Submitted:
24 June 2024
Posted:
25 June 2024
Read the latest preprint version here
Abstract
Keywords:
1. Introduction
2. Materials and Methods
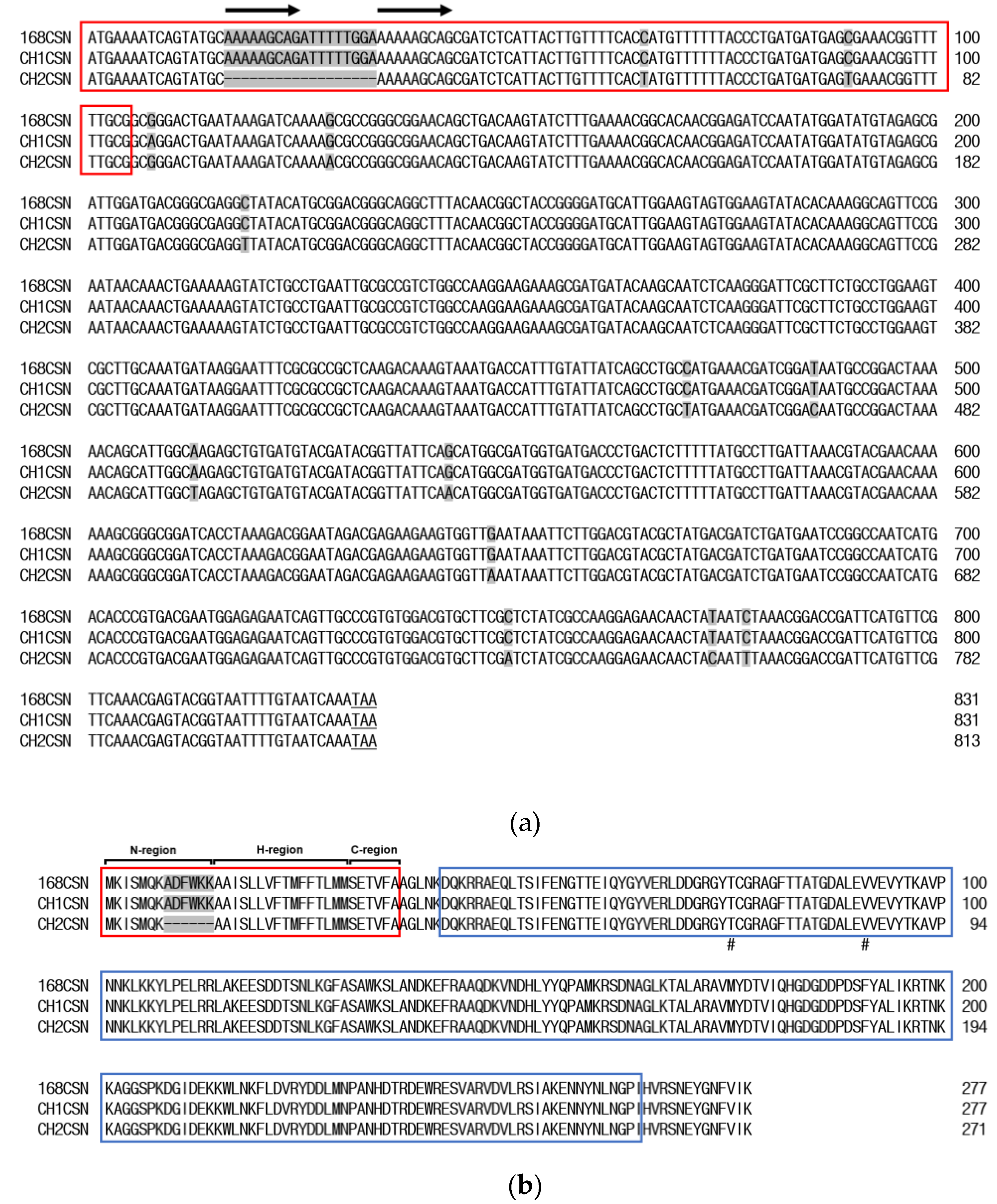
2.1. Molecular Characterization of Chitosanase
2.2. Genomic DNA Extraction
2.3. Cloning of Chitosanase
2.4. Overexpression of Chitosanase in E. coli
2.5. Protein Measurement
2.6. Statistical Analysis
3. Results
3.1. Molecular Characterization of Chitosanase
3.2. Expression and Extracellular Secretion Analysis of Chitosanase in E. coli
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
References
- Yoon, S.H.; Kim, S.K.; Kim, J.F. Secretory production of recombinant proteins in Escherichia coli. Recent Pat Biotechnol 2010, 4, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Baneyx, F.; Mujacic, M. Recombinant protein folding and misfolding in Escherichia coli. Nat Biotechnol 2004, 22, 1399–1408. [Google Scholar] [CrossRef] [PubMed]
- Mergulhao, F.J.; Summers, D.K.; Monteiro, G.A. Recombinant protein secretion in Escherichia coli. Biotechnol Adv 2005, 23, 177–202. [Google Scholar] [CrossRef] [PubMed]
- Mergulhao, F.J.; Monteiro, G.A. Periplasmic targeting of recombinant proteins in Escherichia coli. Methods Mol Biol 2007, 390, 47–61. [Google Scholar] [CrossRef] [PubMed]
- Mierendorf, R.C.; Morris, B.B.; Hammer, B.; Novy, R.E. Expression and Purification of Recombinant Proteins Using the pET System. Methods Mol Med 1998, 13, 257–292. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.H.; Lee, S.Y. Secretory and extracellular production of recombinant proteins using Escherichia coli. Appl Microbiol Biotechnol 2004, 64, 625–635. [Google Scholar] [CrossRef] [PubMed]
- Beckwith, J. The Sec-dependent pathway. Res Microbiol 2013, 164, 497–504. [Google Scholar] [CrossRef] [PubMed]
- Crane, J.M.; Randall, L.L. The Sec System: Protein Export in Escherichia coli. EcoSal Plus 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Yabuki, M.; Uchiyama, A.; Suzuki, K.; Ando, A.; Fujii, T. Purification and Properties of Chitosanase from Bacillus-Circulans Mh-K1. J Gen Appl Microbiol 1988, 34, 255–270. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, W.; Yuan, H.; Wang, Y. Characterization of a novel fungal chitosanase Csn2 from Gongronella sp. JG. Carbohydr Res 2008, 343, 2583–2588. [Google Scholar] [CrossRef]
- Oh, C.-H.; Lee, J.-H. Isolation, purification and characterization of chitosanase from Bacillus subtilis CH1. Journal of Aquaculture 2006, 19, 40–46. [Google Scholar]
- Oh, C.; De Zoysa, M.; Kang, D.H.; Lee, Y.; Whang, I.; Nikapitiya, C.; Heo, S.J.; Yoon, K.T.; Affan, A.; Lee, J. Isolation, purification, and enzymatic characterization of extracellular chitosanase from marine bacterium Bacillus subtilis CH2. J Microbiol Biotechnol 2011, 21, 1021–1025. [Google Scholar] [CrossRef] [PubMed]
- Teufel, F.; Almagro Armenteros, J.J.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nature biotechnology 2022, 40, 1023–1025. [Google Scholar] [CrossRef] [PubMed]
- Freudl, R. Signal peptides for recombinant protein secretion in bacterial expression systems. Microb Cell Fact 2018, 17. [Google Scholar] [CrossRef] [PubMed]
- Brockmeier, U.; Caspers, M.; Freudl, R.; Jockwer, A.; Noll, T.; Eggert, T. Systematic screening of all signal peptides from:: A powerful strategy in optimizing heterologous protein secretion in gram-positive bacteria. Journal of Molecular Biology 2006, 362, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Bzymek, M.; Lovett, S.T. Instability of repetitive DNA sequences: the role of replication in multiple mechanisms. Proc Natl Acad Sci U S A 2001, 98, 8319–8325. [Google Scholar] [CrossRef] [PubMed]
- Rusch, S.L.; Kendall, D.A. Interactions that drive Sec-dependent bacterial protein transport. Biochemistry 2007, 46, 9665–9673. [Google Scholar] [CrossRef] [PubMed]
- Owji, H.; Nezafat, N.; Negandaripour, M.; Hajiebrahimi, A.; Ghasemi, Y. A comprehensive review of signal peptides: Structure, roles, and applications. Eur J Cell Biol 2018, 97, 422–441. [Google Scholar] [CrossRef] [PubMed]
- Vale, F.F.; Lehours, P.; Yamaoka, Y. Editorial: The Role of Mobile Genetic Elements in Bacterial Evolution and Their Adaptability. Frontiers in Microbiology 2022, 13. [Google Scholar] [CrossRef]
- Baldi, L.; Hacker, D.L.; Adam, M.; Wurm, F.M. Recombinant protein production by large-scale transient gene expression in mammalian cells: state of the art and future perspectives. Biotechnology letters 2007, 29, 677–684. [Google Scholar] [CrossRef]
- Beena, K.; Udgaonkar, J.B.; Varadarajan, R. Effect of signal peptide on the stability and folding kinetics of maltose binding protein. Biochemistry 2004, 43, 3608–3619. [Google Scholar] [CrossRef] [PubMed]
- Pechsrichuang, P.; Songsiriritthigul, C.; Haltrich, D.; Roytrakul, S.; Namvijtr, P.; Bonaparte, N.; Yamabhai, M. OmpA signal peptide leads to heterogenous secretion of B. subtilis chitosanase enzyme from E. coli expression system. Springerplus 2016, 5, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Ma, J.; Zhang, Y.; Zhang, M.; Wu, M.; Dai, Z.; Jiang, M. Efficient Secretory Overexpression of Endoinulinase in Escherichia coli and the Production of Inulooligosaccharides. Appl Biochem Biotechnol 2016, 179, 880–894. [Google Scholar] [CrossRef] [PubMed]
- Forster, S.; Brandt, M.; Mottok, D.S.; Zschuttig, A.; Zimmermann, K.; Blattner, F.R.; Gunzer, F.; Pohlmann, C. Secretory expression of biologically active human Herpes virus interleukin-10 analogues in Escherichia coli via a modified Sec-dependent transporter construct. BMC Biotechnol 2013, 13, 82. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Liu, H.; Gao, S.; Weng, Y.; Zhu, L. Enhanced Extracellular Production of IsPETase in Escherichia coli via Engineering of the pelB Signal Peptide. J Agric Food Chem 2021, 69, 2245–2252. [Google Scholar] [CrossRef] [PubMed]
- Miksch, G.; Ryu, S.; Risse, J.M.; Flaschel, E. Factors that influence the extracellular expression of streptavidin in Escherichia coli using a bacteriocin release protein. Applied microbiology and biotechnology 2008, 81, 319–326. [Google Scholar] [CrossRef]
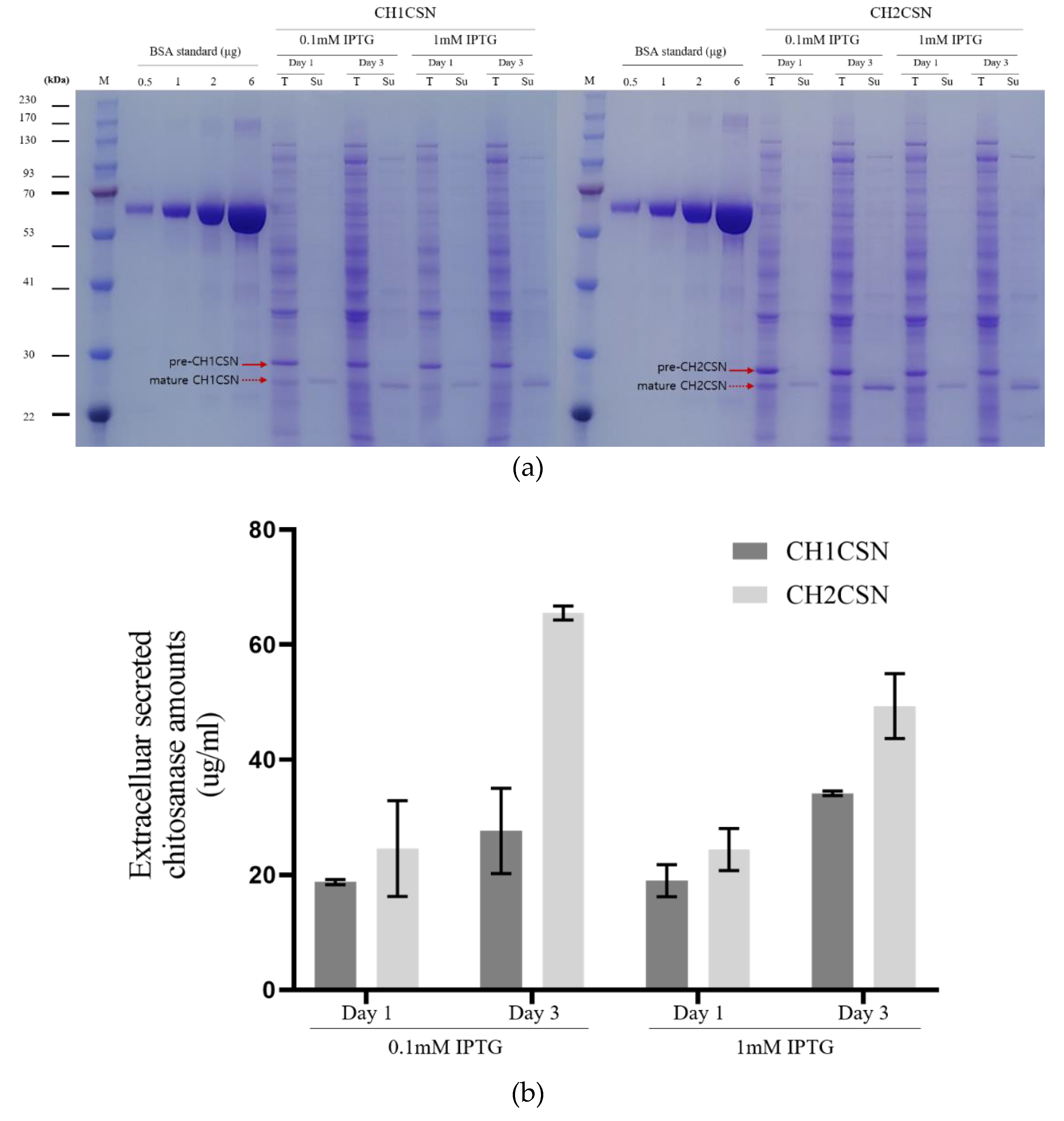
| 0.1mM IPTG | 1mM IPTG | ||||
|---|---|---|---|---|---|
| day 1 | day 3 | day 1 | day 3 | ||
|
CH1CSN (ug/ml) |
Cytoplasm | 48.04 ± 0.87 | 38.80 ± 2.87 | 47.08 ± 1.45 | 33.51 ± 0.51 |
| Periplasm | 16.75 ± 0.39 | 9.73 ± 2.03 | 6.79 ± 5.99 | 6.28 ± 3.75 | |
| Supernatant | 18.75 ± 0.44 | 27.60 ± 7.39 | 18.95 ± 2.77 | 34.12 ± 0.38 | |
|
CH2CSN (ug/ml) |
Cytoplasm | 60.70 ± 1.92 | 51.80 ± 1.93 | 41.08 ± 1.90 | 28.49 ± 2.82 |
| Periplasm | 22.58 ± 1.48 | 10.78 ± 1.25 | 15.54 ± 1.51 | 5.96 ± 1.21 | |
| Supernatant | 24.52 ± 8.29 | 65.48 ± 1.22 | 24.37 ± 3.65 | 49.31 ± 5.65 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





