Submitted:
03 October 2023
Posted:
04 October 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Correlation between biomarkers and pathological responses to NAC
2.2. Correlation between biomarkers and DFS
2.3. Correlation between biomarkers and OS
3. Discussion
4. Patients and Methods
4.1. Characteristics of the study subjects
4.2. Biomarker measurements and evaluations
4.3. Statistical analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Korde, L.A.; Somerfield, M.R.; Carey, L.A.; Crews, J.R.; Denduluri, N.; Hwang, E.S.; Khan, S.A.; Loibl, S.; Morris, E.A.; Perez, A.; et al. Neoadjuvant Chemotherapy, Endocrine Therapy, and Targeted Therapy for Breast Cancer: ASCO Guideline. J. Clin. Oncol. 2021, 39, 1485–1505. [Google Scholar] [CrossRef]
- von Minckwitz, G.; Untch, M.; Blohmer, J.-U.; Costa, S.D.; Eidtmann, H.; Fasching, P.A.; Gerber, B.; Eiermann, W.; Hilfrich, J.; Huober, J.; et al. Definition and Impact of Pathologic Complete Response on Prognosis After Neoadjuvant Chemotherapy in Various Intrinsic Breast Cancer Subtypes. J. Clin. Oncol. 2012, 30, 1796–1804. [Google Scholar] [CrossRef]
- Symmans, W.F.; Wei, C.; Gould, R.; Yu, X.; Zhang, Y.; Liu, M.; Walls, A.; Bousamra, A.; Ramineni, M.; Sinn, B.; et al. Long-Term Prognostic Risk After Neoadjuvant Chemotherapy Associated With Residual Cancer Burden and Breast Cancer Subtype. J. Clin. Oncol. 2017, 35, 1049–1060. [Google Scholar] [CrossRef]
- Mittendorf, E.A.; Jeruss, J.S.; Tucker, S.L.; Kolli, A.; Newman, L.A.; Gonzalez-Angulo, A.M.; Buchholz, T.A.; Sahin, A.A.; Cormier, J.N.; Buzdar, A.U.; et al. Validation of a Novel Staging System for Disease-Specific Survival in Patients With Breast Cancer Treated With Neoadjuvant Chemotherapy. J. Clin. Oncol. 2011, 29, 1956–1962. [Google Scholar] [CrossRef] [PubMed]
- Mittendorf, E.A.; Vila, J.; Tucker, S.L.; Chavez-MacGregor, M.; Smith, B.D.; Symmans, W.F.; Sahin, A.A.; Hortobagyi, G.N.; Hunt, K.K. The Neo-Bioscore Update for Staging Breast Cancer Treated With Neoadjuvant Chemotherapy. JAMA Oncol. 2016, 2, 929–936. [Google Scholar] [CrossRef] [PubMed]
- Lan, A.; Li, H.; Chen, J.; Shen, M.; Jin, Y.; Dai, Y.; Jiang, L.; Dai, X.; Peng, Y.; Liu, S. Nomograms for Predicting Disease-Free Survival Based on Core Needle Biopsy and Surgical Specimens in Female Breast Cancer Patients with Non-Pathological Complete Response to Neoadjuvant Chemotherapy. J. Pers. Med. 2023, 13, 249. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Wu, S.; Xing, H.; Han, M.; Li, J.; Liu, Y. Development and Validation of a Novel Model for Predicting Prognosis of Non-PCR Patients After Neoadjuvant Therapy for Breast Cancer. Front. Oncol. 2021, 11, 675533. [Google Scholar] [CrossRef]
- Hou, N.; Wu, J.; Xiao, J.; Wang, Z.; Song, Z.; Ke, Z.; Wang, R.; Wei, M.; Xu, M.; Wei, J.; et al. Development, verification, and comparison of a risk stratification model integrating residual cancer burden to predict individual prognosis in early-stage breast cancer treated with neoadjuvant therapy. ESMO Open 2021, 6, 100269. [Google Scholar] [CrossRef]
- Tao, M.; Chen, S.; Zhang, X.; Zhou, Q. Ki-67 labeling index is a predictive marker for a pathological complete response to neoadjuvant chemotherapy in breast cancer. Medicine 2017, 96, e9384. [Google Scholar] [CrossRef]
- Waks, A.G.; Winer, E.P. Breast Cancer Treatment: A Review. JAMA 2019, 321, 288–300. [Google Scholar] [CrossRef]
- Dieci, M.V.; Miglietta, F.; Guarneri, V. Immune Infiltrates in Breast Cancer: Recent Updates and Clinical Implications. Cells 2021, 10, 223. [Google Scholar] [CrossRef] [PubMed]
- Kurosumi, M.; Akashi-Tanaka, S.; Akiyama, F.; Komoike, Y.; Mukai, H.; Nakamura, S.; Tsuda, H. ; (Committee for Production of Histopathological Criteria for Assessment of Therapeutic Response of the Japanese Breast Cancer Society) Histopathological criteria for assessment of therapeutic response in breast cancer (2007 version). Breast Cancer 2007, 15, 5–7. [Google Scholar] [CrossRef] [PubMed]
- Yau, C.; Osdoit, M.; van der Noordaa, M.; Shad, S.; Wei, J.; de Croze, D.; Hamy, A.-S.; Laé, M.; Reyal, F.; Sonke, G.S.; et al. Residual cancer burden after neoadjuvant chemotherapy and long-term survival outcomes in breast cancer: a multicentre pooled analysis of 5161 patients. Lancet Oncol. 2021, 23, 149–160. [Google Scholar] [CrossRef]
- Xie, L.-Y.; Wang, K.; Chen, H.-L.; Shi, Y.-X.; Zhang, Y.-Q.; Lin, H.-Y.; Liang, Y.-K.; Xiao, Y.-S.; Wu, Z.-Y.; Yuan, Z.-Y.; et al. Markers Associated With Tumor Recurrence in Patients With Breast Cancer Achieving a Pathologic Complete Response After Neoadjuvant Chemotherapy. Front. Oncol. 2022, 12, 860475. [Google Scholar] [CrossRef]
- Wu, Z.; Zhang, L.; Xu, S.; Lin, Y.; Yin, W.; Lu, J.; Sha, R.; Sheng, X.; Zhou, L.; Lu, J. Predictive and prognostic value of ZEB1 protein expression in breast cancer patients with neoadjuvant chemotherapy. Cancer Cell Int. 2019, 19, 78. [Google Scholar] [CrossRef] [PubMed]
- Elzamly, S.; Badri, N.; Padilla, O.; Dwivedi, A.K.; A Alvarado, L.; Hamilton, M.; Diab, N.; Rock, C.; Elfar, A.; Teleb, M.; et al. Epithelial-Mesenchymal Transition Markers in Breast Cancer and Pathological Responseafter Neoadjuvant Chemotherapy. Breast Cancer: Basic Clin. Res. 2018, 12. [Google Scholar] [CrossRef] [PubMed]
- Denkert, C.; Von Minckwitz, G.; Darb-Esfahani, S.; Lederer, B.; Heppner, B.I.; Weber, K.E.; Budczies, J.; Huober, J.; Klauschen, F.; Furlanetto, J.; et al. Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: A pooled analysis of 3771 patients treated with neoadjuvant therapy. Lancet Oncol. 2018, 19, 40–50. [Google Scholar] [CrossRef]
- Chen, S.; Wang, R.-X.; Liu, Y.; Yang, W.-T.; Shao, Z.-M. PD-L1 expression of the residual tumor serves as a prognostic marker in local advanced breast cancer after neoadjuvant chemotherapy. Int. J. Cancer 2017, 140, 1384–1395. [Google Scholar] [CrossRef]
- Liu, Y.L.; Saraf, A.; Lee, S.M.; Zhong, X.; Hibshoosh, H.; Kalinsky, K.; Connolly, E.P. Lymphovascular invasion is an independent predictor of survival in breast cancer after neoadjuvant chemotherapy. Breast Cancer Res. Treat. 2016, 157, 555–564. [Google Scholar] [CrossRef]
- Hamy, A.-S.; Lam, G.-T.; Laas, E.; Darrigues, L.; Balezeau, T.; Guerin, J.; Livartowski, A.; Sadacca, B.; Pierga, J.-Y.; Vincent-Salomon, A.; et al. Lymphovascular invasion after neoadjuvant chemotherapy is strongly associated with poor prognosis in breast carcinoma. Breast Cancer Res. Treat. 2018, 169, 295–304. [Google Scholar] [CrossRef]
- Ryu, Y.J.; Kang, S.J.; Cho, J.S.; Yoon, J.H.; Park, M.H. Lymphovascular invasion can be better than pathologic complete response to predict prognosis in breast cancer treated with neoadjuvant chemotherapy. Medicine 2018, 97, e11647. [Google Scholar] [CrossRef] [PubMed]
- Karihtala, P.; Auvinen, P.; Kauppila, S.; Haapasaari, K.-M.; Jukkola-Vuorinen, A.; Soini, Y. Vimentin, zeb1 and Sip1 are up-regulated in triple-negative and basal-like breast cancers: association with an aggressive tumour phenotype. Breast Cancer Res. Treat. 2013, 138, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Tanabe, Y.; Tsuda, H.; Yoshida, M.; Yunokawa, M.; Yonemori, K.; Shimizu, C.; Yamamoto, S.; Kinoshita, T.; Fujiwara, Y.; Tamura, K. Pathological features of triple-negative breast cancers that showed progressive disease during neoadjuvant chemotherapy. Cancer Sci. 2017, 108, 1520–1529. [Google Scholar] [CrossRef] [PubMed]
- Grasset, E.M.; Dunworth, M.; Sharma, G.; Loth, M.; Tandurella, J.; Cimino-Mathews, A.; Gentz, M.; Bracht, S.; Haynes, M.; Fertig, E.J.; et al. Triple-negative breast cancer metastasis involves complex epithelial-mesenchymal transition dynamics and requires vimentin. Sci. Transl. Med. 2022, 14, eabn7571. [Google Scholar] [CrossRef]
- Winter, M.; Meignan, S.; Völkel, P.; Angrand, P.-O.; Chopin, V.; Bidan, N.; Toillon, R.-A.; Adriaenssens, E.; Lagadec, C.; Le Bourhis, X. Vimentin Promotes the Aggressiveness of Triple Negative Breast Cancer Cells Surviving Chemotherapeutic Treatment. Cells 2021, 10, 1504. [Google Scholar] [CrossRef] [PubMed]
- Denkert, C.; Loibl, S.; Noske, A.; Roller, M.; Müller, B.M.; Komor, M.; Budczies, J.; Darb-Esfahani, S.; Kronenwett, R.; Hanusch, C.; et al. Tumor-Associated Lymphocytes As an Independent Predictor of Response to Neoadjuvant Chemotherapy in Breast Cancer. J. Clin. Oncol. 2010, 28, 105–113. [Google Scholar] [CrossRef]
- Williamson, C.W.; Sherer, M.V.; Zamarin, D.; Sharabi, A.B.; Dyer, B.A.; Mell, L.K.; Mayadev, J.S. Immunotherapy and radiation therapy sequencing: State of the data on timing, efficacy, and safety. Cancer 2021, 127, 1553–1567. [Google Scholar] [CrossRef]
- Ying-Rui, M.; Bu-Fan, B.; Deng, L.; Rong, S.; Qian-Mei, Z. Targeting the stimulator of interferon genes (STING) in breast cancer. Front. Pharmacol. 2023, 14, 1199152. [Google Scholar] [CrossRef]
- Pratelli, G.; Carlisi, D.; Di Liberto, D.; Notaro, A.; Giuliano, M.; D’anneo, A.; Lauricella, M.; Emanuele, S.; Calvaruso, G.; De Blasio, A. MCL1 Inhibition Overcomes the Aggressiveness Features of Triple-Negative Breast Cancer MDA-MB-231 Cells. Int. J. Mol. Sci. 2023, 24, 11149. [Google Scholar] [CrossRef]
- Hashemi, M.; Arani, H.Z.; Orouei, S.; Fallah, S.; Ghorbani, A.; Khaledabadi, M.; Kakavand, A.; Tavakolpournegari, A.; Saebfar, H.; Heidari, H.; et al. EMT mechanism in breast cancer metastasis and drug resistance: Revisiting molecular interactions and biological functions. Biomed. Pharmacother. 2022, 155, 113774. [Google Scholar] [CrossRef]
- Cho, E.S.; Kang, H.E.; Kim, N.H.; Yook, J.I. Therapeutic implications of cancer epithelial-mesenchymal transition (EMT). Arch. Pharmacal Res. 2019, 42, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Salgado, R.; Denkert, C.; Demaria, S.; Sirtaine, N.; Klauschen, F.; Pruneri, G.; Wienert, S.; Van den Eynden, G.; Baehner, F.L.; Penault-Llorca, F.; et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: Recommendations by an International TILs Working Group 2014. Ann. Oncol. 2015, 26, 259–271. [Google Scholar] [CrossRef] [PubMed]
- .Zhang, W.; Shi, X.; Peng, Y.; Wu, M.; Zhang, P.; Xie, R.; Wu, Y.; Yan, Q.; Liu, S.; Wang, J. HIF-1α Promotes Epithelial-Mesenchymal Transition and Metastasis through Direct Regulation of ZEB1 in Colorectal Cancer. PLOS ONE 2015, 10, e0129603. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Ma, X.; Li, Y.; He, Y.; Huang, D.; Cai, S.; Peng, J. The Characteristics and Prognostic Effect of E-Cadherin Expression in Colorectal Signet Ring Cell Carcinoma. PLOS ONE 2016, 11, e0160527. [Google Scholar] [CrossRef]
- Tian, W.; Yang, Y.; Qin, Q.; Zhang, L.; Wang, Z.; Su, L.; Zeng, L.; Chen, H.; Hu, L.; Hong, J.; et al. Vimentin and tumor–stroma ratio for neoadjuvant chemoradiotherapy response prediction in locally advanced rectal cancer. Cancer Sci. 2022, 114, 619–629. [Google Scholar] [CrossRef]
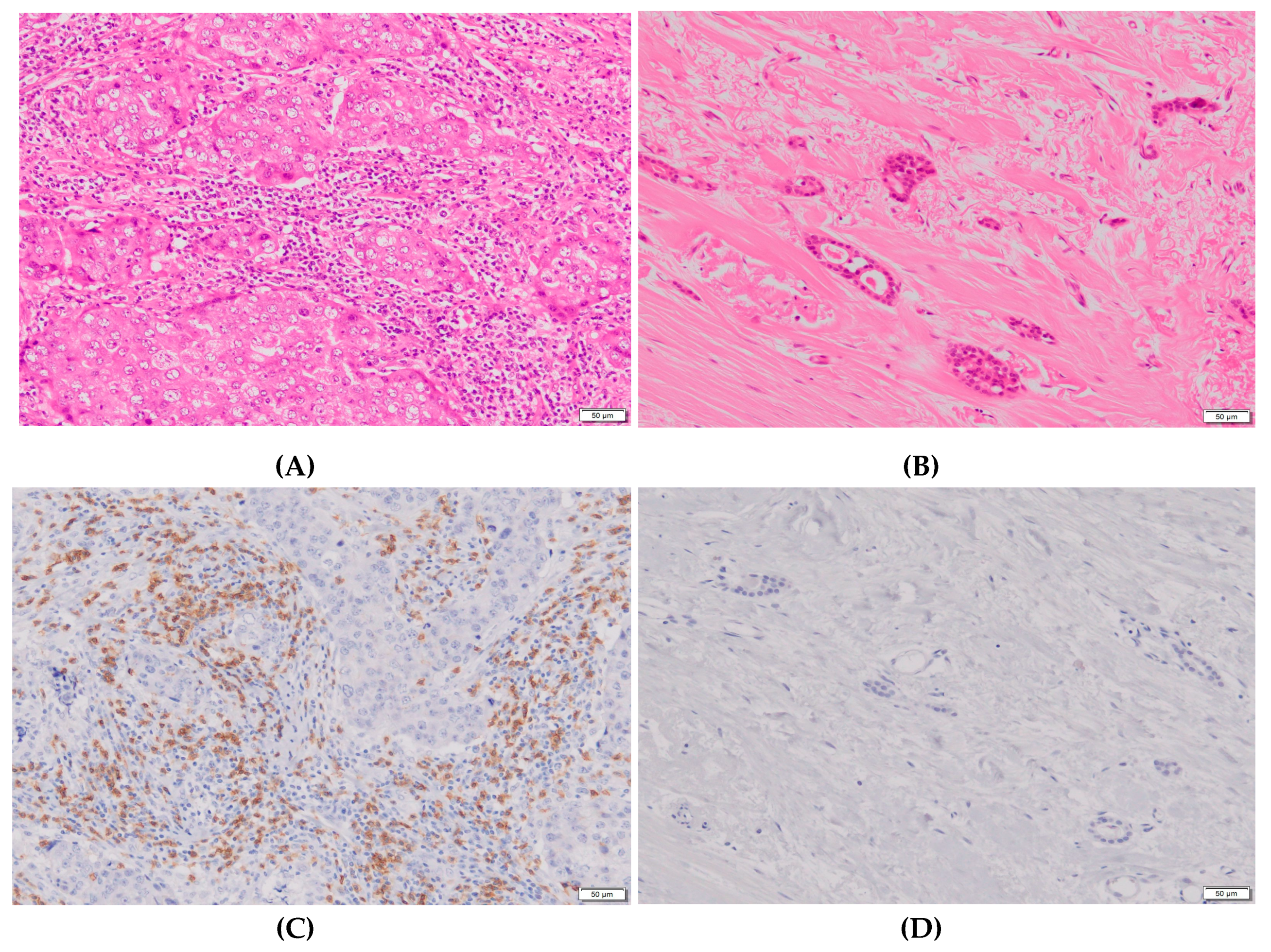
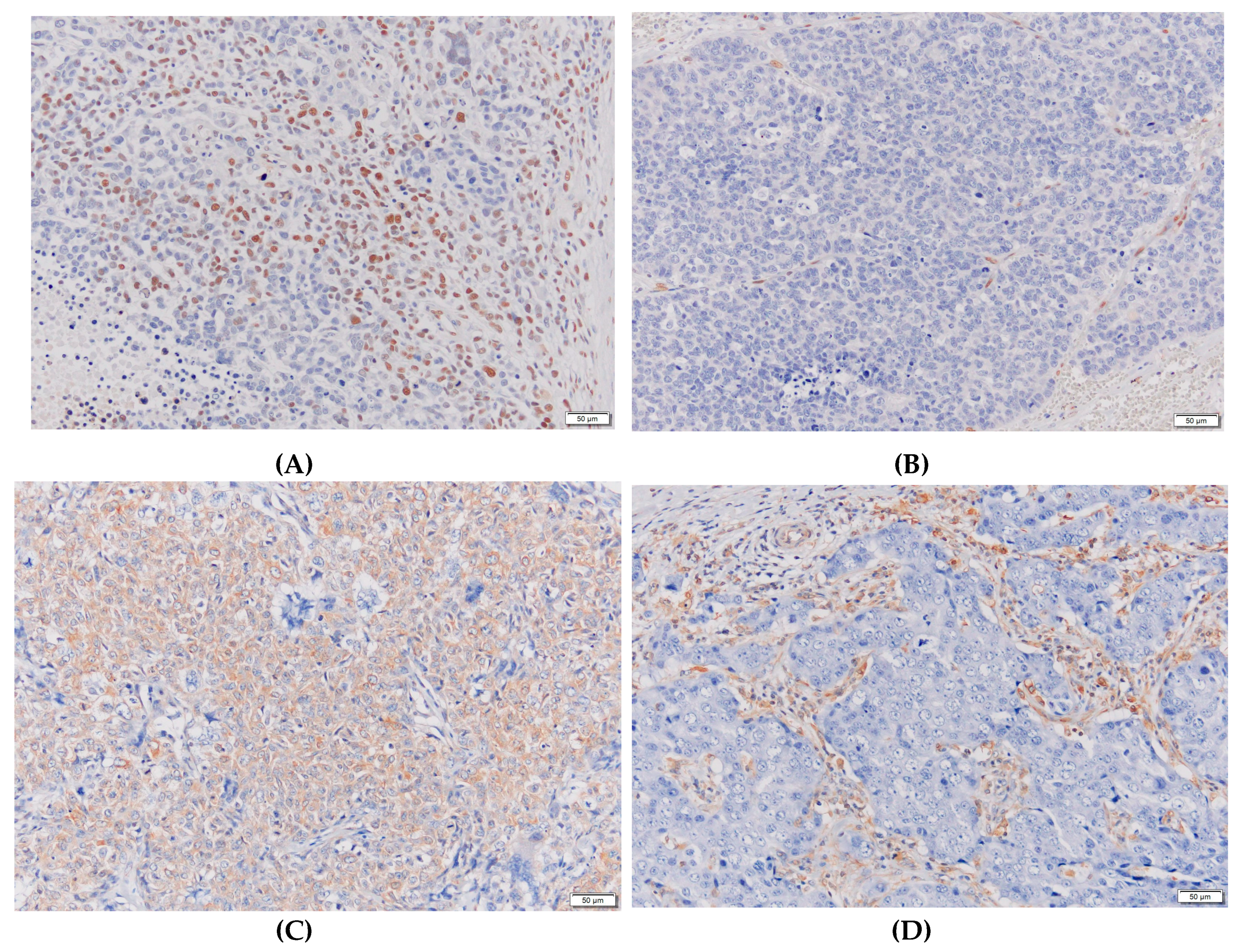
| Biomarkers | Classification | Number of cases | Rate of grade 2 | P-value |
|---|---|---|---|---|
| Ki67LI | < 20% | 8 | 25% | NS* |
| > 20% | 15 | 27% | ||
| Apoptosis | < 1% | 7 | 14% | NS |
| > 1% | 17 | 29% | ||
| ZEB1 | < 10% | 24 | 25% | NA** |
| > 10% | 0 | |||
| Vimentin | < 10% | 24 | 25% | NA |
| > 10% | 0 | |||
| E-cadherin | < 25% | 3 | 0% | NS |
| > 25% | 21 | 29% | ||
| Bmi-1 | < 50% | 5 | 0% | NS |
| > 50% | 19 | 32% | ||
| ALDH | < 10% | 20 | 25% | NS |
| > 10% | 4 | 25% | ||
| TILs | < 50% | 21 | 19% | 0. 0748 (trend) |
| > 50% | 3 | 67% | ||
| PD-L1 | Scores 0–3 | 15 | 13% | 0.0884 (trend) |
| Score 4 | 9 | 44% | ||
| PD-1 | Score 1 or 2 | 19 | 11% | 0.0014 |
| Score 3 | 5 | 80% | ||
| CD8 | Score 1 or 2 | 20 | 15% | 0.0114 |
| Score 3 | 4 | 75% | ||
| CD4 | Score 1 or 2 | 21 | 14% | 0.0053 |
| Score 3 | 3 | 100% | ||
| FoxP3 | Score 1 or 2 | 21 | 19% | 0.0748 (trend) |
| Score 3 | 3 | 67% |
| Biomarkers | Classification | Number of cases | Rate of grade 2 | P-value |
|---|---|---|---|---|
| Ki67LI | < 20% | 9 | 33% | NS* |
| > 20% | 15 | 20% | ||
| Apoptosis | < 1% | 10 | 30% | NS |
| > 1% | 13 | 23% | ||
| ZEB1 | < 10% | 23 | 26% | NS |
| > 10% | 1 | 0% | ||
| Vimentin | < 10% | 21 | 29% | NS |
| > 10% | 3 | 0% | ||
| E-cadherin | < 25% | 7 | 14% | NS |
| > 25% | 17 | 29% | ||
| Bmi-1 | < 50% | 5 | 0% | NS |
| > 50% | 19 | 32% | ||
| ALDH | < 10% | 15 | 27% | NS |
| > 10% | 9 | 22% | ||
| TILs | < 50% | 21 | 19% | 0.0748 (trend) |
| > 50% | 3 | 67% | ||
| PD-L1 | Scores 0–3 | 17 | 12% | 0.0196 |
| Score 4 | 7 | 57% | ||
| PD-1 | Score 1 or 2 | 21 | 24% | NS |
| Score 3 | 3 | 33% | ||
| CD8 | Score 1 or 2 | 20 | 15% | 0.0114 |
| Score 3 | 4 | 75% | ||
| CD4 | Score 1 or 2 | 24 | 25% | NA** |
| Score 3 | 0 | |||
| FoxP3 | Score 1 or 2 | 22 | 23% | NS |
| Score 3 | 2 | 50% |
| Biomarkers | Classification | Number of cases | DFS | OS |
|---|---|---|---|---|
| P-value | P-value | |||
| Ki67LI | < 20% | 8 | NS* | NS |
| > 20% | 15 | |||
| Apoptosis | < 1% | 7 | NS | NS |
| > 1% | 17 | |||
| ZEB1 | < 10% | 24 | NA** | NA |
| > 10% | 0 | |||
| Vimentin | < 10% | 24 | NA | NA |
| > 10% | 0 | |||
| E-cadherin | < 25% | 3 | NE*** | NE |
| > 25% | 21 | |||
| Bmi-1 | < 50% | 5 | NS | NS |
| > 50% | 19 | |||
| ALDH | < 10% | 20 | NS | NS |
| > 10% | 4 | |||
| TILs | < 50% | 21 | NE | NE |
| > 50% | 3 | |||
| PD-L1 | Scores 0–3 | 15 | NS | NS |
| Score 4 | 9 | |||
| PD-1 | Score 1 or 2 | 19 | NS | NS |
| Score 3 | 5 | |||
| CD8 | Score 1 or 2 | 20 | NS | NS |
| Score 3 | 4 | |||
| CD4 | Score 1 or 2 | 21 | NS | NS |
| Score 3 | 3 | |||
| FoxP3 | Score 1 or 2 | 21 | NE | NE |
| Score 3 | 3 |
| Biomarkers | Classification | Number of cases | DFS | OS |
|---|---|---|---|---|
| P-value | P-value | |||
| Ki67LI | < 20% | 9 | 0.0831 | NS* |
| > 20% | 15 | |||
| Apoptosis | < 1% | 10 | NS | NS |
| > 1% | 13 | |||
| ZEB1 | < 10% | 23 | 0.0014 | 0.0014 |
| > 10% | 1 | |||
| Vimentin | < 10% | 20 | 0.0255 | 0.005 |
| > 10% | 4 | |||
| E-cadherin | < 25% | 7 | NS | NS |
| > 25% | 17 | |||
| Bmi-1 | < 50% | 5 | NS | NS |
| > 50% | 19 | |||
| ALDH | < 10% | 15 | NS | NS |
| > 10% | 9 | |||
| TILs | < 50% | 21 | NE*** | NE |
| > 50% | 3 | |||
| PD-L1 | Scores 0–3 | 17 | NS | NS |
| Score 4 | 7 | |||
| PD-1 | Score 1 or 2 | 21 | NS | NE |
| Score 3 | 3 | |||
| CD8 | Score 1 or 2 | 20 | NE | NE |
| Score 3 | 4 | |||
| CD4 | Score 1 or 2 | 24 | NA** | NA |
| Score 3 | 0 | |||
| FoxP3 | Score 1 or 2 | 21 | NE | NS |
| Score 3 | 3 | |||
| ly | Negative | 6 | NE | NE |
| Positive | 18 | |||
| v | Negative | 20 | 0.0019 | 0.0032 |
| Positive | 4 | |||
| NG | 1 or 2 | 11 | 0.0446 | 0.0250 |
| 3 | 12 | |||
| HG | 1 or 2 | 12 | 0.0924 (trend) | 0.0696 (trend) |
| 3 | 11 | |||
| pN | Negative | 11 | NS | NS |
| Positive | 13 |
| Biomarkers | Classification | Number of cases | DFS | OS |
|---|---|---|---|---|
| P-value | P-value | |||
| Ki67LI | Increase | 10 | NS* | NS |
| Apoptosis | Increase | 12 | NS | NS |
| ZEB1 | Increase | 1 | 0.0014 | 0.0014 |
| Vimentin | Increase | 3 | 0.0255 | 0.0050 |
| E-cadherin | Increase | 19 | NS | NS |
| Bmi-1 | Increase | 5 | NS | NS |
| ALDH | Increase | 8 | NS | NS |
| TILs | Increase | 6 | NS | NS |
| PD-L1 | Increase | 5 | NS | NS |
| PD-1 | Increase | 5 | NS | NS |
| CD8 | Increase | 5 | NS | NS |
| CD4 | Increase | 1 | NE*** | NE |
| FoxP3 | Increase | 3 | NS | NS |
| Biomarkers | Providers | Clone | Conditions |
|---|---|---|---|
| Ki67 | Dako | MIB-1 | Following provider's recommendation |
| TUNEL | Exalpha | Not applicable | Following provider's recommendation |
| ZEB1 | SantaCruz | H-102 | Dilution, ×200; incubation, 4 °C, overnight |
| E-Cadherin | Dako | NCH-38 | Dilution, ×100; incubation, room temperature, 30min |
| Vimentin (Ready to Use) | Dako | V9 | Dilution, ×1; incubation, room temperature, 30 min |
| Bmi-1 | abcam | EPR3745(2) | Dilution, ×400; incubation, room temperature, 60 min |
| ALDH | BD | 44/ALDH | Dilution, ×100; incubation, 4 °C, overnight |
| CD4 | Thermo | 4B12 | Dilution, ×10; incubation, room temperature, 60 min |
| CD8 | Thermo | SP16 | Dilution, ×50; incubation, 4 °C, overnight |
| FoxP3 | abcam | 236A/E7 | Dilution, ×100; incubation, 4 °C, overnight |
| PD-L1 | abcam | 28-8 | Dilution, ×200; incubation, room temperature, 60 min |
| PD-1 | abcam | NAT105 | Dilution, ×100; mechanical staining |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




