 | ||||||||||||||||||||
| EL6 | 0.30 | 0.60 | 0.46 | 0.54 | 0.27 | 1.00 | |||||||||||||||||||
| EL7 | 0.28 | 0.33 | 0.56 | 0.49 | 0.29 | 0.42 | 1.00 | ||||||||||||||||||
| EL8 | 0.28 | 0.40 | 0.60 | 0.74 | 0.26 | 0.64 | 0.57 | 1.00 | |||||||||||||||||
| EL9 | 0.28 | 0.29 | 0.53 | 0.70 | 0.31 | 0.53 | 0.43 | 0.68 | 1.00 | ||||||||||||||||
| EL10 | 0.31 | 0.38 | 0.66 | 0.67 | 0.34 | 0.53 | 0.66 | 0.64 | 0.64 | 1.00 | |||||||||||||||
| EL11 | 0.16 | 0.37 | 0.57 | 0.77 | 0.26 | 0.48 | 0.49 | 0.71 | 0.52 | 0.55 | 1.00 | ||||||||||||||
| EL12 | 0.28 | 0.30 | 0.58 | 0.61 | 0.31 | 0.52 | 0.51 | 0.62 | 0.59 | 0.56 | 0.52 | 1.00 | |||||||||||||
| EL13 | 0.27 | 0.38 | 0.49 | 0.49 | 0.33 | 0.52 | 0.59 | 0.60 | 0.58 | 0.56 | 0.46 | 0.56 | 1.00 | ||||||||||||
| EL14 | 0.22 | 0.37 | 0.39 | 0.43 | 0.35 | 0.37 | 0.36 | 0.53 | 0.58 | 0.46 | 0.40 | 0.61 | 0.57 | 1.00 | |||||||||||
| EL15 | 0.31 | 0.20 | 0.56 | 0.69 | 0.25 | 0.47 | 0.48 | 0.57 | 0.63 | 0.54 | 0.52 | 0.60 | 0.62 | 0.33 | 1.00 | ||||||||||
| EL16 | 0.24 | 0.44 | 0.58 | 0.74 | 0.33 | 0.58 | 0.73 | 0.72 | 0.68 | 0.68 | 0.71 | 0.66 | 0.59 | 0.52 | 0.61 | 1.00 | |||||||||
| EL17 | 0.29 | 0.24 | 0.60 | 0.70 | 0.23 | 0.45 | 0.56 | 0.55 | 0.58 | 0.67 | 0.50 | 0.58 | 0.54 | 0.37 | 0.75 | 0.62 | 1.00 | ||||||||
| EL18 | 0.32 | 0.39 | 0.55 | 0.69 | 0.33 | 0.59 | 0.54 | 0.58 | 0.64 | 0.60 | 0.59 | 0.73 | 0.57 | 0.48 | 0.75 | 0.76 | 0.62 | 1.00 | |||||||
| EL19 | 0.25 | 0.36 | 0.52 | 0.74 | 0.34 | 0.49 | 0.54 | 0.67 | 0.68 | 0.73 | 0.74 | 0.59 | 0.50 | 0.58 | 0.54 | 0.75 | 0.61 | 0.67 | 1.00 | ||||||
| EL20 | 0.36 | 0.25 | 0.59 | 0.66 | 0.24 | 0.56 | 0.55 | 0.60 | 0.55 | 0.52 | 0.53 | 0.58 | 0.62 | 0.38 | 0.83 | 0.61 | 0.72 | 0.71 | 0.52 | 1.00 | |||||
| EL21 | 0.09 | 0.40 | 0.53 | 0.69 | 0.25 | 0.44 | 0.52 | 0.60 | 0.54 | 0.66 | 0.72 | 0.44 | 0.48 | 0.36 | 0.52 | 0.70 | 0.72 | 0.51 | 0.70 | 0.52 | 1.00 | ||||
| EL22 | 0.23 | 0.48 | 0.57 | 0.69 | 0.32 | 0.54 | 0.53 | 0.67 | 0.55 | 0.61 | 0.72 | 0.52 | 0.52 | 0.39 | 0.56 | 0.72 | 0.63 | 0.52 | 0.67 | 0.54 | 0.79 | 1.00 | |||
| EL23 | 0.22 | 0.44 | 0.54 | 0.60 | 0.32 | 0.48 | 0.52 | 0.58 | 0.49 | 0.55 | 0.63 | 0.48 | 0.46 | 0.33 | 0.52 | 0.68 | 0.60 | 0.48 | 0.58 | 0.47 | 0.69 | 0.90 | 1.00 | ||
| EL24 | 0.22 | 0.23 | 0.65 | 0.79 | 0.25 | 0.53 | 0.48 | 0.65 | 0.69 | 0.63 | 0.62 | 0.73 | 0.54 | 0.49 | 0.81 | 0.73 | 0.72 | 0.78 | 0.63 | 0.77 | 0.58 | 0.59 | 0.52 | 1.00 | |
| EL25 | EL26 | EL27 | EL28 | EL29 | EL30 | EL31 | EL32 | EL33 | EL34 | EL35 | EL36 | EL37 | EL38 | EL39 | EL40 | EL41 | EL42 | EL43 | EL44 | EL45 | EL46 | EL47 | EL48 | EL49 | |
| EL25 | 1.00 | ||||||||||||||||||||||||
| EL26 | 0.92 | 1.00 | |||||||||||||||||||||||
| EL27 | 0.74 | 0.83 | 1.00 | ||||||||||||||||||||||
| EL28 | 0.62 | 0.65 | 0.67 | 1.00 | |||||||||||||||||||||
| EL29 | 0.70 | 0.69 | 0.62 | 0.66 | 1.00 | ||||||||||||||||||||
| EL30 | 0.50 | 0.49 | 0.55 | 0.66 | 0.67 | 1.00 | |||||||||||||||||||
| EL31 | 0.70 | 0.62 | 0.52 | 0.56 | 0.67 | 0.57 | 1.00 | ||||||||||||||||||
| EL32 | 0.53 | 0.52 | 0.58 | 0.69 | 0.70 | 0.67 | 0.53 | 1.00 | |||||||||||||||||
| EL33 | 0.56 | 0.58 | 0.57 | 0.52 | 0.63 | 0.62 | 0.49 | 0.59 | 1.00 | ||||||||||||||||
| EL34 | 0.61 | 0.55 | 0.51 | 0.55 | 0.64 | 0.53 | 0.56 | 0.72 | 0.49 | 1.00 | |||||||||||||||
| EL35 | 0.52 | 0.45 | 0.51 | 0.65 | 0.53 | 0.49 | 0.46 | 0.66 | 0.45 | 0.60 | 1.00 | ||||||||||||||
| EL36 | 0.53 | 0.52 | 0.52 | 0.62 | 0.54 | 0.43 | 0.37 | 0.63 | 0.43 | 0.53 | 0.46 | 1.00 | |||||||||||||
| EL37 | 0.58 | 0.57 | 0.56 | 0.67 | 0.61 | 0.51 | 0.41 | 0.58 | 0.47 | 0.45 | 0.47 | 0.78 | 1.00 | ||||||||||||
| EL38 | 0.61 | 0.60 | 0.52 | 0.57 | 0.58 | 0.41 | 0.44 | 0.44 | 0.37 | 0.45 | 0.40 | 0.68 | 0.86 | 1.00 | |||||||||||
| EL39 | 0.54 | 0.53 | 0.49 | 0.60 | 0.58 | 0.41 | 0.37 | 0.64 | 0.43 | 0.56 | 0.57 | 0.61 | 0.69 | 0.62 | 1.00 | ||||||||||
| EL40 | 0.47 | 0.46 | 0.39 | 0.43 | 0.44 | 0.30 | 0.40 | 0.50 | 0.36 | 0.44 | 0.36 | 0.47 | 0.47 | 0.58 | 0.47 | 1.00 | |||||||||
| EL41 | 0.65 | 0.63 | 0.59 | 0.70 | 0.80 | 0.68 | 0.71 | 0.65 | 0.63 | 0.62 | 0.48 | 0.55 | 0.59 | 0.56 | 0.52 | 0.52 | 1.00 | ||||||||
| EL42 | 0.63 | 0.56 | 0.56 | 0.56 | 0.63 | 0.57 | 0.66 | 0.49 | 0.56 | 0.59 | 0.51 | 0.43 | 0.49 | 0.52 | 0.43 | 0.37 | 0.69 | 1.00 | |||||||
| EL43 | 0.72 | 0.65 | 0.67 | 0.61 | 0.66 | 0.56 | 0.56 | 0.59 | 0.48 | 0.66 | 0.61 | 0.46 | 0.47 | 0.50 | 0.50 | 0.46 | 0.60 | 0.65 | 1.00 | ||||||
| EL44 | 0.56 | 0.49 | 0.48 | 0.43 | 0.51 | 0.69 | 0.47 | 0.44 | 0.49 | 0.53 | 0.40 | 0.31 | 0.38 | 0.41 | 0.35 | 0.41 | 0.52 | 0.54 | 0.55 | 1.00 | |||||
| EL45 | 0.55 | 0.48 | 0.53 | 0.67 | 0.55 | 0.52 | 0.48 | 0.68 | 0.48 | 0.62 | 0.98 | 0.45 | 0.46 | 0.39 | 0.56 | 0.35 | 0.47 | 0.50 | 0.60 | 0.42 | 1.00 | ||||
| EL46 | 0.46 | 0.45 | 0.47 | 0.59 | 0.52 | 0.40 | 0.46 | 0.46 | 0.34 | 0.51 | 0.42 | 0.40 | 0.35 | 0.38 | 0.38 | 0.34 | 0.53 | 0.45 | 0.54 | 0.38 | 0.44 | 1.00 | |||
| EL47 | 0.43 | 0.42 | 0.38 | 0.35 | 0.41 | 0.30 | 0.43 | 0.26 | 0.29 | 0.33 | 0.26 | 0.36 | 0.37 | 0.43 | 0.40 | 0.26 | 0.35 | 0.34 | 0.32 | 0.28 | 0.29 | 0.54 | 1.00 | ||
| EL48 | 0.41 | 0.41 | 0.37 | 0.38 | 0.39 | 0.29 | 0.41 | 0.22 | 0.28 | 0.29 | 0.22 | 0.35 | 0.35 | 0.45 | 0.35 | 0.32 | 0.37 | 0.33 | 0.31 | 0.27 | 0.25 | 0.55 | 0.94 | 1.00 | |
| EL49 | 0.40 | 0.39 | 0.39 | 0.33 | 0.41 | 0.30 | 0.43 | 0.27 | 0.30 | 0.33 | 0.26 | 0.33 | 0.34 | 0.41 | 0.37 | 0.23 | 0.35 | 0.34 | 0.33 | 0.28 | 0.29 | 0.54 | 0.98 | 0.92 | 1.00 |
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siamabele, B. Soya Beans Production in Zambia: Opportunities and Challenges. Am J Agric Biol Sci 2019, 14, 55–60. [CrossRef]
- Tufa, A.H.; Alene, A.D.; Manda, J.; Akinwale, M.G.; Chikoye, D.; Feleke, S.; Wossen, T.; Manyong, V. The Productivity and Income Effects of Adoption of Improved Soybean Varieties and Agronomic Practices in Malawi. World Dev 2019, 124, 104631. [CrossRef]
- 2011; 3. Technoserve Southern Africa Regional Soybean Roadmap Final Report; 2011;
- Khojely, D.M.; Ibrahim, S.E.; Sapey, E.; Han, T. History, Current Status, and Prospects of Soybean Production and Research in Sub-Saharan Africa. Crop J 2018, 6, 226–235. [CrossRef]
- FAOSTAT Available online: https://www.fao.org/faostat/en/#home.
- Omondi, J.O.; Mkuhlani, S.; Mugo, J.; Chibeba, A.M.; Chiduwa, M.S.; Chigeza, G.; Kyei-Boahen, S.; Masikati, P.; Nyagumbo, I. Closing the Yield Gap of Soybean (Glycine Max (L.) Merril) in Southern Africa: A Case of Malawi, Zambia, and Mozambique. Frontiers in Agronomy 2023, 5, 1219490. [CrossRef]
- Santos, M.; Da, M.; Santos, F. SOYBEAN VARIETIES IN SUB-SAHARAN AFRICA. Afr. J. Food Agric. Nutr. Dev 2019, 19, 15136–15139. [CrossRef]
- Mukuze, C.; Tukamuhabwa, P.; Maphosa, M.; Dari, S.; Dramadri, I.O.; Obua, T.; Kongai, H.; Rubaihayo, P. Genetic Diversity Analysis among Soybean Genotypes Using SSR Markers in Uganda. African Journal of Biotechnology 2020, 19, 439–448. [CrossRef]
- Jo, H.; Lee, J.Y.; Cho, H.; Choi, H.J.; Son, C.K.; Bae, J.S.; Bilyeu, K.; Song, J.T.; Lee, J.D. Genetic Diversity of Soybeans (Glycine Max (L.) Merr.) with Black Seed Coats and Green Cotyledons in Korean Germplasm. Agronomy 2021, Vol. 11, Page 581 2021, 11, 581. [CrossRef]
- Gwata, E.T.; Wofford, D.S.; Boote, K.J.; Blount, A.R.; Pfahler, P.L. Inheritance of Promiscuous Nodulation in Soybean. Crop Sci 2005, 45, 635–638. [CrossRef]
- Gwata, E.T.; Nziramasanga, N. Seed Protein and Oil Content in Zimbabwean Soyabean (Glycine Max L.) Varieties. Plant Varieties and Seeds 2001, 14, 125–128.
- Nassiuma, D.; Wasike, W. Stability Assessment of Soybean Varieties in Kenya. Afr Crop Sci J 2002, 10. [CrossRef]
- Reyna, N.; Sneller, C.H. Evaluation of Marker-Assisted Introgression of Yield QTL Alleles into Adapted Soybean. Crop Sci 2001, 41, 1317–1321. [CrossRef]
- Gizlice, Z.; Carter Jnr, T.E.; Burton, J.W. Genetic Base for North American Public Soybean Cultivars Released between 1947 and 1988. Crop Sci 1994, 34, 1143–1151. [CrossRef]
- Junyi, G.; Tuanjie, Z. The Core Ancestors of Soybean Cultivars in China. Journal of Nanjing Agricultural University 2001, 24, 20–23.
- Bhat, S.; Basavaraja, G.; Salimath, P. Analysis of Variability in Segregating Generation of Soybean (Glycine Max (L.) Merrill). Karnataka Journal of Agricultural Sciences 2012.
- Mulato, B.M.; Möller, M.; Zucchi, M.I.; Quecini, V.; Pinheiro, J.B. Genetic Diversity in Soybean Germplasm Identified by SSR and EST-SSR Markers. Pesqui Agropecu Bras 2010, 45, 276–283.
- Tantasawat, P.; Trongchuen, J.; Prajongjai, T.; Jenweerawat, S.; Chaowiset, W. SSR Analysis of Soybean (“Glycine Max” (L.) Merr.) Genetic Relationship and Variety Identification in Thailand. Aust J Crop Sci 2011.
- Baruah, J.; Pandey, S.K.; Sarmah, N.; Lal, M. Assessing Molecular Diversity among High Capsaicin Content Lines of Capsicum Chinense Jacq. Using Simple Sequence Repeat Marker. Ind Crops Prod 2019, 141, 111769. [CrossRef]
- Behera, T.K.; Sharma, P.; Singh, B.K.; Kumar, G.; Kumar, R.; Mohapatra, T.; Singh, N.K. Assessment of Genetic Diversity and Species Relationships in Eggplant (Solanum Melongena L.) Using STMS Markers. Sci Hortic 2006, 107, 352–357. [CrossRef]
- Franco, J.; Crossa, J.; Ribaut, J.M.; Bertran, J.; Warburton, M.L.; Khairallah, M. A Method for Combining Molecular Markers and Phenotypic Attributes for Classifying Plant Genotypes. Theoretical and Applied Genetics 2001, 103, 944–952. [CrossRef]
- Manjarrez-Sandoval, P.; Carter, T.E.; Webb, D.M.; Burton, J.W. RFLP Genetic Similarity Estimates and Coefficient of Parentage as Genetic Variance Predictors for Soybean Yield. Crop Sci 1997, 37, 698–703. [CrossRef]
- Mushoriwa, H.; Mathew, I.; Gwata, E.T.; Tongoona, P.; Derera, J. Grain Yield Potential and Stability of Soybean Genotypes of Different Ages across Diverse Environments in Southern Africa. Agronomy 2022, Vol. 12, Page 1147 2022, 12, 1147. [CrossRef]
- Cregan, P.B.; Jarvik, T.; Bush, A.L.; Shoemaker, R.C.; Lark, K.G.; Kahler, A.L.; Kaya, N.; VanToai, T.T.; Lohnes, D.G.; Chung, J.; et al. An Integrated Genetic Linkage Map of the Soybean Genome. Crop Sci 1999, 39, 1464–1490. [CrossRef]
- Gogoi, A.; Munda, S.; Paw, M.; Begum, T.; Siddiqui, M.H.; Gaafar, A.R.Z.; Kesawat, M.S.; Lal, M. Molecular Genetic Divergence Analysis amongst High Curcumin Lines of Golden Crop (Curcuma Longa L.) Using SSR Marker and Use in Trait-Specific Breeding. Scientific Reports 2023 13:1 2023, 13, 1–18. [CrossRef]
- Rongwen, J.; Akkaya, M.S.; Bhagwat, A.A.; Lavi, U.; Cregan, P.B. The Use of Microsatellite DNA Markers for Soybean Genotype Identification. Theoretical and Applied Genetics 1995, 90, 43–48.
- Guichoux, E.; Lagache, L.; Wagner, S.; Chaumeil, P.; Léger, P.; Lepais, O.; Lepoittevin, C.; Malausa, T.; Revardel, E.; Salin, F.; et al. Current Trends in Microsatellite Genotyping. Mol Ecol Resour 2011, 11, 591–611. [CrossRef]
- Dice, L.R. Measures of the Amount of Ecologic Association Between Species. Ecology 1945, 26, 297–302. [CrossRef]
- Rohlf, F.J.; Applied Biostatistics, Inc.; Exeter Software (Firm) NTSYS-Pc : Numerical Taxonomy and Multivariate Analysis System. 2009.
- Liu, K.; Muse, S. V PowerMarker: An Integrated Analysis Environment for Genetic Marker Analysis. BIOINFORMATICS APPLICATIONS NOTE 2005, 21, 2128–2129. [CrossRef]
- Nei, M. Analysis of Gene Diversity in Subdivided Populations. Proc Natl Acad Sci U S A 1973, 70, 3321–3323. [CrossRef]
- Priolli, R.H.G.; Pinheiro, J.B.; Zucchi, M.I.; Bajay, M.M.; Vello, N.A. Genetic Diversity among Brazilian Soybean Cultivars Based on SSR Loci and Pedigree Data. Brazilian Archives of Biology and Technology 2010, 53, 519–531. [CrossRef]
- Powell, W.; Morgante, M.; Andre, C.; Hanafey, M.; Vogel, J.; Tingey, S.; Rafalski, A. The Comparison of RFLP, RAPD, AFLP and SSR (Microsatellite) Markers for Germplasm Analysis. Molecular breeding 1996, 2, 225–238. [CrossRef]
- Thompson, J.A.; Nelson, R.L.; Vodkin, L.O. Identification of Diverse Soybean Germplasm Using RAPD Markers. Crop Sci 1998, 38, 1348–1355. [CrossRef]
- Ude, G.N.; Kenworthy, W.J.; Costa, J.M.; Cregan, P.B.; Alvernaz, J. Genetic Diversity of Soybean Cultivars from China, Japan, North America, and North American Ancestral Lines Determined by Amplified Fragment Length Polymorphism. Crop Sci 2003, 43, 1858–1867. [CrossRef]
- Thovhogi, F.; Gwata, E.T.; McHau, G.R.A.; Safodien, S.S.; Koopman, T. Molecular Characterization of Spider Plant (Cleome Gynandra) Accessions Using SSR Markers. Agronomy 2021, 11, 2206. [CrossRef]
- Biswas, M.K.; Mondal, M.A.A.; Hossain, M.; Islam, R. Utilization of Genetic Diversity and Its Association with Heterosis for Progeny Selection in Potato Breeding Programs. American-Eurasian J. Agric. Env. Sci 2008, 3, 882–887.
| Genotype Code | Designated Name | Growth Habit | Year of Release / Registration | Country |
|---|---|---|---|---|
| EL1 | Hernon 147 | Indeterminate | 1940 | Zimbabwe |
| EL2 | Rhosa | Indeterminate | 1966 | Zimbabwe |
| EL3 | Bragg | Determinate | 1972 | Zimbabwe |
| EL4 | Oribi | Determinate | 1973 | Zimbabwe |
| EL5 | Buffalo | Determinate | 1974 | Zimbabwe |
| EL6 | Impala | Indeterminate | 1977 | Zimbabwe |
| EL7 | Kudu | Determinate | 1977 | Zimbabwe |
| EL8 | Sable | Indeterminate | 1980 | Zimbabwe |
| EL9 | Duiker | Indeterminate | 1982 | Zimbabwe |
| EL10 | Roan | Determinate | 1985 | Zimbabwe |
| EL11 | Gazelle | Indeterminate | 1988 | Zimbabwe |
| EL12 | Nyala | Indeterminate | 1989 | Zimbabwe |
| EL13 | Nondo | Determinate | 1992 | Zimbabwe |
| EL14 | Soma | Indeterminate | 1992 | Zimbabwe |
| EL15 | SCS1 | Determinate | 1995 | Zambia, Zimbabwe |
| EL16 | Sonnet | Determinate | 1994 | Zimbabwe |
| EL17 | Sonata | Determinate | 1997 | Zimbabwe |
| EL18 | Viking | Indeterminate | 1999 | Zimbabwe |
| EL19 | Solitaire | Indeterminate | 1998 | Zambia, Malawi, Zimbabwe |
| EL20 | Soprano | Determinate | 1998 | Zambia, Malawi, Zimbabwe |
| EL21 | Bimha | Determinate | 1999 | Zimbabwe |
| EL22 | Mhofu | Determinate | 1999 | Zimbabwe |
| EL23 | Nyati | Indeterminate | 1999 | Zimbabwe |
| EL24 | Scorpio | Indeterminate | 2000 | Zimbabwe |
| EL25 | Storm | Determinate | 2000 | Zambia, Malawi, Zimbabwe |
| EL26 | Safari | Indeterminate | 2001 | Zambia, Zimbabwe |
| EL27 | Score | Indeterminate | 2003 | Zambia, Zimbabwe |
| EL28 | Santa | Determinate | 2005 | Zambia, Zimbabwe |
| EL29 | Siesta | Determinate | 2005 | Zimbabwe |
| EL30 | Serenade | Indeterminate | 2005 | Malawi, Zimbabwe |
| EL31 | Sirocco | Indeterminate | 2006 | Zambia, Malawi, Zimbabwe |
| EL32 | Scribe | Indeterminate | 2007 | Zambia, Zimbabwe |
| EL33 | Satellite | Indeterminate | 2007 | Zambia, Zimbabwe |
| EL34 | Squire | Determinate | 2007 | Malawi, Zimbabwe |
| EL35 | Saga | Determinate | 2008 | Zimbabwe |
| EL36 | Sequel | Indeterminate | 2008 | Zimbabwe |
| EL37 | Sputnik | Indeterminate | 2008 | Zimbabwe |
| EL38 | Sovereign | Indeterminate | 2008 | Zimbabwe |
| EL39 | S810/6/10 | Determinate | 2010 | Experimental line |
| EL40 | Semeki | Indeterminate | 2012 | Zambia, Zimbabwe |
| EL41 | Spike | Determinate | 2012 | Zambia, Zimbabwe |
| EL42 | PN 1867 | Determinate | 2012 | Zimbabwe |
| EL43 | PN 1856 | Determinate | 2013 | Malawi, Zambia, Zimbabwe |
| EL44 | PN 891 | Indeterminate | 2005 | Zimbabwe |
| EL45 | Duocrop | Indeterminate | 1999 | Exotic line |
| EL46 | Tulumayo | Determinate | 2013 | Zimbabwe |
| EL47 | SB88 | Indeterminate | 2013 | Exotic line |
| EL48 | SB90 | Determinate | 2013 | Exotic line |
| EL49 | SB92 | Determinate | 2013 | Exotic line |
| Primer name | Forward 5’  3 3 |
Reverse 3’  5 5 |
|---|---|---|
| Satt012 | GCAATTAGTTTTAAAATGTTTC | AGAATAGAGCCTACATATAATCATA |
| Satt148 | AATCCGGGAC-GCAAAATTATTATTAA | TGCAAATTCCCTAATTAACACCCTTTATAC |
| Satt156 | CGCACCCCTCATCCTATGTA | CCAACTAATCCCAGGGACTTACTT |
| Satt172 | AGCCTCCGGTATCACAG | CCTCCTTTCTCCCATTTT |
| Satt180 | TCGCGTTTGTCAGC | TTGATTGAAACCCAACTA |
| Satt182 | GGTCCACATGAAATGAAGGT | TCTCAGCCTGCAAAGAAAA |
| Satt184 | GCGCTATGTAGAT-TATCCAAATTACGC | GCCACTTACTGTTACTCAT |
| Satt215 | GCGCCTTCTTCTGCTAAATCA | CCCATTCAATTGAGATCCAAAATTAC |
| Satt242 | GCGTTGATCAGGTCGAT-TTTTATTTGT | GCGAGTGCCAACTAACTACTTTTATGA |
| Satt294 | GCGGGTCAAATGCAAATTATTTTT | GCGCTCAGTGTGAAAGTTGTTTCTAT |
| Satt372 | CAGAAAAGGAATAA-TAACAACATCAC | GCGAAAACATAATTCACACAAAAGACAG |
| Satt387 | GCGTTACGTTTCAC-TATTTATTTAACAT | GCGGCAGGCTAGCTACATCAAGAG |
| Satt394 | GCGTTTTTTCAATTTAAAGA-GAATTGAC | GCGTAACTTGCATGTGGTATATCGAGATG |
| Satt397 | TCTCGGGATCCTTGTTAGAT | GCGAAGAAGAAGAGAACATGTGAA |
| Satt414 | GCGTATTCCTAGTCACATGC-TATTTCA | GCGTCATAATAATGCCTAGAACATAAA |
| Satt429 | GCGAC-CATCATCTAATCACAATCTACTA | TCCCCATCATTTATCGAAAATAATAATT |
| Satt434 | GCGTTCCGATATACTATA-TAATCCTAAT | GCGGGGTTAGTCTTTTTATTTAACTTAA |
| Satt441 | AAACCCACCCTCAAAAATAAAAA | AAATGCACCCATCAATCACA |
| Satt459 | TCGTGTTAGAT-TTTTACTGTCACATT | AACTGCATACCCTTTGTTTGAA |
| Satt477 | GTTGGGAAAAGGTTACTAC-CATATC | GGTCCGTATGCAATTCTTGACTAATA |
| Satt490 | GCGGCACGAG-TCAACTTTCTGTTTCCT | GCGGAAGAAGATTTTCGTTTTTAT |
| Satt509 | GCGCTACCGTGTGGTGGTGTGC-TACCT | GCGCAAGTGGCCAGCTCATCTATT |
| Satt511 | GCGACTTTACTGAAAACCTGGAAA | GCTTCAAACCAACAAACAACTTA |
| Satt522 | GCGAACTGCCTAGGTTAAAA | TTAGGCGAAATCAACAAT |
| Satt530 | CATGCATATTGACTTCATTATT | CCAAGCGGGTGAAGAGGTTTTT |
| Satt577 | CAAGCTTAAGTCTTGGTCTTCTCT | GGCCTGACCCAAAACTAAGGGAAGTG |
| Satt590 | GCGCG-CATTTTTTAAGTTAATGTTCT | GCGCGAGTTAGCGAATTATTTGTC |
| Satt598 | CGATTTGAATATACTTACCGTC-TATA | CACAATACCTGTGGCTGTTATACTAT |
| Sct_034 | AATTCTCACTCTCACAACTTC | CCATGGGAATAGTTGGGT |
| Sct_067 | CTCCCCATCTCTCTCTAAC | GGATTTTGTTATTTATTTATTTGA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





