Submitted:
23 April 2024
Posted:
24 April 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
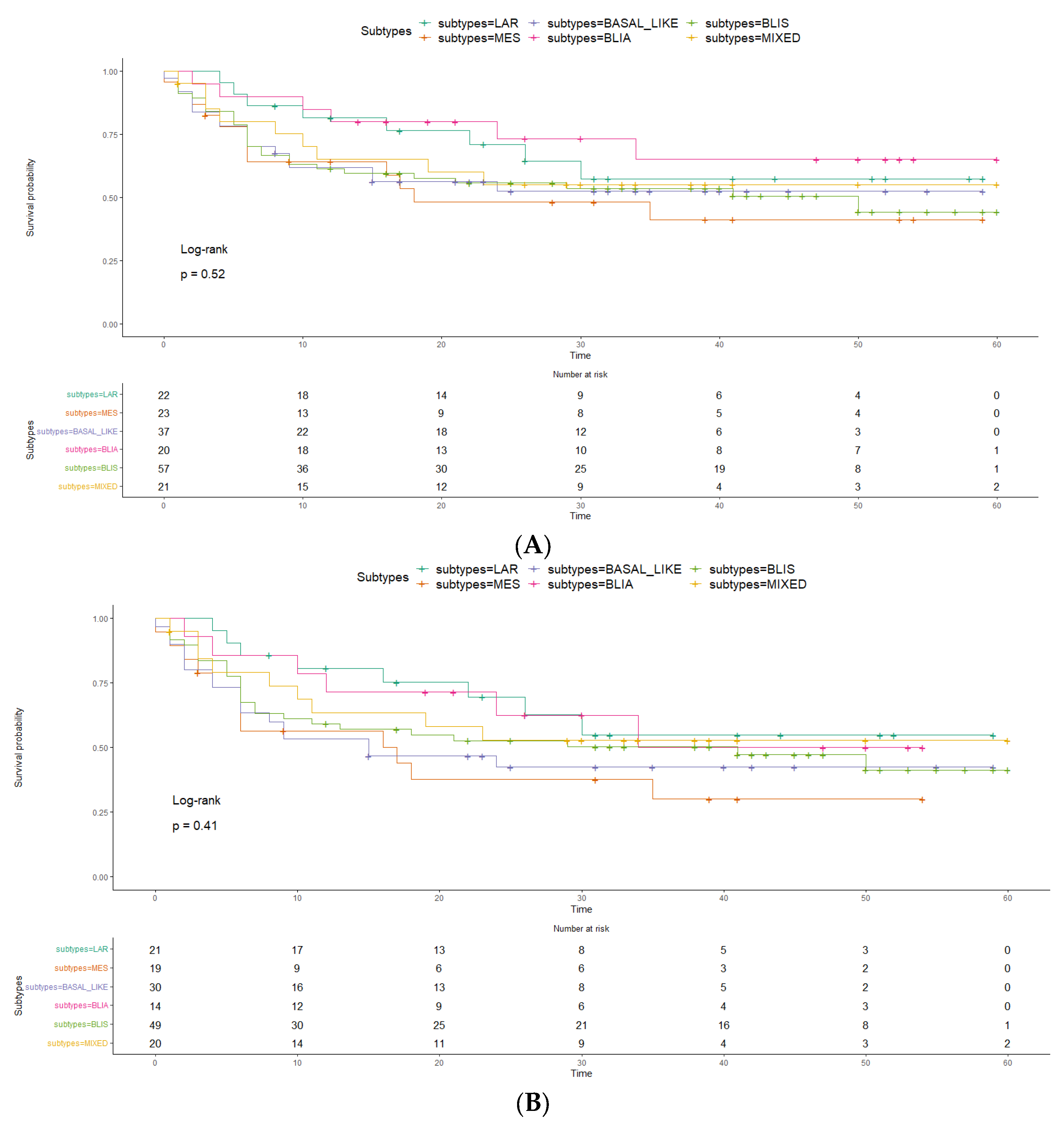
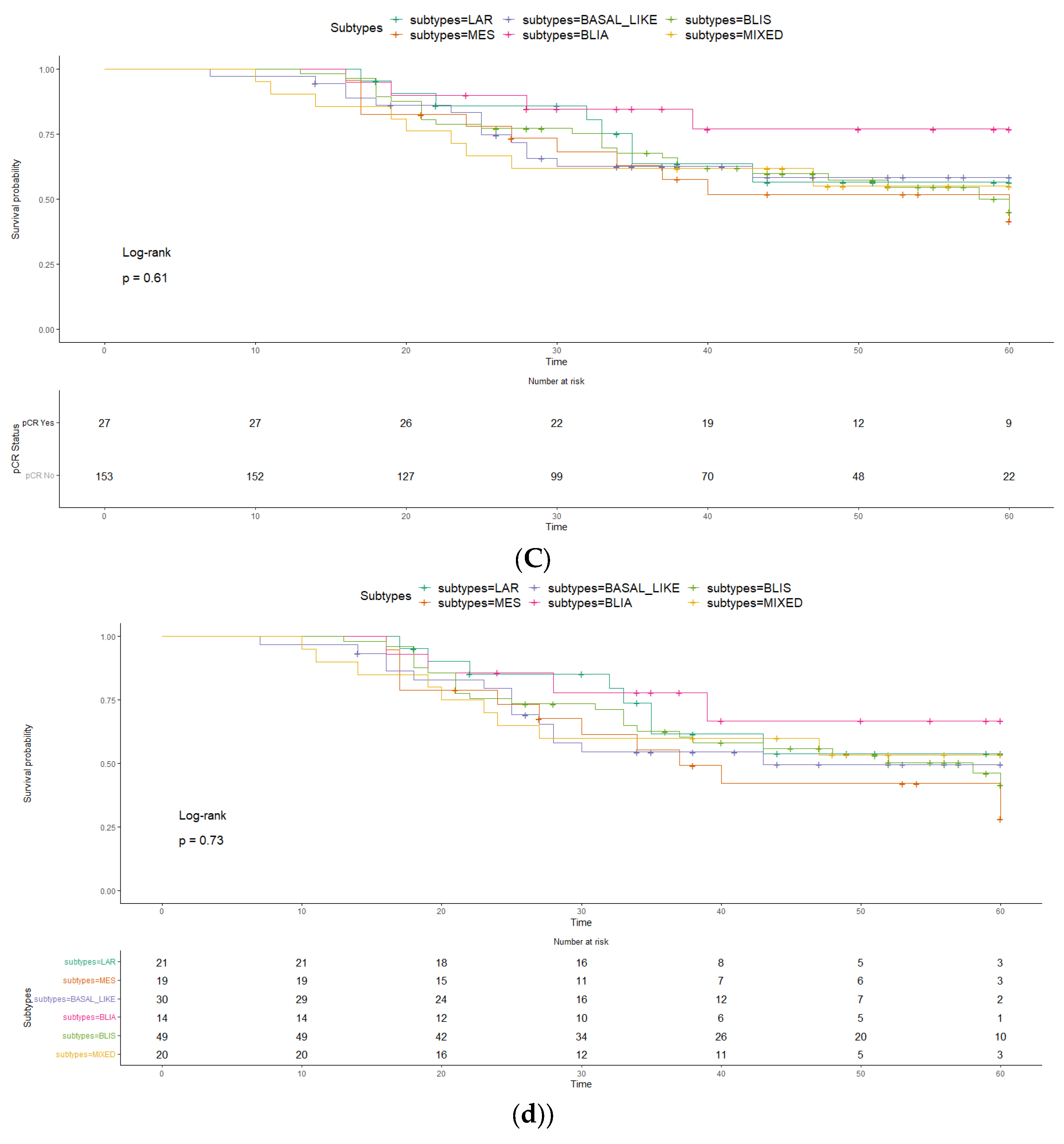
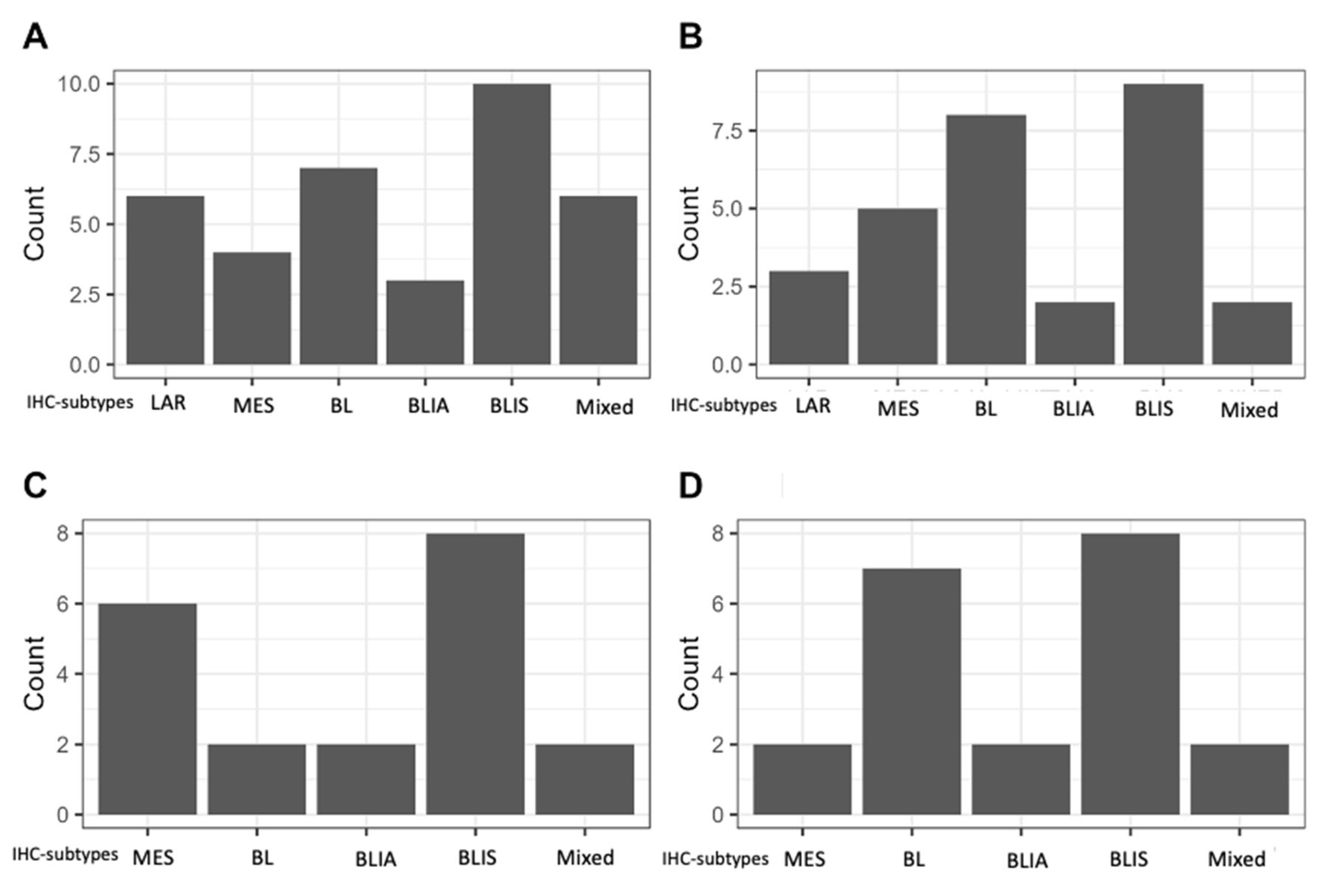
2. Results
3. Discussion
4. Materials and Methods
4.1. Sample Selection and Outcome Definitions
2.2. IHC Analysis and Interpretation
| IHC-subtype | Antibody results | |||||
|---|---|---|---|---|---|---|
| IDO-1 | FOX-C1 | AR | Claudin-3 | |||
| Basal-like unspecific (BL) | IDO-1 ≤ 10% and FOX-C1 < 4or IDO > 10% and FOX-C1 ≥ 4 | < 10% | Weak (W), M or I | |||
| Basal like immune-suppressed (BLIS) | ≤ 10% | ≥ 4 | < 10% | |||
| Basal like immune-activated (BLIA) | > 10% | < 4 | < 10% | |||
| Luminal androgen receptor (LAR) | ≤ 10% | < 4 | ≥ 10% | |||
| Mesenchymal (MES) | any | any | any | 0 | ||
| Mixed (criteria for ≥ 2 subtypes) | BLIS | LAR | ≤ 10% | ≥ 4 | ≥ 10% | W, M or I |
| BLIA | LAR | > 10% | < 4 | ≥ 10% | ||
| BL | LAR | IDO-1 ≤ 10% and FOX-C1 < 4or IDO > 10% and FOX-C1 ≥ 4 | ≥ 10% | |||
| MES | LAR | any | any | ≥ 10% | 0 | |
| Unclassifiable | ≤ 10% | ≤ 2 | < 5% | W, M or I | ||
2.3. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cancer Research UK, https://www.cancerresearchuk.org/about-cancer/breast-cancer/types/triple-negative-breast-cancer, Accessed [March [2023]. Publications: Cancer Research UK ([y2023]), Triple Negative Breast Cancer, Cancer Research UK. [Internet]. Cancer Research UK. 2023 [cited 2024 Mar 3]. Available from: https://www.cancerresearchuk.org/about-cancer/breast-cancer/types/triple-negative-breast-cancer.
- de Paula B, Kieran R, Koh SSY, Crocamo S, Abdelhay E, Muñoz-Espín D. Targeting Senescence as a Therapeutic Opportunity for Triple-Negative Breast Cancer. Mol Cancer Ther [Internet]. 2023, 22, 583–598. [CrossRef] [PubMed]
- Rala de Paula, B.H.; Kumar, S.; Morosini, F.M.; Calábria Cardoso, D.E.M.; Moreira de Sousa, C.A.; Crocamo, S. Real-world assessment of the effect of impact of tumor size on pathological complete response rates in triple negative breast cancer after neoadjuvant chemotherapy. Chinese Clin Oncol. 2020, 9, 1–9. [Google Scholar] [CrossRef]
- Marra, A.; Trapani, D.; Viale, G.; Criscitiello, C.; Curigliano, G. Practical classification of triple-negative breast cancer: intratumoral heterogeneity, mechanisms of drug resistance, and novel therapies. npj Breast Cancer [Internet]. 2020, 6, 54. [Google Scholar] [CrossRef] [PubMed]
- Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest [Internet]. 2011, 121, 2750–2767. [CrossRef]
- Burstein MD, Tsimelzon A, Poage GM, Covington KR, Contreras A, Fuqua SAW, et al. Comprehensive Genomic Analysis Identifies Novel Subtypes and Targets of Triple-Negative Breast Cancer. Clin Cancer Res [Internet]. 2015, 21, 1688–1698. [CrossRef] [PubMed]
- Liu Y-R, Jiang Y-Z, Xu X-E, Yu K-D, Jin X, Hu X, et al. Comprehensive transcriptome analysis identifies novel molecular subtypes and subtype-specific RNAs of triple-negative breast cancer. Breast Cancer Res [Internet]. 2016, 18, 33. [CrossRef]
- Bianchini, G.; De Angelis, C.; Licata, L.; Gianni, L. Treatment landscape of triple-negative breast cancer — expanded options, evolving needs. Nat Rev Clin Oncol [Internet]. 2022, 19, 91–113. [Google Scholar] [CrossRef]
- Yang, R.; Li, Y.; Wang, H.; Qin, T.; Yin, X.; Ma, X. Therapeutic progress and challenges for triple negative breast cancer: targeted therapy and immunotherapy. Mol Biomed [Internet]. 2022, 3, 8. [Google Scholar] [CrossRef] [PubMed]
- Rossi C, Fraticelli S, Fanizza M, Ferrari A, Ferraris E, Messina A, et al. Concordance of immunohistochemistry for predictive and prognostic factors in breast cancer between biopsy and surgical excision: a single-centre experience and review of the literature. Breast Cancer Res Treat [Internet]. 2023, 198, 573–582. [CrossRef]
- Choi, J.; Jung, W.-H.; Koo, J.S. Clinicopathologic features of molecular subtypes of triple negative breast cancer based on immunohistochemical markers. Histol Histopathol. 2012, 27, 1481–1493. [Google Scholar]
- Kim, S.; Moon, B.I.; Lim, W.; Park, S.; Cho, M.S.; Sung, S.H. Feasibility of Classification of Triple Negative Breast Cancer by Immunohistochemical Surrogate Markers. Clin Breast Cancer. 2018, 18, e1123–32. [Google Scholar] [CrossRef] [PubMed]
- Kumar S, Bal A, Das A, Bhattacharyya S, Laroiya I, Khare S, et al. Molecular Subtyping of Triple Negative Breast Cancer by Surrogate Immunohistochemistry Markers [Internet]. 2020. Available from: www.appliedimmunohist.com.
- Zhao S, Ma D, Xiao Y, Li X-M, Ma J-L, Zhang H, et al. Molecular Subtyping of Triple-Negative Breast Cancers by Immunohistochemistry: Molecular Basis and Clinical Relevance. Oncologist. 2020, 25, e1481–91. [CrossRef] [PubMed]
- Yoo, T.-K.; Kang, J.; Lee, A.; Chae, B.J. A triple-negative breast cancer surrogate subtype classification that correlates with gene expression subtypes. Breast Cancer Res Treat [Internet]. 2022, 191, 599–610. [Google Scholar] [CrossRef] [PubMed]
- Lian J, Ma H xia, Xu EW, Bu P, Yun KM, Xi YF. Subclassifying triple-negative breast cancers and its potential clinical utility. Virchows Arch. 2022, 481, 13–21. [CrossRef]
- Leeha, M.; Kanokwiroon, K.; Laohawiriyakamol, S.; Thongsuksai, P. Immunohistochemistry-based molecular subtyping of triple-negative breast cancer and its prognostic significance. Pathol Oncol Res [Internet]. 2023, 29. Available from: https://www.por-journal.com/articles/10.3389/pore.2023.1611162.
- Hu H, Tong K, Tsang JY, Ko CW, Tam F, Loong TC, et al. Subtyping of triple-negative breast cancers: its prognostication and implications in diagnosis of breast origin. ESMO Open [Internet] 2024, 9. [CrossRef]
- Oakman, C.; Viale, G.; Di Leo, A. Management of triple negative breast cancer. The Breast [Internet]. 2010, 19, 312–321. [Google Scholar] [CrossRef]
- Lluch A, Barrios CH, Torrecillas L, Ruiz-Borrego M, Bines J, Segalla J, et al. Phase III Trial of Adjuvant Capecitabine After Standard Neo-/Adjuvant Chemotherapy in Patients With Early Triple-Negative Breast Cancer (GEICAM/2003-11\_CIBOMA/2004-01). J Clin Oncol [Internet]. 2020, 38, 203–213. [CrossRef]
- Mayer IA, Zhao F, Arteaga CL, Symmans WF, Park BH, Burnette BL, et al. Randomized Phase III Postoperative Trial of Platinum-Based Chemotherapy Versus Capecitabine in Patients With Residual Triple-Negative Breast Cancer Following Neoadjuvant Chemotherapy: ECOG-ACRIN EA1131. J Clin Oncol [Internet]. 2021, 39, 2539–2551. [CrossRef]
- Han, H.S.; Vikas, P.; Costa, R.L.B.; Jahan, N.; Taye, A.; Stringer-Reasor, E.M. Early-Stage Triple-Negative Breast Cancer Journey: Beginning, End, and Everything in Between. Am Soc Clin Oncol Educ B [Internet], 2023; e390464. [Google Scholar] [CrossRef]
- Yin, L.; Duan, J.-J.; Bian, X.-W.; Yu, S. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res [Internet]. 2020, 22, 61. [Google Scholar] [CrossRef]
- Xu M, Yuan Y, Yan P, Jiang J, Ma P, Niu X, et al. Prognostic Significance of Androgen Receptor Expression in Triple Negative Breast Cancer: A Systematic Review and Meta-Analysis. Clin Breast Cancer [Internet]. 2020, 20, e385–96. [CrossRef]
- Fizazi K, Foulon S, Carles J, Roubaud G, McDermott R, Fléchon A, et al. Abiraterone plus prednisone added to androgen deprivation therapy and docetaxel in de novo metastatic castration-sensitive prostate cancer (PEACE-1): a multicentre, open-label, randomised, phase 3 study with a 2 , × , 2 factorial design. Lancet [Internet]. 2022, 399, 1695–1707. [CrossRef] [PubMed]
- Aldrich, J.; Canning, M.; Bhave, M. Monitoring of Triple Negative Breast Cancer After Neoadjuvant Chemotherapy. Clin Breast Cancer [Internet]. 2023, 23, 832–834. [Google Scholar] [CrossRef] [PubMed]
- Asleh, K.; Riaz, N.; Nielsen, T.O. Heterogeneity of triple negative breast cancer: Current advances in subtyping and treatment implications. J Exp Clin Cancer Res [Internet]. 2022, 41, 265. [Google Scholar] [CrossRef] [PubMed]
| Choi J et al, 201211 | Kim S et al, 201812 | Kumar S et al, 202013 | Zhao S et al, 202014 | Yoo T-K et al, 202115 | Lian J et al, 202216 | Leeha M et al, 202317 | Hu H. et al18 | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Total subjects | 122 | 200 | 245 | 210 | 183 | 214 | 145 | 93 | 195 | 123 |
| Methods used for subtyping | IHC | IHC | IHC | RNA and Gene expression and IHC staining | RNA and Gene expression and IHC staining | IHC | RNA and Gene expression and IHC staining | IHC | ||
| Adjuvant treatment | Chemo or radiation based on staging | Not reported | Not reported | ~93% (taxane 75.7% and non-taxane 17.6%) | ~90% (62.8% and 26.8%) | ~91% (84.1% and 7.0%) | Not reported | Not reported | ~ 90% (regimen not disclosed) | Not reported |
| IHC-Apocrine/LAR definition | AR and/or GGT1> 10% | AR > 1% | AR ≥1% | AR ≥ 10% | AR Allred score 8 (5 + 3) | AR ≥ 10% | AR Allred score 8 (5 + 3) | AR≥10%, regardless of the expressionof other markers) | ||
| IHC-BLIS definition | - | FOXC1 ≥ 4 and IDO-1 ≤ 10% | - | AR –, CD8 -, FOXC1> 10% | TIL low; AR <10% ; CD8 <20% ; FOXC1 >= 10% | AR < 10%, CD8 TIL < 20%, FOX-C1 <10%, and regardless of DCLK1 values | ||||
| IHC-BLIA or IM definition | - | IDO-1 > 10% and FOX-C1 < 4 | - | AR – and CD8 activated ³20% | LAR-negative and TIL score > 70% | AR <10% ; TIL high ; CD8 ³ 20% | LAR-negative and TIL score > 70% | AR < 10%, CD8 TIL ≥ 20%, and regardless of FOX-C1 and DCLK1 values | ||
| IHC-Basal definition | CK 5/6 > 10% and/or EGFR moderate or intense | CK 5/6 and/or EGFR > 1% | BL1: EGFR <4, CK5/6 ≥ 4 and/or CK4/14 ≥4 | - | LAR -, IM -, M -, and with diffuse and strong p16 staining | - | - | CK 5/6 and or EGFR + | ||
| BL2: EGFR ≥ 4, irrespective of CK5/6 and/or CK 4/14 result | ||||||||||
| IHC-Claudin-low / Mesenchymal definition | Claudin 3, 4, 7 negative and/or e-cadherin negative | Claudin-3 negative and or e-Cadherin negative | E-cadherin, Claudin 3 and 7 ≥4, Vimentin ≥ 4 | AR – CD8- FOXC1- DCLK1≥10% | LAR negative and TIL score < 20% | Metaplastic features; AR <10% ; CD8 <20% ; FOXC1 <10% | LAR negative and TIL score < 20% | AR < 10%, CD8 TIL < 20%, FOX-C1 <10% and DCLK1 ≥10%, | ||
| IHC-Mixed definition | 2 characteristics of 2 different subtypes | 2 or 3 different tumours | ≥ 2 of other categories | Not reported | Not reported | |||||
| IHC-Unclassifiable definition | Not belonging to any subtype | None of the above features | Did not fit in any category | AR – CD8- FOXC1- DCLK1- | All other manifestations | Not reported | All other manifestations | AR < 10%, CD8 TIL < 20%, FOX-C1 <10% and DCLK1 <10% | ||
| IHC-LAR rate | 12 (9.8%) | 22 (11%) | 41 (16.7%) | 60 (28.6%) | 42 (23%) | 53 (24.8%) | 26 (17.9%) | 23 (24.7%) | 37 (18.9%) | 28 (28.6%) |
| IHC-BLIS rate | 11 (5.5%) | - | 80 (38.1%) | 71 (38.8%) | 90 (42.1%) | 39 (41.9%) | 103 (52.8%) | 20 (20.4%) | ||
| IHC-BLIA or IM rate | 27 (13.5%) | - | 40 (19.4%) | 34 (18.6%) | 39 (18.2%) | 21 (14.5%) | 24 (25.8%) | 34 (17.4%) | 39 (39.8%) | |
| IHC-Basal rate | 27 (22.1%) | 85 (42.5%) | BL 36 (14.6%) ; BL1 32 (13.1%) ; BL2 4 (1.6%) | 120 (57.1%) | 105 (57.4%) | 129 (60.3%) | BL1 27 (18.6%) | 63 (67.7%) | 137 (70.2%) | |
| IHC-Mesenchymal rate | 28 (23%) | 23 (11.5%) | 70 (28.6%) | 16 (7.6%) | 18 (9.8%) | 17 (7.9%) | 44 (30.3%) | 7 (7.5%) | 1 (0.5%) | 11 (11.2%) |
| IHC-Mixed rate | 23 (18.9%) | 60 (30%)LAR+MES 8 (4%) ; LAR+BL 27 (13.5%) ; MES+BL 19 (9.5%) ; LAR+MES+BL 6 (3%) | 37 (15.1%) | 0 | 0 | 0 | NR | 0 | NR | NR |
| IHC-Unclassifiable rate | 32 (26.2%) | 10 (5%) | 61 (24.9%) | 14(6.7%) | 18 (9.8%) | 15 (7%) | 18 (12.4%) | 0 | 20 (10.2%) | 25 (20.3%) |
| Confirmation method | no | no | mRNA | mRNA | no | mRNA | No | |||
| follow-up median | 59.5 months | 41 m (0-64) | 40 m (12-58) | 40.95 m (IQR 23.48-89.22) | 62 m (IQR 43-105) | |||||
| Disease free survival (DFS) | Basal like and unclassifiable show less favourable prognosis, mesenchymal and mixed intermediate and AR showed a better prognosisCk 5/6 and Claudin positivity worse DFS | BLIS worse prognosis.LAR, BLIA, BL and NOS favourableThis was also true for Burstein (4 subtypes)FOXC1 – worse prognosis | - | IM (HR=0.07), LAR (HR=0.18), BLIS (HR=0.26) better RFS than MES | MES worse RFS | Significantly worse DFS for M subtype according to surrogate subtypes and although not clinically significant IM tends to be better survival | No significant differenceLow recurrence 11 cases (11.83%) | 5 y 64.7% and no subtypes difference | IM-inflamed better DFS compared to others and BLIS the worse survival | |
| Overall Survival (OS) | Discohesiviness worse survival | Mesenchymal and unclassified shorted OS (68.2 and 69.2m) | T+N+IHC was superior to T+N categories in time dependent AUC | - | - | 5 y OS = 65.0%IM significantly better OS but other did not differentiate between each other | IM-inflamed better breast specific survival and BLIS the worse compared to others | |||
| Variables | |||||
|---|---|---|---|---|---|
| Age (Mean (SD) | 52.42 | (12.79) | |||
| Tumor size (Mean (SD) | 47.64 | (53.61) | |||
| Number of positive lymph nodes (Mean (SD) | 2.14 | (4.01) | |||
| T1-T2 | 58 | (31.0%) | |||
| Clinical Tumor(T) stage (N (%) | T3 | 57 | (30.5%) | ||
| T4 | 72 | (38.5%) | |||
| Clinical Lymph node(N) stage (N (%) | N0 | 79 | (42.2%) | ||
| N1 | 78 | (41.7%) | |||
| N2 | 28 | (15.0%) | |||
| N3 | 2 | (1.1%) | |||
| Histologic subtype (N (%) | Invasive ductal carcinoma | 176 | (94.2%) | ||
| other | 10 | (5.3%) | |||
| NA | 1 | (0.5%) | |||
| Tumor Grade (N (%) | 1 | 2 | (1.1%) | ||
| 2 | 59 | (31.6%) | |||
| 3 | 118 | (63.1%) | |||
| NA | 8 | 4.8% | |||
| ki67 (Mean (SD) | 65.47 | (24.49) | |||
| Angiolymphatic invasion (N (%) | Yes | 98 | (52.4%) | ||
| no | 39 | (20.9%) | |||
| Not-assessed | 50 | 26.7% | |||
| Lymphatic infiltrate (N (%) | Yes | 32 | (17.1%) | ||
| No | 100 | (53.5%) | |||
| Not-assessed | 55 | 29.4% | |||
| IHC-subtype (N (%) | Basal-like (BL) | unspecified | 36 | 19.6% | |
| Immunosuppressed (IS) | 57 | 31.0% | |||
| Imunoactivated (IA) | 20 | 10.9% | |||
| Luminal Androgen Receptor | 22 | 12.0% | |||
| Mesenchymal | 23 | 12.5% | |||
| Mixed | 21 | 11.4% | |||
| Unclassifiable | 4 | 2.2% | |||
| Not amendable of subtyping | 3 | (1.6%) | |||
| Neoadjuvant chemotherapy (N (%) | ACx4 + TXTx4 | 144 | 77% | ||
| ACx4 + wPacx12 | 19 | 10.2% | |||
| TCx4 | 5 | 2.7% | |||
| Other regimens | 19 | 10.2% | |||
| Surgery modality (N (%) | Modified radical mastectomy | 120 | (64.2%) | ||
| Conventional mastectomy | 17 | (9.1%) | |||
| Conservative surgery | 50 | (26.7%) | |||
| Pathologic response (N (%) | Complete | 28 | (15.0%) | ||
| Noncomplete | 159 | (85.0%) | |||
| Pathologic complete response according to IHC-subtype (N (%) | Basal-like (BL) | unspecified | 7 | 19.4% | |
| Immunosuppressed (IS) | 8 | 14.0% | |||
| Imunoactivated (IA) | 6 | 30.0% | |||
| Luminal Androgen Receptor | 1 | 4.5% | |||
| Mesenchymal | 4 | 17.4% | |||
| Mixed | 1 | 4.8% | |||
| Unclassifiable | 1 | 25.0% | |||
| Subject number | Age | Clinical stage (TNM) | histological subtype | Grade | Ki67(%) | Neoadjuvant regimen (x number of cycles) | IHC-subtype |
|---|---|---|---|---|---|---|---|
| Patient 1 | 66 | T4N1M0 | IDC | 2 | 90 | Other – Acx3 + wPacx3 | BLIS |
| Patient 2 | 63 | T3N0M0 | IDC | 3 | 5 | ACx4 + TXTx4 | unclassifiable |
| Patient 3 | 54 | T3N1M0 | IDC | 3 | 70 | ACx4 + TXTx4 | MES |
| Patient 4 | 67 | TxN2M0 | IDC | NR | 70 | ACx4 + TXTx4 | LAR |
| Patient 5 | 47 | T3N0M0 | IDC | 3 | 95 | ACx4 + TXTx4 | BLIS |
| Patient 6 | 60 | T4N2M0 | IDC | 3 | 80 | ACx4 + TXTx4 | BL-unspecific |
| Patient 7 | 47 | T4N1M0 | IDC | 3 | 80 | ACx4 + TXTx4 | BL-unspecific |
| Patient 8 | 52 | T4N1M0 | IDC | 3 | 90 | ACx4 + TXTx4 | LAR/BLIS |
| Patient 9 | 56 | T3N0M0 | IDC | 2 | 50 | ACx4 + TXTx4 | MES |
| Patient 10 | 44 | T3N1M0 | IDC | 3 | 95 | ACx4 + TXTx4 | BL-unspecific |
| Patient 11 | 61 | T4N2M0 | IDC | 2 | 70 | ACx4 + TXTx4 | BLIA |
| Patient 12 | 39 | T3N0M0 | Other | NR | 95 | ACx4 + TXTx4 | BLIS |
| Patient 13 | 47 | T2N0M0 | IDC | 2 | 95 | ACx4 + TXTx4 | BLIA |
| Patient 14 | 46 | T4N3M0 | IDC | 2 | 60 | ACx4 + TXTx4 | BL-unspecific |
| Patient 15 | 44 | T3N0M0 | IDC | 3 | 90 | ACx4 + TXTx4 | BLIS |
| Patient 16 | 68 | T4N0M0 | IDC | 2 | 75 | ACx4 + TXTx4 | BLIA |
| Patient 17 | 64 | T4N2M0 | IDC | 3 | 70 | Other - TCx4 + ACx6 | BL-unspecific |
| Patient 18 | 41 | T3N0M0 | IDC | 3 | 70 | ACx4 + TXTx4 | BLIA |
| Patient 19 | 63 | T4N0M0 | IDC | 2 | 15 | ACx4 + TXTx4 | BLIA |
| Patient 20 | 34 | T4N0M0 | IDC | 3 | 100 | ACx4 + TXTx4 | BLIS |
| Patient 21 | 37 | T3N0M0 | IDC | 3 | 50 | ACx4 + TXTx4 | BLIS |
| Patient 22 | 71 | T2N1M0 | IDC | 2 | 50 | ACx4 + wPacx4 | BLIA |
| Patient 23 | 43 | T3N0M0 | IDC | 3 | 70 | ACx4 + TXTx4 | BLIS |
| Patient 24 | 47 | T2N0M0 | IDC | 2 | 5 | ACx4 + TXTx4 | MES |
| Patient 25 | 62 | T1N0M0 | IDC | 3 | 80 | ACx4 + TXTx4 | BL-unspecific |
| Patient 26 | 51 | T2N1M0 | IDC | 3 | 80 | ACx4 + TXTx4 | MES |
| Patient 27 | 27 | T2N0M0 | IDC | 3 | 95 | ACx4 + TXTx4 | BLIS |
| Patient 28 | 59 | T4N0M0 | IDC | 3 | 60 | ACx4 + wPacx4 | BL-unspecific |
| Antibody | Definition of positivity expression | Brand | Dilution | Control |
|---|---|---|---|---|
| IDO-1 | > 10% of tumor cells | Abcam | 1:1000 | Breast |
| Claudin-3 | Allred score ≥ 4 | Abcam | 1:200 | Breast |
| Androgen Receptor | ≥ 20% of tumor cells | Cell Marque | 1:400 | Gallbladder |
| FOX-C1 | Moderate (M) or intense (I) | Abcam | 1:300 | Breast |
| Ki-67 | ≥1% of tumor cells | Ventana-Roche | Ready to use | Breast |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





