Submitted:
07 August 2023
Posted:
08 August 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
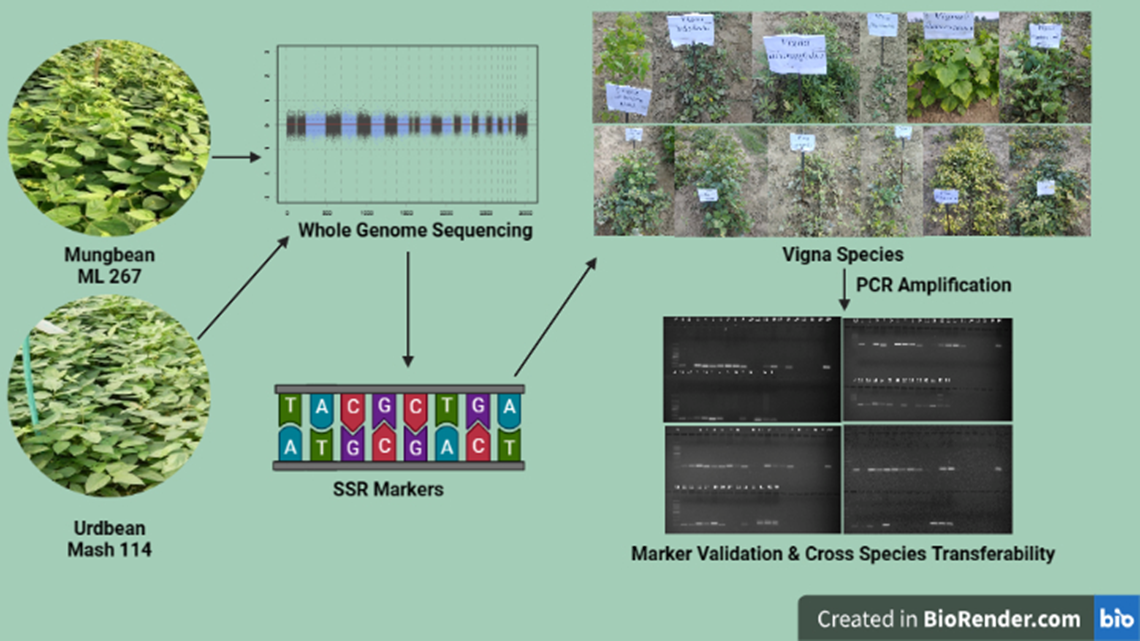
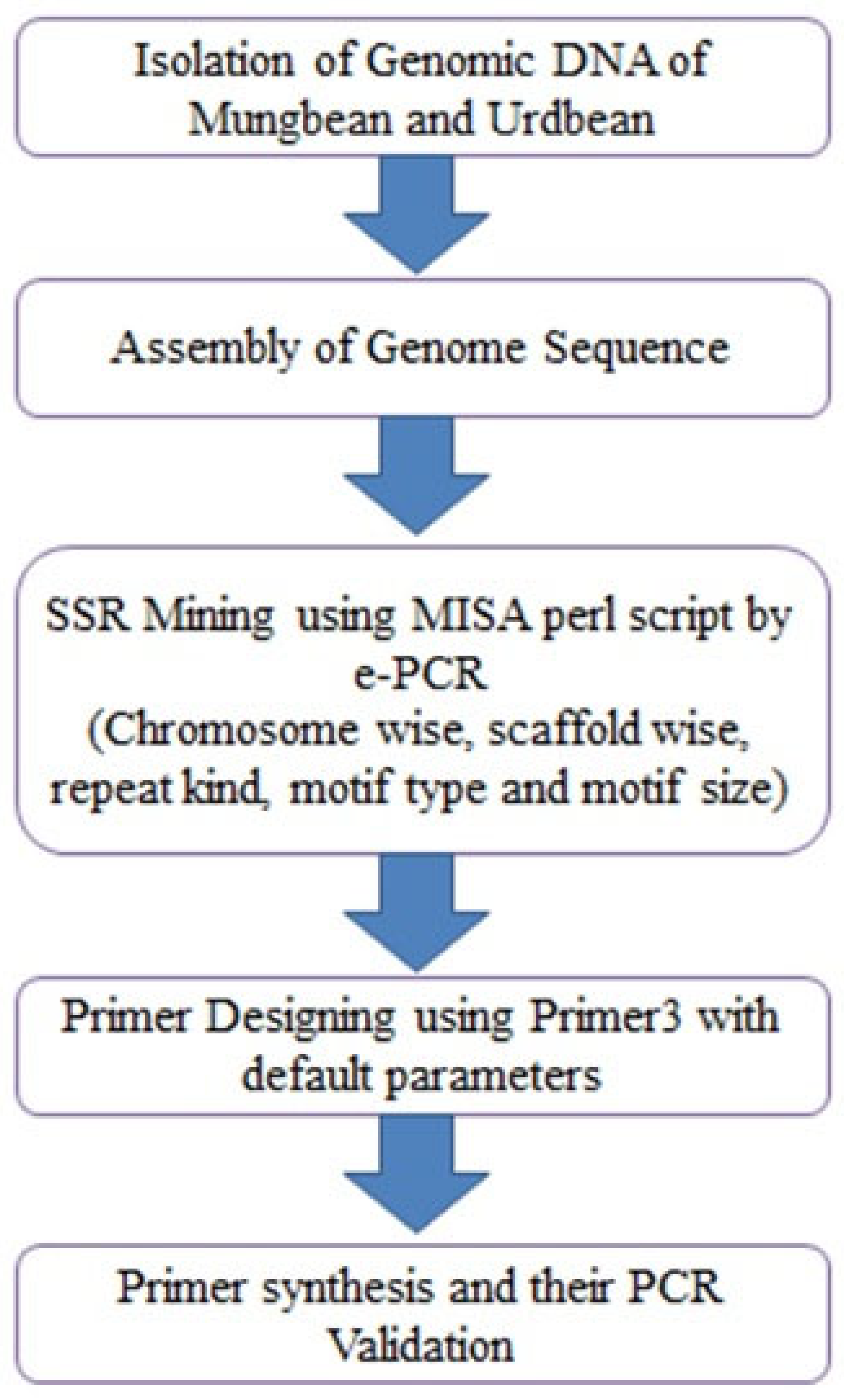
2. Materials and Methods
2.1. Plant Material
2.2. DNA Extraction and Quantification
2.3. SSR Markerdesigning
2.4. SSR Validation
2.5. GeneticDiversity, AMOVA and PCoA in Vigna Species
2.6. Population Structure Analysis
2.7. Data Analysis
2.7.1. Polymorphic Information Content (PIC)
2.7.2. Effective Multiplex Ratio (EMR)
2.7.3. Marker Index (MI)
2.7.4. Resolving Power (RP)
3. Results
3.1. WGS Based SSR Markers Development
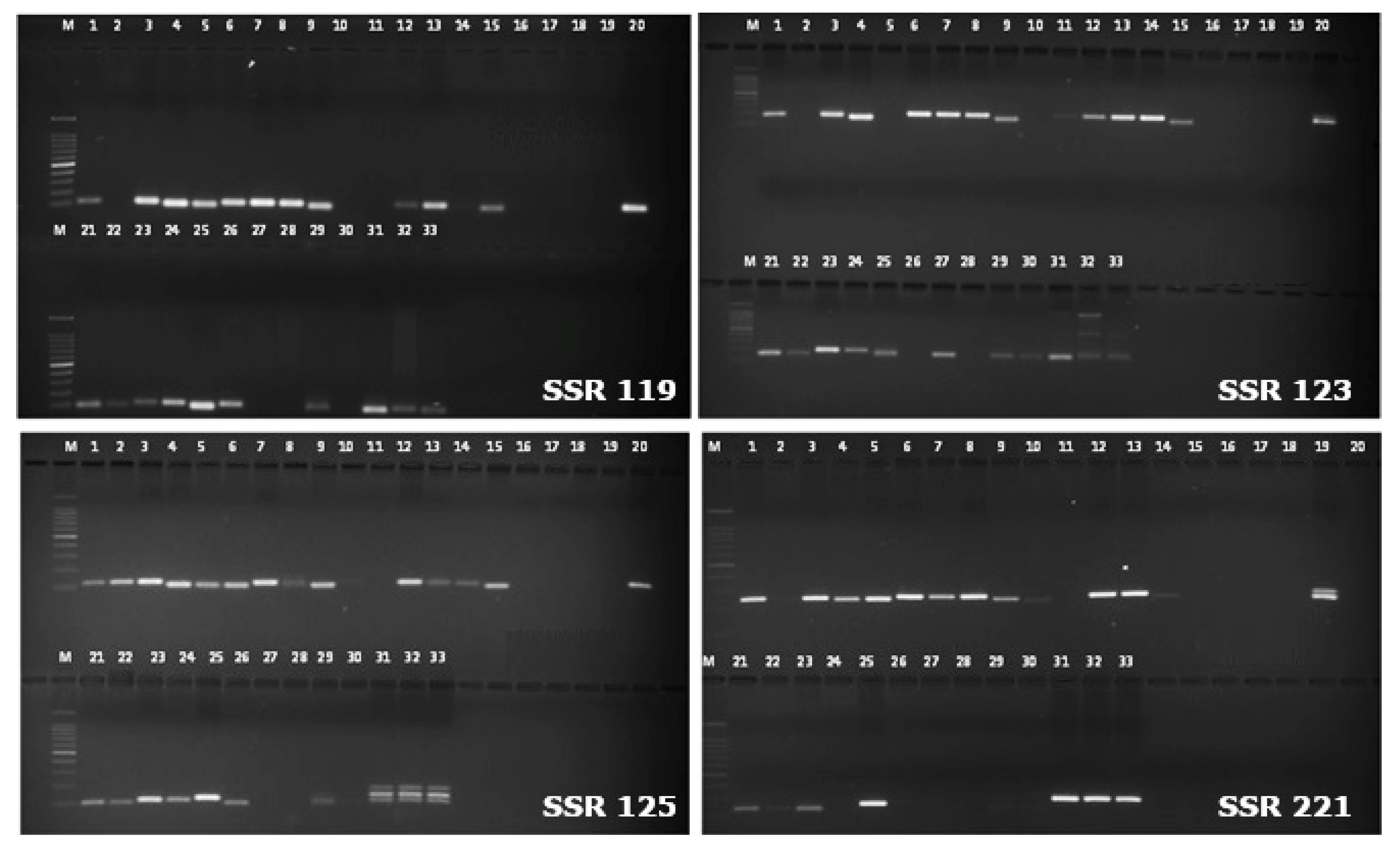
3.2. Validation of SSR Markers on Vigna Species Accession for Transferability Studies
3.3. SSR Marker Analysis
3.4. Genetic Diversity Andrelationship Among the Different Vigna Species
3.5. Population Structure Analysis
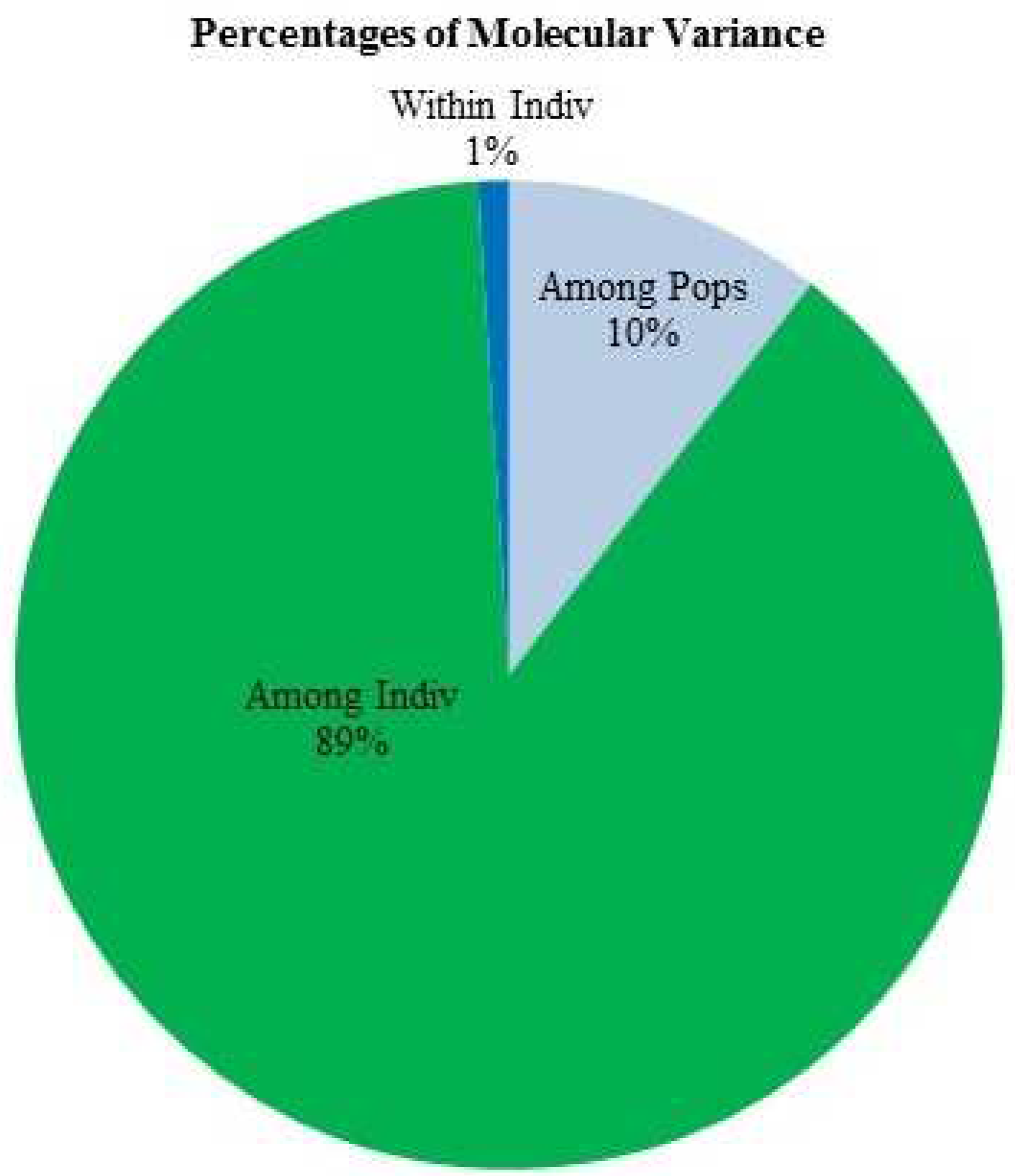
3.6. Analysis of Molecular Variance (AMOVA) and Principal Co-Ordinate Analysis (PCoA)
4. Discussion
5. Conclusions
Authors' Contributions
Declaration of Competing Interest
Acknowledgments
References
- Schrire, B.D. Tribe Phaseoleae. In Legumes of the world. Royal Botanic Gardens; Lewis, G., Schrire, B., Mackinder, B.; Lock, M. Eds.; 2005, pp. 393–431.
- Dikshit, H.K.; Singh, D.; Singh, A.; Jain, N.; Kumari, J.; Sharma, T.R. Utility of adzuki bean [Vigna angularis (Willd.) Ohwi & Ohashi] simple sequence repeat (SSR) markers in genetic analysis of mungbean and related Vigna spp. Afr. J. Biotechnol. 2012, 11, 13261–13268. [Google Scholar] [CrossRef]
- Tomooka, N.; Yoon, M.S.; Doi, K.; Kaga, A.; Vaughan, D. AFLP analysis in diploid species in the genus Vignasub-genus Ceratotropis. Genet. Resour. Crop Evol. 2002, 49, 521–530. [Google Scholar] [CrossRef]
- Takahashi, Y.; Somta, P.; Muto, C.; Iseki, K.; Naito, K.; Pandiyan, M.; Natesan, S.; Tomooka, N. Novel Genetic Resources in the Genus Vigna Unveiled from Gene Bank Accessions. PLoS ONE 2016, 11, e0147568. [Google Scholar] [CrossRef]
- Tomooka, N.; Isemura, T.; Naito, K.; Kaga, A.; Vaughan, D. Vigna species. In Broadening of Genetic Base of Grain Legumes; Singh, M., et al., Eds. Springer: India, 2014; pp. 175–208. [Google Scholar] [CrossRef]
- Bisht, I.S.; Bhat, K.V.; Lakhanpaul, S.; Latha, M.; Jayan, P.K.; Biswas, B.K.; Singh, A.K. Diversity and genetic resources of wild Vigna species in India. Genet. Resour. Crop Evol. 2005, 52, 53–68. [Google Scholar] [CrossRef]
- Akbar, W.; Akhtar, M.A.; Murtaza, G.; Hussain, A.; Javed, H.M.; Arshad, M.; Maqbool, M.A. Mungbean Yellow Mosaic Disease and its Management. J. Agric. Basic Sci. 2019, 4, 34–44. [Google Scholar]
- Kang, Y.J.; Kim, S.K.; Kim, M.Y.; Lestari, P.; Kim, K.H.; Ha, B.K.; Jun, T.H.; et al. Genome sequence of mungbean and insights into evolution within Vigna species. Nat. Commun. 2014, 5, 5443. [Google Scholar] [CrossRef] [PubMed]
- Rohilla, V.; Yadav, R.K.; Poonia. A.; Sheoran, R.; Kumari, G.; Shanmugavadivel, P.S.; Pratap, A. Association Mapping for Yield Attributing Traits and Yellow Mosaic Disease Resistance in Mung Bean [Vignaradiata (L.)Wilczek]. Front. Plant Sci. 2022, 12, 749439. [Google Scholar] [CrossRef] [PubMed]
- Garg, D.D.; Arya, R.S.; Sharma, T.; Dhuria, R.K. Effect of replacement of sewan straw (Lasiruss indicus) by moong (Phaseolus aureus) chara on rumen and haemato-biochemical parameters in sheep. Vet. Pract. 2004, 5, 70–73. [Google Scholar]
- Agboola, A.A.; Fayemi, A.A.A. Fixation and excretion of nitrogen by tropical legumes. Agron. J. 1972, 64, 409–412. [Google Scholar] [CrossRef]
- Chen, X.; Sorajjapinun, W.; Reiwthongchum, S.; Srinives, P. Identification of parental mungbean lines for production of hybrid varieties. Chiang Mai Univ. J. 2003, 2, 97–105. [Google Scholar]
- Yasmeen, T.; Hameed, S.; Tariq, M.; Ali, S. Significance of arbuscularmycorrhizal and bacterial symbionts in a tripartite association with Vignaradiata. Acta Physiologiae Plantarum 2012, 34, 1519–1528. [Google Scholar] [CrossRef]
- Huttner, E. The international mungbean improvement network. In Souvenir International Conference Pulses as the Climate Smart Crops: Challenges and Opportunities at International Convention Centre (Minto Hall); Bhopal, India, 2020, pp. 33–36.
- Nair, R.M.; MahbubulAlam, A.K.M.; Douglas, C.; Gowda, A.; Aditya Pratap; Win, M.M.; Karimi, R.; Emmanuel, M.K.; Binagwa, P.; Boddepalli, V.N.; Chaudhari, S.; et al. Establishing the International Mungbean Improvement Network. Australian Centre for International Agricultural Research. FR2021-086. 2022, Available Online:. Available online: https://www.aciar.gov.au/sites/default/files/2022-03/CIM-2014-079-final-report_0.pdf (accessed on 17 January 2023).
- 16. Annonymous. Project Coordinator’s Report, All India Coordinated Research Project on MULLARP.ICAR-Indian Institute of Pulses Research, Kanpur, 2018.
- Baraki, F.; Gebregergis, Z.; Belay, Y.; Berhe, M.; Zibelo, H. Genotype x environment interaction and yield stability analysis of mungbean (Vigna radiata (L.) Wilczek) genotypes in Northern Ethiopia. Cogent Food Agric. 2020, 6, 1729581. [Google Scholar] [CrossRef]
- Chauhan, Y.S.; Douglas, C.; Rachaputi, R.C.N.; Agius, P.; Martin, W.; King, K.; et al. Physiology of mungbean and development of the mungbean crop model. In: Proceedings of the Ist Australian Summer Grains Conference, Gold Coast, QL. 2010.
- Pratap, A.; Gupta, S.; Basu, P.S.; Tomar, R.; Dubey, S.; Rathore, M.; et al. Towards development of climate smart mungbean: Challenges and opportunities. In Genomic Designing of Climate-Smart Pulse Crops; Springer: Cham, Switzerland, 2019; pp. 235–264. [Google Scholar] [CrossRef]
- Hinz, R.; Sulser, T.B.; Hüfner, R.; Mason-D’Croz, D.; Dunston, S.; Nautiyal, S.; et al. Agricultural development and land use change in India: A scenario analysis of trade-offs between UN sustainable development goals (SDGs). Earth’s Future 2020, 8, e2019EF001287. [Google Scholar] [CrossRef]
- Saini, P. Inheritance Studies and Mapping of Yellow Mosaic Disease Resistance in an Interspecific Cross of Mungbean (Vigna radiata (L.) Wilczek) and Urdbean (Vigna mungo (L.) Hepper). Ph.D. Dissertation, Punjab Agricultural University, Ludhiana, India, 2020; pp. 1–150. [Google Scholar]
- Somta, P.; Musch, W.; Kongsamai, B.; Chanprame, S.; Nakasathien, S.; Toojinda, T.; Sorajjapinun, W.; Seehalak, W.; Tragoonrung, S.; Srinives, P. New microsatellite markers isolated from mungbean (Vignaradiata(L.) Wilczek). Mol. Ecol. Resour. 2008, 8, 1155–1157. [Google Scholar] [CrossRef]
- Lestari, P.; Kim, S.K. ; Reflinur; Kang, Y.J.; Dewi, N.; Lee Suk-Ha. Genetic diversity of mungbean (Vignaradiata L.) germplasm in Indonesia. Plant Genet. Resour. 2014, 12, S91–S94. [Google Scholar] [CrossRef]
- Savithramma, D.L.; Ramakrishnan, C.K.D. Development and characterization of newly developed genomic SSR markers in Mung bean (Vignaradiata(L. In ) Wilczek). In Proceedings of the 2nd International Conference on Genetic & Protein Engineering, Atlanta, GA, USA, 14–16 November 2016; 43p. [Google Scholar]
- Somta, P.; Srinives, P. Genome research in mungbean [Vignaradiata (L.)Wilczek] and blackgram [V. mungo (L.) Hepper]. Science Asia 2007, 1, 69–74. [Google Scholar] [CrossRef]
- Chaitieng, B.; Kaga, A.; Tomooka, N.; Isemura, T.; Kuroda, Y.; Vaughan, D.A. Development of a black gram [Vigna mungo (L.)Hepper] linkage map and its comparison with an azuki bean [Vigna angularis (Willd.) Ohwi and Ohashi] linkage map. Theor. Appl. Genet. 2006, 113, 1261–1269. [Google Scholar] [CrossRef]
- Gupta, S.K.; Gopalakrishna, T. Development of unigene-derived SSR markers in cowpea (Vignaunguiculata) and their transferability to other Vigna species. Genome 2010, 53, 508–523. [Google Scholar] [CrossRef]
- Singh, B.; Das, A.; Parihar, A.K.; Bhagawati, B.; Singh, D.; Pathak, K.N.; Dwivedi, K.; Das, N.; Keshari, N.; Midha, R.L.; et al. Delineation of Genotype-by-Environment interactions for identifcation and validation of resistant genotypes in root-knot nematode (Meloidogyne incognita) using GGE biplot. Sci. Rep. 2020, 10, 4108. [Google Scholar] [CrossRef]
- Kumar, S.V.; Tan, S.G.; Quah, S.C.; Yusoff, K. Isolation of microsatellite markers in mungbean, Vigna radiata. Mol. Ecol. Notes 2002, 2, 96–98. [Google Scholar] [CrossRef]
- Kumar, S.V.; Tan, S.G.; Quah, S.C.; Yusoff, K. Isolation and characterization of seven tetranucleotide microsatellite loci in mungbean, Vigna radiata. Mol. Ecol. Notes 2002, 2, 293–295. [Google Scholar] [CrossRef]
- Miyagi, M.; Humphry, M.; Ma, Z.Y.; Lambrides, C.J.; Bateson, M.; Liu, C.J. Construction of bacterial artificial chromosome libraries and their application in developing PCR-based markers closely linked to a major locus conditioning bruchid resistance in Mungbean [Vigna radiata (L.) Wilczek]. Theor. Appl. Genet. 2004, 110, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Gwag, J.G.; Chung, W.K.; Chung, H.K.; Lee, J.H.; Ma, K.H.; Dixit, A.; Park, Y.J.; Cho, E.G.; Kim, T.S.; Lee, S.H. Characterization of new microsatellite markers in mungbean. Mol. Ecol. Notes 2006, 6, 1132–1134. [Google Scholar] [CrossRef]
- Tangphatsornruang, S.; Somta, P.; Uthaipaisanwong, P.; Chanprasert, P.; Sangsrakru, D.; Seehalak, W.; Sommanas, W.; Tragoonrung, S.; Srinives, P. Characterization of microsatellites and gene contents fromgenome shotgun sequences of mungbean (Vignaradiata(L.) Wilczek). BMC Plant Biology 2009, 9, 137. [Google Scholar] [CrossRef]
- Singh, N. Development of cost efficient and rapid method for developing homozygous SSR markers in mungbean Vignaradiata (L.)Wilczek and its validation in Vigna species. M.Sc. Thesis, . Tamil Nadu Agricultural University, Tamil Nadu, India, 2011. [Google Scholar]
- Singh, N.; Singh, H.; Nagarajan, P. Development of SSR markers in mung bean, Vigna radiata (L.) Wilczek using in silico methods. J. Crop Weed 2013, 9, 69–74. [Google Scholar]
- Shrivastava, D.; Verma, P.; Bhatia, S. Expanding the repertoire of microsatellite markers for polymorphism studies in Indian accessions of mungbean (Vigna radiata L. Wilczek). Mol. Biol. Rep. 2014, 41, 5669–5680. [Google Scholar] [CrossRef]
- Bangar, P.; Chaudhary, A.; Umdale, S.; Kumari, R.; Tiwari, B.; Kumar, S.; Gaikwad, A.B.; Bhat, K.V. Detection and characterization of polymorphic simple sequence repeats markers for the analysis of genetic diversity in Indian mungbean [Vigna radiata (L.) Wilczek]. Indian J. Genet. 2018, 78, 111–117. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Thakur, S. Whole genome de novo assembly of Vigna mungo and Vigna radiata and In silico comparative analysis for marker development. Ph.D. Dissertation, Punjab Agricultural University, Ludhiana, India, 2018. [Google Scholar]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research: An update. Bioinformatics 2012, 28, 2537e2539. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Earl, D.A. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Perrier, X.; Jacquemoud-Collet, J.P. DARwin software: http://darwin. cirad. fr/darwin. 5 edn.Cirad, Montpellier. 2006.
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphism. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar] [PubMed]
- Powell, W.; Morgante, M.; Andre, C.; Hanafey, M.; Vogel, J.; Tingey, S.; Rafalski, A. The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol. Breed. 1996, 2, 225–238. [Google Scholar] [CrossRef]
- Prevost, A.; Wilkinson, M.J. A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Ther. Appl. Genet. 1999, 98, 107–112. [Google Scholar] [CrossRef]
- Nayak, S.N.; Song, J.; Villa, A.; Pathak, B.; Ayala-Silva, T.; Yang, X.; Todd, J.; Glynn, N.C.; Kuhn, D.N.; Glaz, B.; et al. Promoting utilization of Saccharum spp. genetic resources through genetic diversity analysis and core collection construction. PLoS ONE 2014, 9, 110856. [Google Scholar] [CrossRef]
- Nass, L.L.; Paterniani, E. Pre-breeding: A link between genetic resources and maize breeding. Scientia Agricola 2000, 57, 581–587. [Google Scholar] [CrossRef]
- Haussmann, B.I.G.; Parzies, H.K.; Presterl, T.; Susic, Z.; Mieddaner, T. Plant genetic resources in crop improvement. Plant Genet. Resour. 2004, 2, 3–21. [Google Scholar] [CrossRef]
- Acosta-Gallegos, J.; Kelly, J.D.; Gepts, P. Prebreeding in common bean and use of genetic diversity from wild germplasm. Crop Sci. 2007, 47, 44–59. [Google Scholar] [CrossRef]
- Shimelis, H.; Laing, M. Timelines in conventional crop improvement: Pre-breeding and breeding procedures. Aus. J. Crop Sci. 2012, 11, 1542–1549. [Google Scholar]
- Sharma, S.; Upadhyaya, H.D.; Varshney, R.K.; Gowda, C.L.L. Pre-breeding for diversification of primary gene pool and genetic enhancement of grain legume. Front. Plant Sci. 2013, 4, 1–14. [Google Scholar] [CrossRef]
- Kumar, V.; Shukla, Y.M. Pre-breeding: Its applications in crop improvement. Research News For U (RNFU) 2014, 16, 199–202. [Google Scholar]
- Meena, A.K.; Gurjar, D.; Kumhar, B.L. Pre-breeding is a bridge between wild species and improved genotypes-a review. Chem. Sci. Rev. Lett. 2017, 6, 1141–1151. [Google Scholar]
- Jain, S.K. ; Omprakash Pre-breeding: A Bridge between Genetic Resources and Crop Improvement. Int. J. Curr. Microbiol. App. Sci. 2019, 8, 1998–2007. [Google Scholar] [CrossRef]
- Singh, R.B.; Singh, B.; Singh, R.K. Evaluation of genetic diversity in Saccharum species clones and commercial varieties employing molecular (SSRs) and physiological markers. Indian J. Plant Genet. Resour. 2018, 31, 17–26. [Google Scholar] [CrossRef]
- Wang, M.L.; Barkley, N.A.; Gillaspie, G.A.; Pederson, G.A. Phylogenetic relationships and genetic diversity of the USDA Vigna germplasm collection revealed by gene-derived markers and sequencing. Genet. Res. 2008, 90, 467–480. [Google Scholar] [CrossRef]
- Blair, M.W.; Hurtado, N.; Chavarro, C.M.; Munoz-Torres, M.C.; Pedraza, M.F.; Tomins, J.; Wing, R. Gene-based SSR markers for common bean (Phaseolus vulgaris L.) derived from root and leaf tissue ESTs: An integration of the BMC series. BMC Plant Biol. 2011, 11, 50. [Google Scholar] [CrossRef]
- Gupta, S.K.; Bansal, R.; Gopalakrishna, T. Development and characterization of genic SSR markers for mungbean (Vigna radiate (L.) Wilczek). Euphytica 2014, 195, 245–258. [Google Scholar] [CrossRef]
- Chen, H.; Qiao, L.; Wang, L.; Wang, S.; Blair, M.W.; Cheng, X. Assessment of genetic diversity and population structure of mungbean (Vigna radiata) germplasm using EST-based and genomic SSR markers. Gene 2015, 566, 175–183. [Google Scholar] [CrossRef]
- Gong, Y.M.; Xu, S.C.; Mao, W.H.; Hu, Q.Z.; Zhang, G.W.; Ding, J.; Li, Y.D. Developing new SSR markers from ESTs of pea (Pisum sativum L.). J. Zhejiang Univ. Sci. B. 2010, 11, 702–707. [Google Scholar] [CrossRef]
- Choudhary, S.; Sethy, N.K.; Shokeen, B.; Bhatia, S. Development of chickpea EST-SSR markers and analysis of allelic variation across related species. Theor. Appl. Genet. 2009, 118, 591–608. [Google Scholar] [CrossRef]
- Blair, M.W.; Hurtado, N. EST-SSR markers from five sequenced cDNA libraries of common bean (Phaseolus vulgaris L.) comparing three bioinformatic algorithms. Mol. Ecol. Resour. 2013, 13, 688–695. [Google Scholar] [CrossRef]
- Bhardwaj, J.; Chauhan, R.; Swarnkar, M.K.; Chahota, R.K.; Singh, A.K.; Shankar, R.; Yadav, S.K. Comprehensive transcriptomic study on horse gram (Macrotylomauniflorum): De novo assembly, functional characterization and comparative analysis in relation to drought stress. BMC Genomics 2013, 14, 647. [Google Scholar] [CrossRef] [PubMed]
- Jasrotia, R.S.; Yadav, P.K.; Iquebal, M.A.; Bhatt, S.B.; Arora, V.; Angadi, U.B.; Tomar, R.S.; Jaiswal, S.; Rai, A.; Kumar, D. VigSatDB: Genome-wide microsatellite DNA marker database of three species of Vigna for germplasm characterization and improvement. Database 2019, 1–13. [Google Scholar] [CrossRef]
- Chen, H.; Liu, L.; Wang, L.; Wang, S.; Somta, P.; Cheng, X. Development and validation of EST SSR markers from the transcriptome of adzuki bean (Vigna angularis). PLoS ONE 2015, 10, e0131939. [Google Scholar] [CrossRef] [PubMed]
- Luro, F.L.; Constantino, G.; Terol, J.; Argout, X.; Allario, T.; Wincker, P.; Talon, M.; Ollitrault, P.; Morillon, R. Transferability of the ESR-SSRs developed on Nules clementine (Citrus Clementine Hort ex Tan) to other Citrus species and their effectiveness for genetic mapping. BMC Genomics 2008, 9, 287. [Google Scholar] [CrossRef] [PubMed]
- Mnejja, M.; Garcia-Mas, J.; Audergon, J.M.; Arús, P. Prunus microsatellite marker transferability across rosaceous crops. Tree Genet. Genomes 2010, 6, 689–700. [Google Scholar] [CrossRef]
- Somta, P.; Seehalak, W.; Srinives, P. Development, characterization and cross-species amplification of mungbean (Vignaradiata) genic microsatellite markers. Conserv.Genet. 2009, 10, 1939–1943. [Google Scholar] [CrossRef]
- Gupta, S.K.; Bansal, R.; Vaidya, U.J.; Gopalakrishna, T. Development of EST-derived microsatellite markers in mungbean [Vigna radiata (L.) Wilczek] and their transferability to other Vignaspecies. Indian J. Genet. 2012, 72, 468–471. [Google Scholar]
- Singh, A.; Dikshit, H.K.; Jain, N.; Singh, D.; Yadav, R.N. Efficiency of SSR, ISSR and RAPD markers in molecular characterization of mungbean and other Vigna species. Indian J. Biotechnol. 2014, 13, 81–88. [Google Scholar]
- Satinder Kaur; Gill, R. K.; Bains, T.S.; Kaur, M.; Thakur, S. Comparative assessment of SSR markers derived from different sources in genetic diversity analysis of Vigna genotypes. Agric. Res. J. 2017, 54, 462–468. [Google Scholar] [CrossRef]
- Simranjit Kaur; Bains, T. S.; Sirari, A.; Kaur, S. Evaluation and molecular characterization of advanced interspecific lines for genetic improvement in mungbean [Vigna radiata (L.) Wilczek]. Legume Res. 2019, 42, 729–735. [Google Scholar] [CrossRef]
- Pratap, A.; Gupta, S.; Malviya, N.; Tomar, R.; Maurya, R.; John, K.J.; et al. Genome scanning of Asiatic Vigna species for discerning population genetic structure based on microsatellite variation. Mol. Breed. 2015, 35, 178. [Google Scholar] [CrossRef]
- GeetaKumari; RoopaLavanya, G. ; Shanmugavadivel, P.S.; Singh, Y.; Singh, P.; Patidar, B.; Madhavan, L.; Gupta, S.; Singh, N.P.; Pratap, A. Genetic diversity and population genetic structure analysis of an extensive collection of wild and cultivated Vigna accessions. Mol. Genet. Genomics, 2021, 296, 1337–1353. [Google Scholar] [CrossRef] [PubMed]
- Dachapak, S.; Somta, P.; Poonchaivilaisak, S.; Yimram,T. ; Srinives, P. Genetic diversity and structure of the zombi pea (Vignavexillata (L.) A. Rich) gene pool based on SSR marker analysis. Genetica 2017, 145, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Sarr, A.; Bodian, A.; Gbedevi, K.M.; et al. Genetic Diversity and population structure analyses of wild relatives and cultivated cowpea (Vigna unguiculata(L.) Walp.)fromsenegal using simple sequence repeat markers. Plant Mol. Biol. Rep. 2020. [Google Scholar] [CrossRef]
- Noble, T.J.; Tao, Y.; Mace, E.S.; Williams, B.; Jordan, D.R.; Douglas, C.A.; Mundree, S.G. Characterization of linkage disequilibrium and population structure in a mungbean diversity panel. Front. Plant Sci. 2018, 8, 2102. [Google Scholar] [CrossRef]
- Singh, R.B.; Mahenderakar, M.D.; Jugran, A.K.; Singh, R.K.; Srivastava, R.K. Assessing genetic diversity and population structure of sugarcane cultivars, progenitor species and genera using microsatellite (SSR) markers. Gene 2020, 753, 144800. [Google Scholar] [CrossRef]
- Xiong, H.; Shi, A.; Mou, B.; Qin, J.; Motes, D.; Lu, W.; Ma, J.; Weng, Y.; Yang, W.; Wu, D. Genetic diversity and population structure of cowpea (Vigna unguiculata L. Walp). PLoS ONE 2016, 11, e0160941. [Google Scholar] [CrossRef]
- Chen, H.; Chen, H.; Hu, L.; Wang, L.; Wang, S.; Wang, M.L.; Cheng, X. Genetic diversity and a population structure analysis of accessions in the Chinese cowpea [Vigna unguiculata(L.) Walp.] germplasm collection. The Crop J. 2017, 5, 363–372. [Google Scholar] [CrossRef]
- Luo, Z.; Brock, J.; Dyer, J.M.; Kutchan, T.; Schachtman, D.; Augustin, M.; Ge, Y.; Fahlgren, N.; Abdel-Haleem, H. Genetic Diversity and Population Structure of a Camelina sativa Spring Panel. Front. Plant Sci. 2019, 10, 184. [Google Scholar] [CrossRef]
- Saxena, S.; Kole, P.R.; Bhat, K.V.; Pandey, S.P. Species diversity and relationship among Vigna radiata (L.) wilczek and its close wild relatives. Bangladesh J. Botany 2016, 45, 567–573. [Google Scholar]
- Pandiyan, M.; Senthil, N.; Ramamoorthi, N.; Muthiah, A.R.; Tomooka, N.; Duncan, V. Interspecifc hybridization of Vigna radiata × 13 wild Vigna species for developing MYMV donor. Electron. J. Plant Breed. 2010, 1, 600–610. [Google Scholar]
- Kumar, S.; Gupta, S.; Chandra, S.; Singh, B.B. How wide is the genetic base of pulse crops. In Pulses in New Perspective; Ali, M., Singh, B.B., Kumar, S., Dhar, V., Eds.; Indian Society of Pulses Research and Development: Kanpur, India, 2004; pp. 211–221. [Google Scholar]
- Frankham, R.; Ballou, J.D.; Briscoe, D.A. Introduction to Conservation Genetics; Cambridge University Press: Cambridge, UK, 2002. [Google Scholar] [CrossRef]
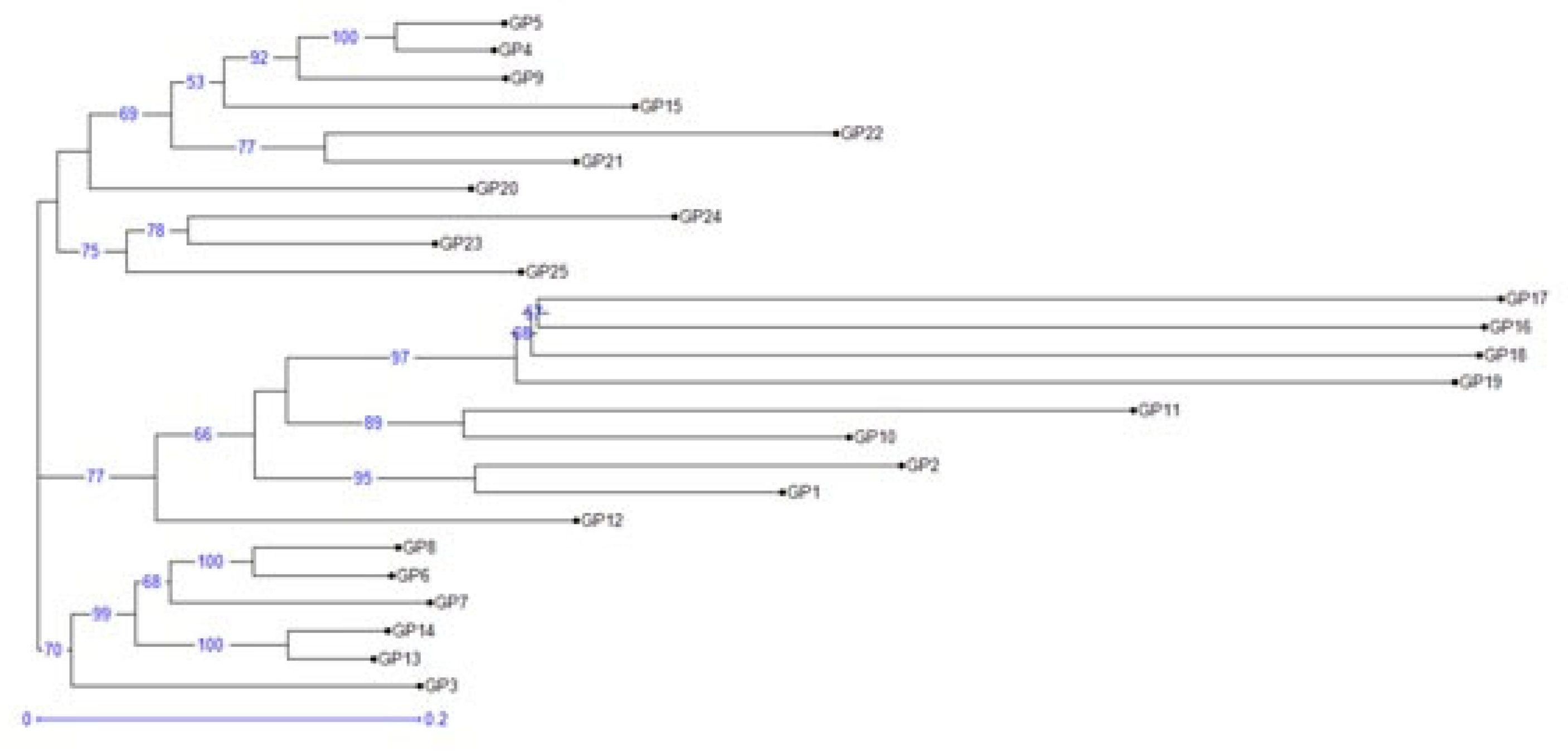
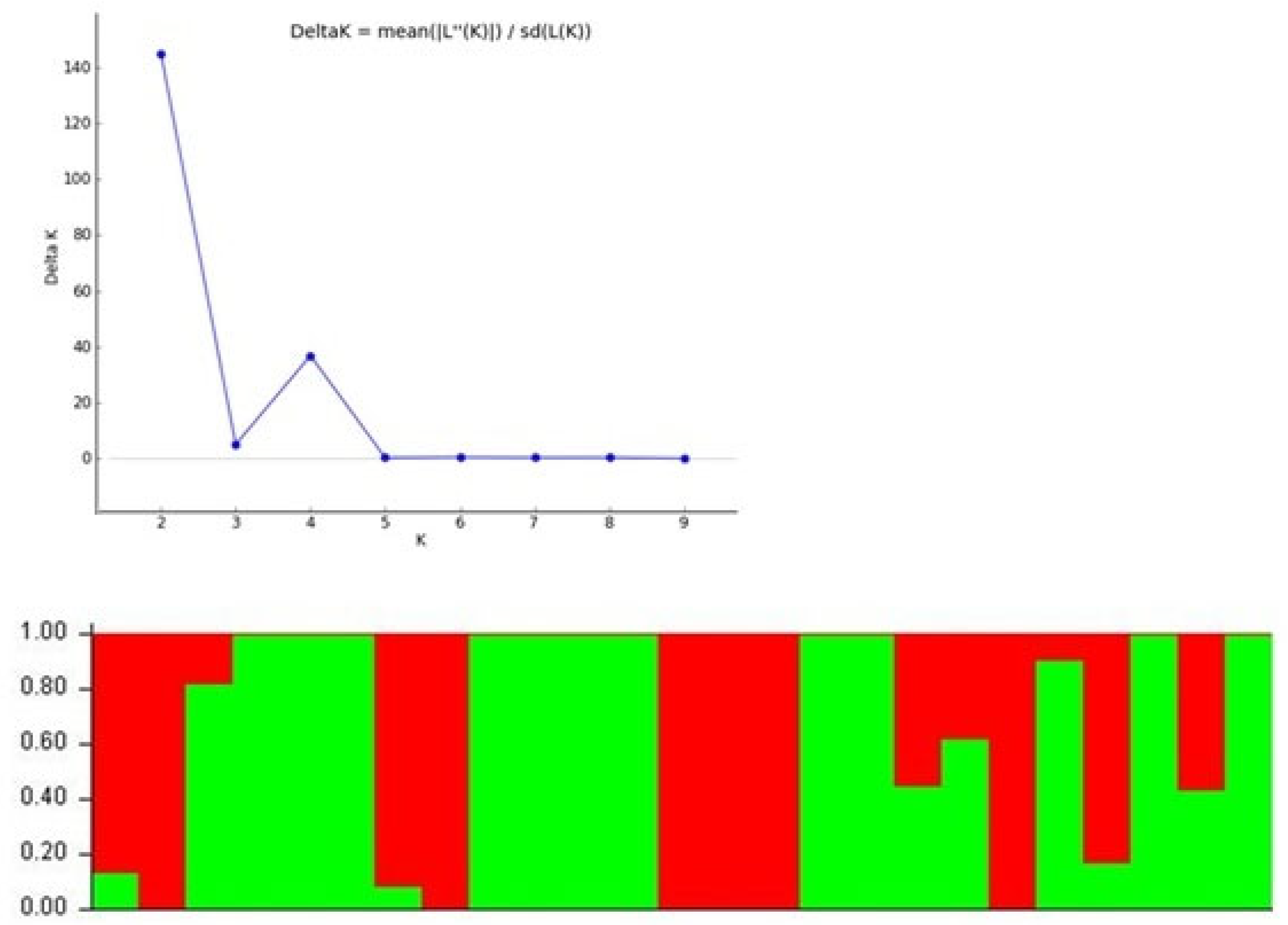
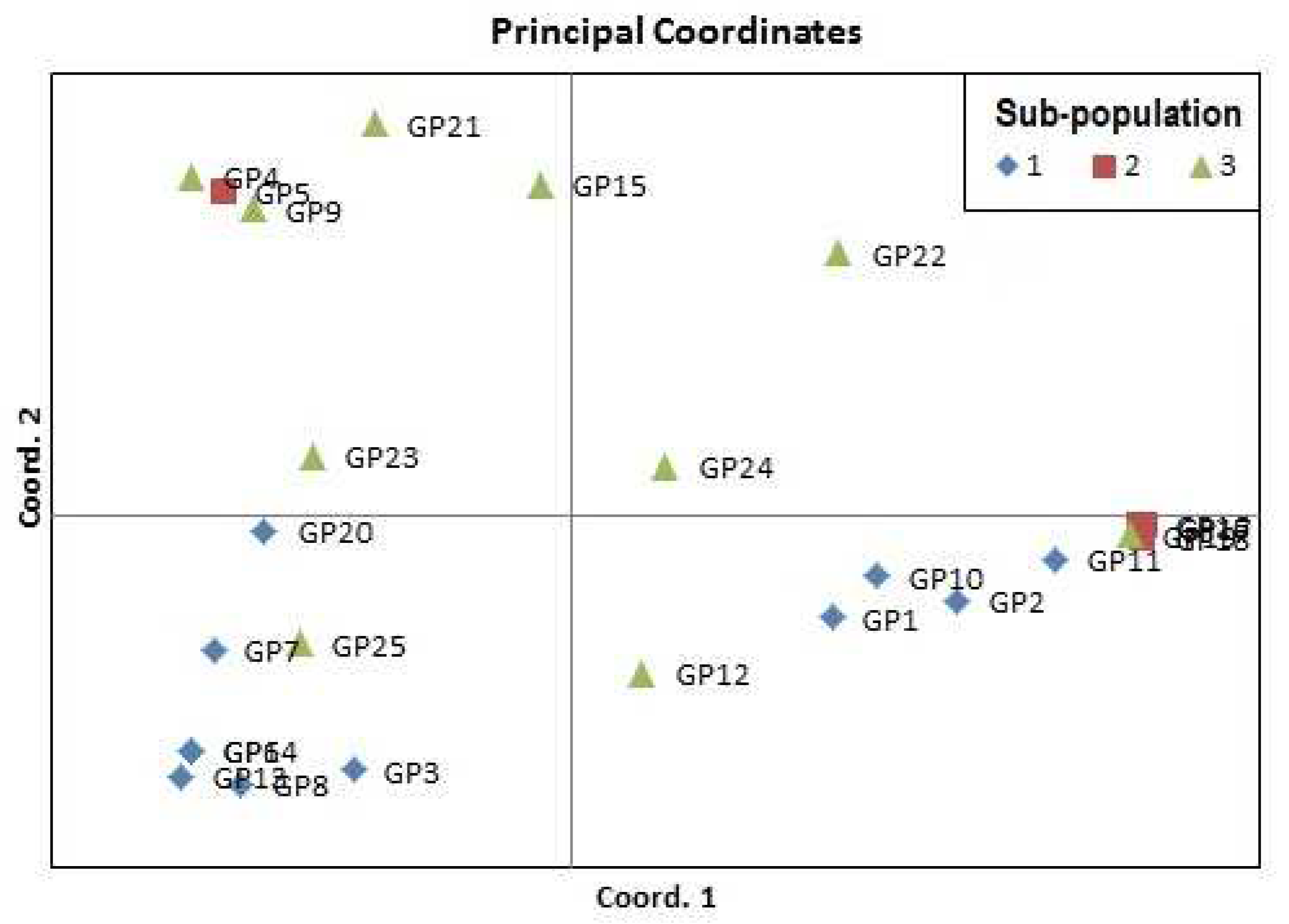
| Designation | Accessions | Designation | Accessions |
|---|---|---|---|
| GP1 | V. umbellata(Cultivated) | GP14 | V. stipulacea |
| GP2 | V.umbellata(Cultivated) | GP15 | V. radiatavar.radiata |
| GP3 | V. umbellata | GP16 | V. radiatavar.mungo |
| GP4 | V. sublobata | GP17 | V. radiatavar.mungo |
| GP5 | V. sublobata | GP18 | V. radiatavar.mungo |
| GP6 | V. trilobata | GP19 | V. slyestris |
| GP7 | V. trilobata | GP20 | V. glabrescence |
| GP8 | V. trilobata | GP21 | V. radiatavar.satulosa |
| GP9 | V. trilobata | GP22 | V. vexillata |
| GP10 | V. aconitifolia | GP23 | V. hainiana |
| GP11 | V. aconitifolia | GP24 | V. dalzelliana |
| GP12 | V. aconitifolia(TMV-1) | GP25 | V. unguiculata |
| GP13 | V. stipulacea | ||
| Parameters | Number of SSR | |
|---|---|---|
| V. radiata cv. ML267 | V. mungo cv. Mash114 | |
| SSR Mining | ||
| SSR sequences examined | 444,059 | 471,725 |
| SSRs identified | 225,359 | 218,508 |
| SSR containing sequences | 130,125 | 126,749 |
| Sequences containing more than 1 SSR | 50,760 | 46,626 |
| SSRs present in compound formation | 16,201 | 15,565 |
| Repeat Typea | ||
| Mononucleotide | 173,536 (77%) | 170,071 (77.83%) |
| Dinucleotide | 29,559 (13.12%) | 27,625 (12.64%) |
| Trinucleotide | 19,732 (8.76%) | 18,490 (8.46%) |
| Tetranucleotide | 1939 (0.86%) | 1794 (0.82%) |
| Pentanucleotide | 410 (0.18%) | 369 (0.17%) |
| Hexanucleotide | 183 (0.08%) | 159 (0.08%) |
| Dinucleotide repeat | n | Number | Percentage |
|---|---|---|---|
| (AT)n | 6, 7, 8, 9, 10, 11, 12, 13, 14, 17 | 69 | 27.6 |
| (AG)n | 6, 7, 8, 9, 10, 13, 14, 16, 17, 22 | 30 | 12.0 |
| (AC)n | 6, 7, 8, 9 | 16 | 6.40 |
| (TA)n | 6, 7, 8, 9, 10, 11, 12, 13, 20 | 62 | 24.8 |
| (TC)n | 6, 7. 8, 9, 10, 11, 13, 14, 16, 17 | 31 | 12.4 |
| (TG)n | 6, 7, 9 | 11 | 4.40 |
| (CT)n | 6, 7, 8, 9, 12, 14 | 13 | 5.20 |
| (CA)n | 6,7 | 02 | 0.80 |
| (GA)n | 6, 7, 8, 10, 12. 19, 20 | 10 | 4.00 |
| (GT)n | 6, 7, 14 | 06 | 2.40 |
| Pop ID | Pop 1 | Pop 2 | Pop 3 |
|---|---|---|---|
| Pop 2 | 0.374 | - | - |
| Pop 3 | 0.208 | 0.442 | - |
| Pop 4 | 0.250 | 0.458 | 0.189 |
| Source of Variation | Df | Sum of square | Mean Sum of square | Estimated Variance | % Variance | F statistics |
|---|---|---|---|---|---|---|
| Among population | 2 | 470.62 | 235.31 | 7.24 | 10 | 0.105 |
| Among individual | 22 | 2706.18 | 123.00 | 61.15 | 89 | 0.989 |
| Within individual | 25 | 17.50 | 0.700 | 0.70 | 1 | 0.990 |
| Total | 49 | 3194.30 | 69.09 | 100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





