Submitted:
21 June 2024
Posted:
24 June 2024
You are already at the latest version
Abstract
Keywords:
Introduction
Results
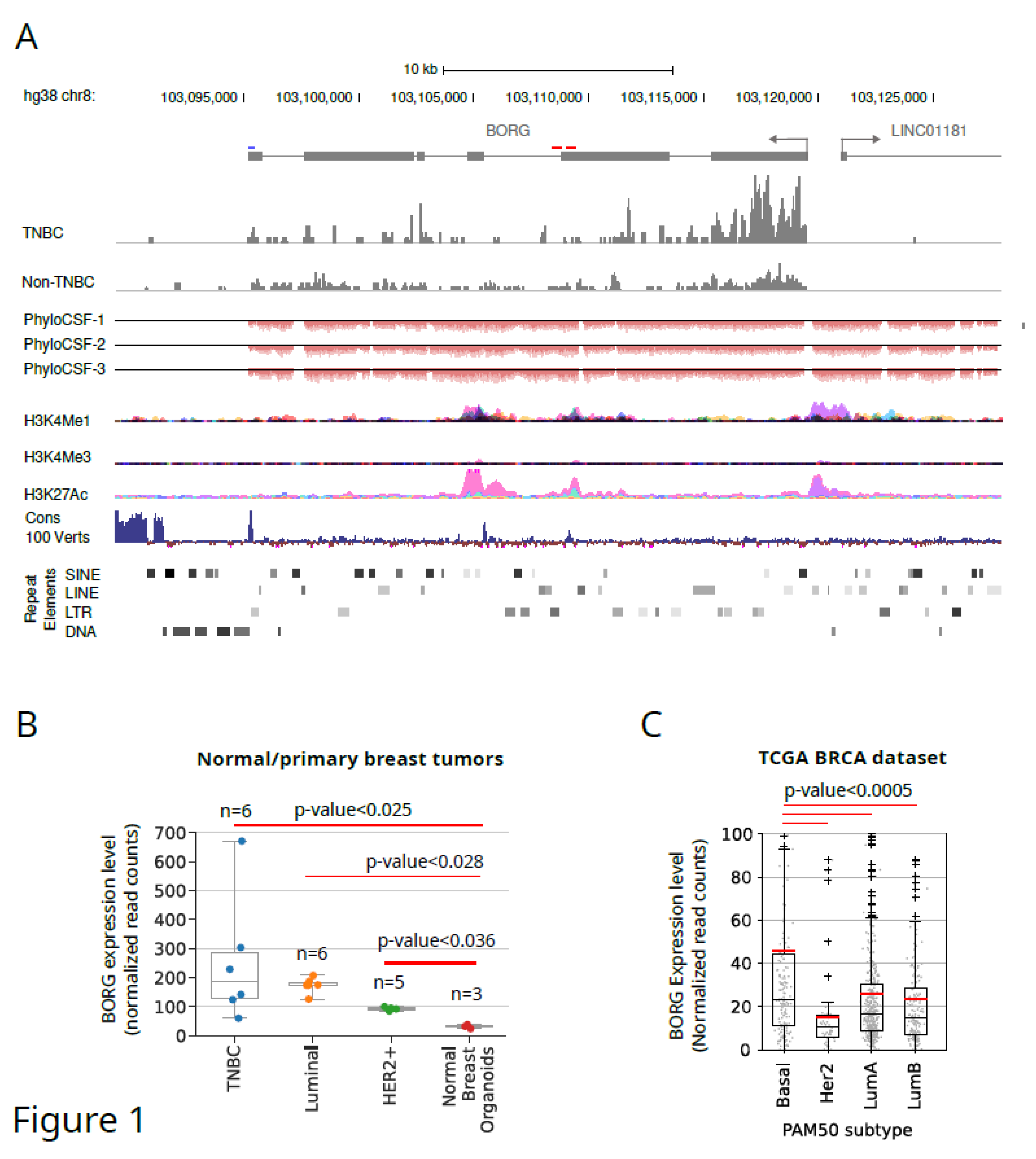
Identification and Architecture of Human BORG Genomic Locus
Human BORG rna Is Strongly Upregulated in a Subset of Human TNBC Tumors
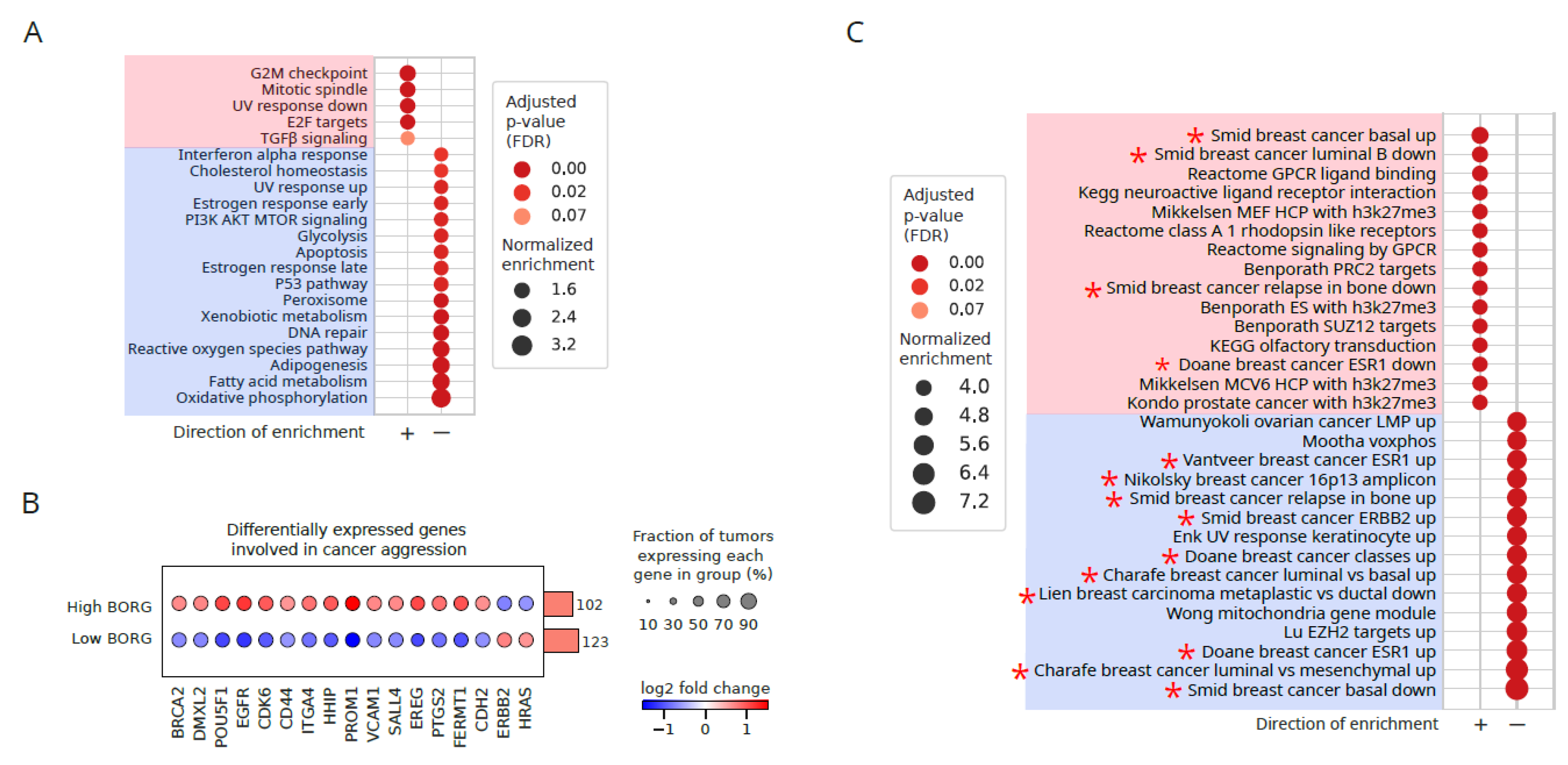
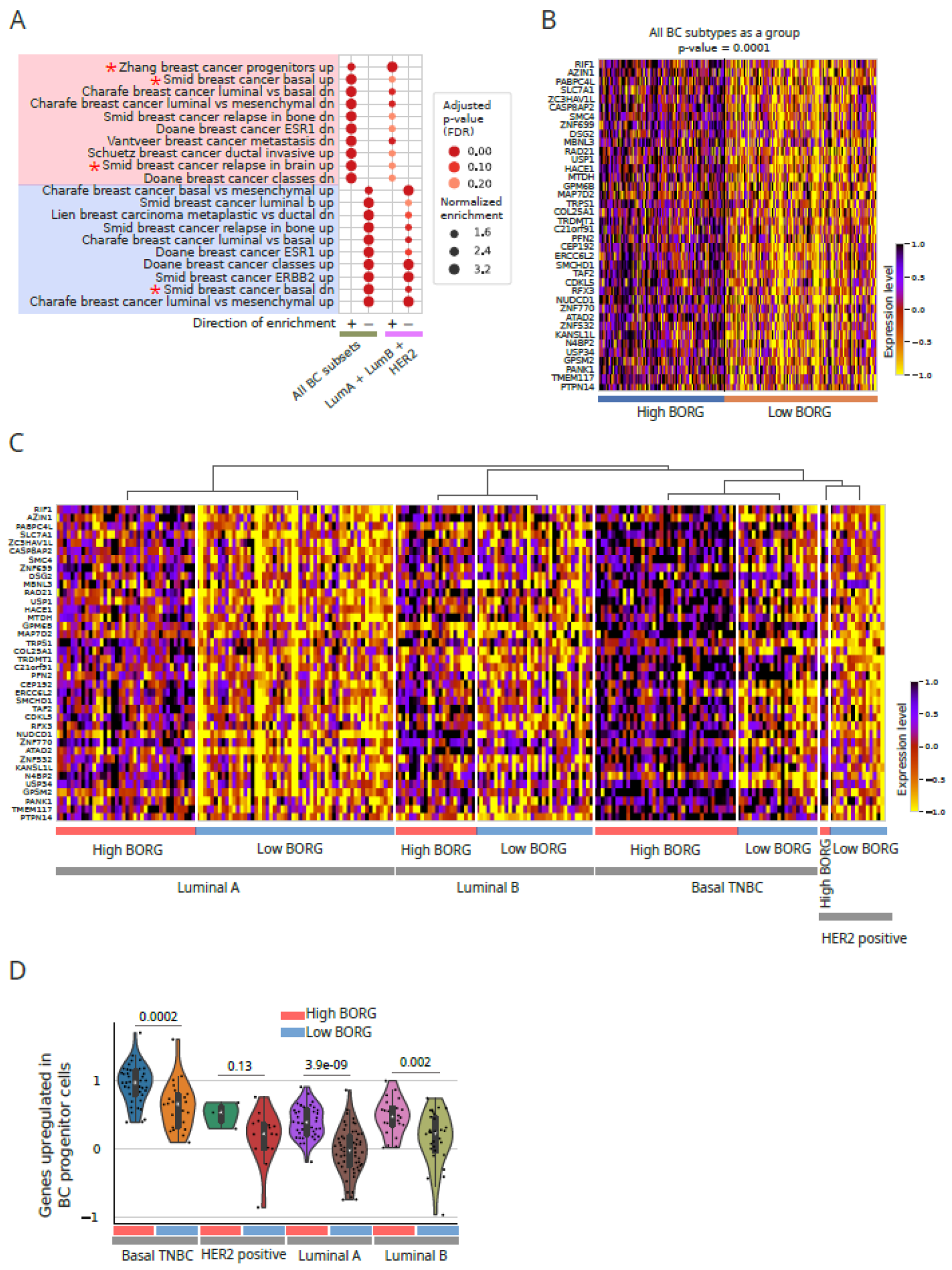
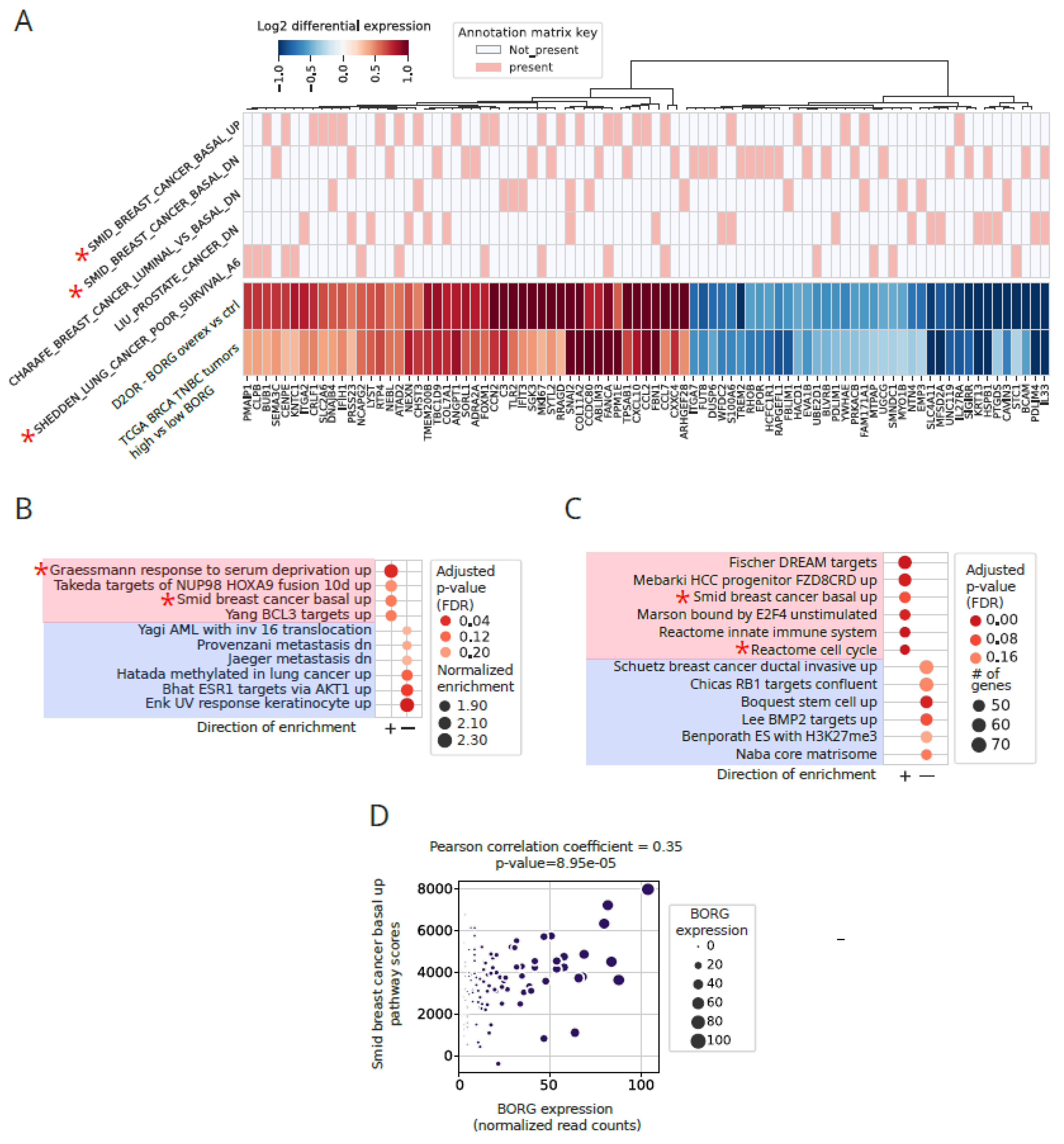
Increased Expression of BORG Is Associated with the Induction of Basal Transcriptomic Signatures
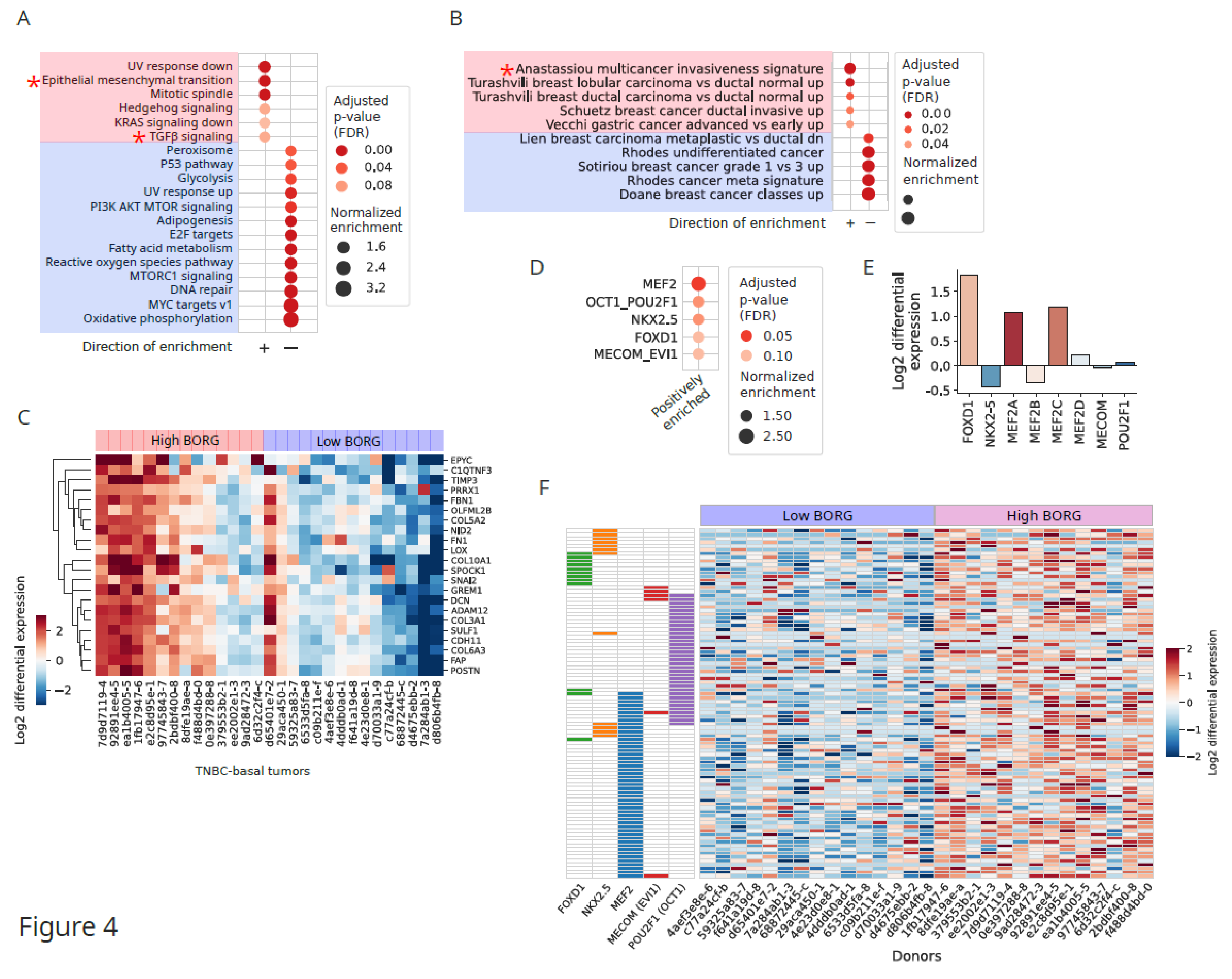
Induction of Invasive Signatures in High BORG-Expressing TNBC Tumors
Aberrant BORG Expression Leads to Induction of Genes Associated with Aggressive Basal Phenotypes
Discussion
Methods
BORG Overexpression Studies in Mouse Cell Lines
Bulk RNA-Seq Analyses
Identification of the Human BORG Locus
Filtering and Preparation of the Study Cohort
Supplementary Materials
Author Contributions
Data Availability Statement
Acknowledgments
Conflicts of Interest
Materials and Correspondence
References
- Siegel, R. L., Miller, K. D., Fuchs, H. E. & Jemal, A. Cancer statistics, 2022. CA Cancer J Clin 72, 7–33 (2022).
- Siegel, R. L., Miller, K. D., Wagle, N. S. & Jemal, A. Cancer statistics, 2023. CA Cancer J Clin 73, 17–48 (2023).
- Sorlie, T. et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A 100, 8418–8423 (2003). [CrossRef]
- Koboldt, D. C. et al. Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012).
- Sørlie, T. et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A 98, 10869–10874 (2001). [CrossRef]
- Gooding, A. J. & Schiemann, W. P. Epithelial-Mesenchymal Transition Programs and Cancer Stem Cell Phenotypes: Mediators of Breast Cancer Therapy Resistance. Mol Cancer Res 18, 1257–1270 (2020). [CrossRef]
- Loizides, S. & Constantinidou, A. Triple negative breast cancer: Immunogenicity, tumor microenvironment, and immunotherapy. Front Genet 13, 1095839 (2022). [CrossRef]
- Anders, C. & Carey, L. A. Understanding and treating triple-negative breast cancer. Oncology (Williston Park) 22, 1233–1239; discussion 1239-1240, 1243 (2008).
- Parker, J. S. et al. Supervised Risk Predictor of Breast Cancer Based on Intrinsic Subtypes. J Clin Oncol 27, 1160–1167 (2009). [CrossRef]
- Lehmann, B. D. et al. Refinement of Triple-Negative Breast Cancer Molecular Subtypes: Implications for Neoadjuvant Chemotherapy Selection. PLoS One 11, e0157368 (2016).
- Castaneda, M., den Hollander, P., Kuburich, N. A., Rosen, J. M. & Mani, S. A. Mechanisms of cancer metastasis. Semin Cancer Biol 87, 17–31 (2022). [CrossRef]
- Morrison, B. J., Schmidt, C. W., Lakhani, S. R., Reynolds, B. A. & Lopez, J. A. Breast cancer stem cells: implications for therapy of breast cancer. Breast Cancer Research 10, 210 (2008). [CrossRef]
- Hu, T., Zhou, R., Zhao, Y. & Wu, G. Integrin α6/Akt/Erk signaling is essential for human breast cancer resistance to radiotherapy. Sci Rep 6, 33376 (2016). [CrossRef]
- Abdoli Shadbad, M. et al. A Systematic Review to Clarify the Prognostic Values of CD44 and CD44+CD24- Phenotype in Triple-Negative Breast Cancer Patients: Lessons Learned and The Road Ahead. Frontiers in Oncology 11, (2021). [CrossRef]
- Honeth, G. et al. The CD44+/CD24- phenotype is enriched in basal-like breast tumors. Breast Cancer Res 10, R53 (2008).
- Olsson, M., Larsson, P., Johansson, J., Sah, V. R. & Parris, T. Z. Cancer stem cells are prevalent in the basal-like 2 and mesenchymal triple-negative breast cancer subtypes in vitro. Frontiers in Cell and Developmental Biology 11, (2023). [CrossRef]
- Shan, N. L., Shin, Y., Yang, G., Furmanski, P. & Suh, N. Breast Cancer Stem Cells: A Review of Their Characteristics and The Agents That Affect Them. Mol Carcinog 60, 73–100 (2021). [CrossRef]
- Bobbitt, J. R., Seachrist, D. D. & Keri, R. A. Chromatin Organization and Transcriptional Programming of Breast Cancer Cell Identity. Endocrinology 164, bqad100 (2023). [CrossRef]
- Prat, A. et al. Molecular Characterization of Basal-Like and Non-Basal-Like Triple-Negative Breast Cancer. Oncologist 18, 123–133 (2013). [CrossRef]
- MacDonald, I., Nixon, N. A. & Khan, O. F. Triple-Negative Breast Cancer: A Review of Current Curative Intent Therapies. Curr Oncol 29, 4768–4778 (2022). [CrossRef]
- Aysola, K. et al. Triple Negative Breast Cancer – An Overview. Hereditary Genet 2013, 001 (2013).
- Keenan, T. E. & Tolaney, S. M. Role of Immunotherapy in Triple-Negative Breast Cancer. J Natl Compr Canc Netw 18, 479–489 (2020). [CrossRef]
- Mattick, J. S. et al. Long non-coding RNAs: definitions, functions, challenges and recommendations. Nat Rev Mol Cell Biol 24, 430–447 (2023). [CrossRef]
- Rinn, J. L. & Chang, H. Y. Long Noncoding RNAs: Molecular Modalities to Organismal Functions. Annu. Rev. Biochem. 89, 283–308 (2020). [CrossRef]
- Amaral, P. et al. The status of the human gene catalogue. Nature 622, 41–47 (2023). [CrossRef]
- Pang, K. C., Frith, M. C. & Mattick, J. S. Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends Genet 22, 1–5 (2006). [CrossRef]
- Ulitsky, I., Shkumatava, A., Jan, C. H., Sive, H. & Bartel, D. P. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 147, 1537–1550 (2011). [CrossRef]
- Gil, N. & Ulitsky, I. Regulation of gene expression by cis-acting long non-coding RNAs. Nat. Rev. Genet. 21, 102–117 (2020). [CrossRef]
- McDonel, P. & Guttman, M. Approaches for Understanding the Mechanisms of Long Noncoding RNA Regulation of Gene Expression. Cold Spring Harb Perspect Biol 11, (2019). [CrossRef]
- Sun, Q., Hao, Q. & Prasanth, K. V. Nuclear Long Noncoding RNAs: Key Regulators of Gene Expression. Trends Genet 34, 142–157 (2018). [CrossRef]
- Richard, J. L. C. & Eichhorn, P. J. A. Deciphering the roles of lncRNAs in breast development and disease. Oncotarget 9, 20179–20212 (2018). [CrossRef]
- Su, J., Deng, L. & Wang, Y.-D. Roles and Mechanisms of Long Non-Coding RNAs in Breast Cancer. Int J Mol Sci 24, 89 (2022). [CrossRef]
- Amelio, I., Bernassola, F. & Candi, E. Emerging roles of long non-coding RNAs in breast cancer biology and management. Semin Cancer Biol 72, 36–45 (2021). [CrossRef]
- Ferrer, J. & Dimitrova, N. Transcription regulation by long non-coding RNAs: mechanisms and disease relevance. Nat Rev Mol Cell Biol (2024). [CrossRef]
- Olivero, C. E. & Dimitrova, N. Identification and characterization of functional long noncoding RNAs in cancer. FASEB J 34, 15630–15646 (2020). [CrossRef]
- Winkler, L. & Dimitrova, N. A mechanistic view of long noncoding RNAs in cancer. Wiley Interdiscip Rev RNA 13, e1699 (2022). [CrossRef]
- Takeda, K. et al. Identification of a novel bone morphogenetic protein-responsive gene that may function as a noncoding RNA. J. Biol. Chem 273, 17079–17085 (1998). [CrossRef]
- Gooding, A. J. et al. The lncRNA BORG Drives Breast Cancer Metastasis and Disease Recurrence. Sci Rep 7, 12698 (2017). [CrossRef]
- Zhang, B. et al. A Novel RNA Motif Mediates the Strict Nuclear Localization of a Long Noncoding RNA. Mol. Cell. Biol. 34, 2318–2329 (2014). [CrossRef]
- Parker, K. A., Gooding, A. J., Valadkhan, S. & Schiemann, W. P. lncRNA BORG:TRIM28 Complexes Drive Metastatic Progression by Inducing α6 Integrin/CD49f Expression in Breast Cancer Stem Cells. Mol Cancer Res 19, 2068–2080 (2021). [CrossRef]
- Aj, G., Ka, P., S, V. & Wp, S. The IncRNA BORG: A novel inducer of TNBC metastasis, chemoresistance, and disease recurrence. Journal of cancer metastasis and treatment 5, (2019).
- Gooding, A. J. et al. The lncRNA BORG facilitates the survival and chemoresistance of triple-negative breast cancers. Oncogene 1 (2018). [CrossRef]
- Joseph, C. et al. Elevated MMP9 expression in breast cancer is a predictor of shorter patient survival. Breast Cancer Res Treat 182, 267–282 (2020). [CrossRef]
- Radisky, E. S. & Radisky, D. C. Matrix metalloproteinases as breast cancer drivers and therapeutic targets. Front Biosci (Landmark Ed) 20, 1144–1163 (2015).
- Liu, H. et al. The role of MMP-1 in breast cancer growth and metastasis to the brain in a xenograft model. BMC Cancer 12, 583 (2012). [CrossRef]
- Ogura, T. et al. OCT1 Is a Poor Prognostic Factor for Breast Cancer Patients and Promotes Cell Proliferation via Inducing NCAPH. Int J Mol Sci 22, 11505 (2021). [CrossRef]
- Galego, S., Kauppila, L. A., Malhó, R., Pimentel, J. & Brito, M. A. Myocyte Enhancer Factor 2C as a New Player in Human Breast Cancer Brain Metastases. Cells 10, 378 (2021). [CrossRef]
- Kumegawa, K., Yang, L., Miyata, K. & Maruyama, R. FOXD1 is associated with poor outcome and maintains tumor-promoting enhancer–gene programs in basal-like breast cancer. Frontiers in Oncology 13, (2023). [CrossRef]
- Long, Y. et al. FOXD1-dependent RalA-ANXA2-Src complex promotes CTC formation in breast cancer. Journal of Experimental & Clinical Cancer Research 41, 301 (2022).
- Hwang-Verslues, W. W. et al. Loss of corepressor PER2 under hypoxia up-regulates OCT1-mediated EMT gene expression and enhances tumor malignancy. Proceedings of the National Academy of Sciences 110, 12331–12336 (2013). [CrossRef]
- Lim, J.-S., Jung, G. Y. & Park, S.-Y. Nkx-2.5 Regulates MDR1 Expression via Its Upstream Promoter in Breast Cancer Cells. J Korean Med Sci 34, e100 (2019). [CrossRef]
- Pradeepa, null et al. EVI1 promotes metastasis by downregulating TIMP2 in metastatic colon and breast cancer cells. Int J Biochem Cell Biol 142, 106118 (2022). [CrossRef]
- Wang, H. et al. Prominent Oncogenic Roles of EVI1 in Breast Carcinoma. Cancer Res 77, 2148–2160 (2017). [CrossRef]
- Asimi, V. et al. Hijacking of transcriptional condensates by endogenous retroviruses. Nat Genet 54, 1238–1247 (2022). [CrossRef]
- Oleksiewicz, U. et al. TRIM28 and Interacting KRAB-ZNFs Control Self-Renewal of Human Pluripotent Stem Cells through Epigenetic Repression of Pro-differentiation Genes. Stem Cell Reports 9, 2065–2080 (2017). [CrossRef]
- Bray, N. L., Pimentel, H., Melsted, P. & Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34, 525–527 (2016). [CrossRef]
- Pimentel, H., Bray, N. L., Puente, S., Melsted, P. & Pachter, L. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat Meth 14, 687–690 (2017). [CrossRef]
- Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [CrossRef]
- Eswaran, J. et al. Transcriptomic landscape of breast cancers through mRNA sequencing. Sci Rep 2, 264 (2012). [CrossRef]
- Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [CrossRef]
- Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [CrossRef]
- Mao, S., Pachter, L., Tse, D. & Kannan, S. RefShannon: A genome-guided transcriptome assembler using sparse flow decomposition. PLOS ONE 15, e0232946 (2020). [CrossRef]
- Roberts, A., Pimentel, H., Trapnell, C. & Pachter, L. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics 27, 2325–2329 (2011). [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).



