Submitted:
20 June 2024
Posted:
21 June 2024
Read the latest preprint version here
Abstract
Keywords:
1. Introduction
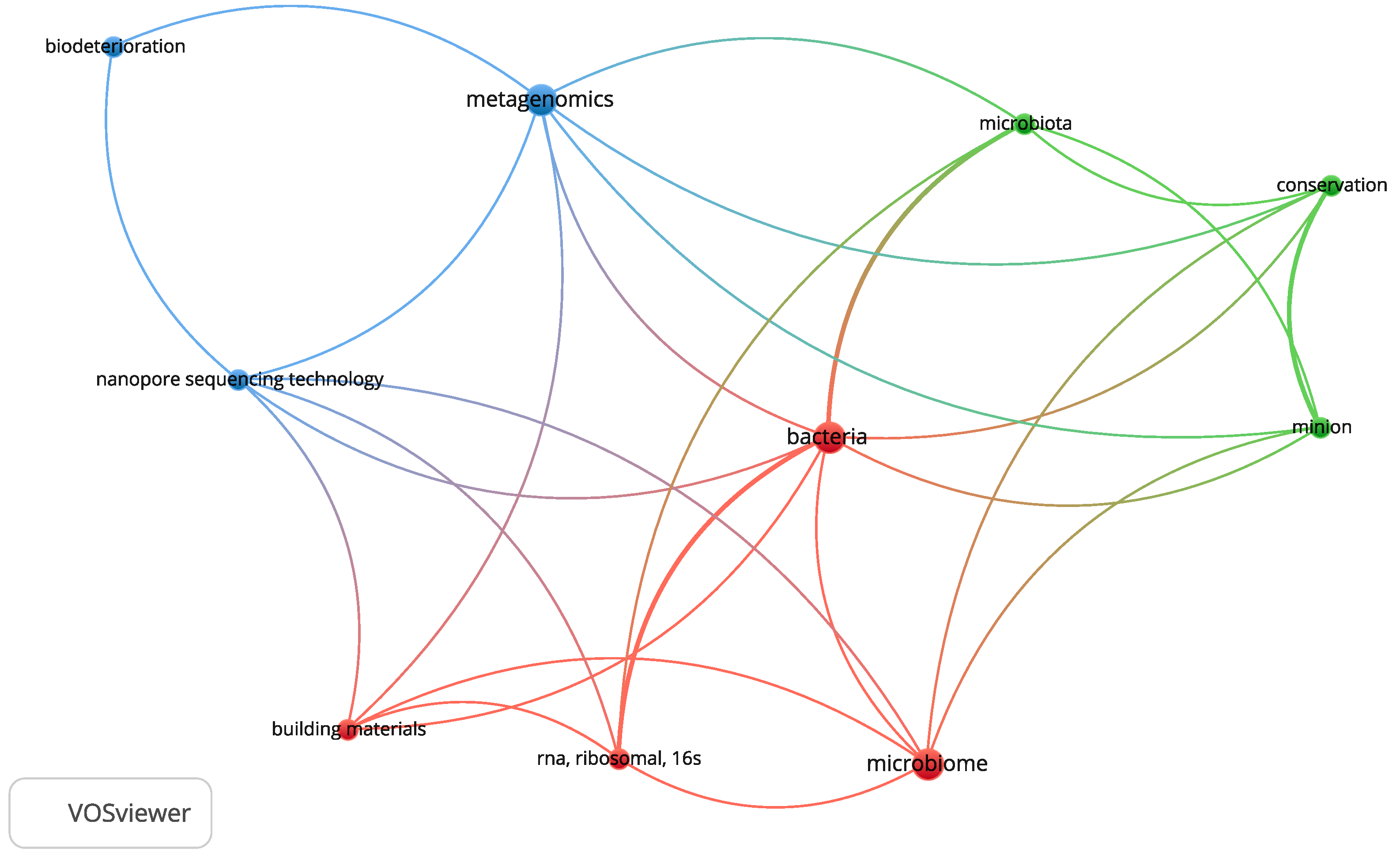
2. Application of Oxford Nanopore® in Cultural Heritage BIODETERIORATION studies
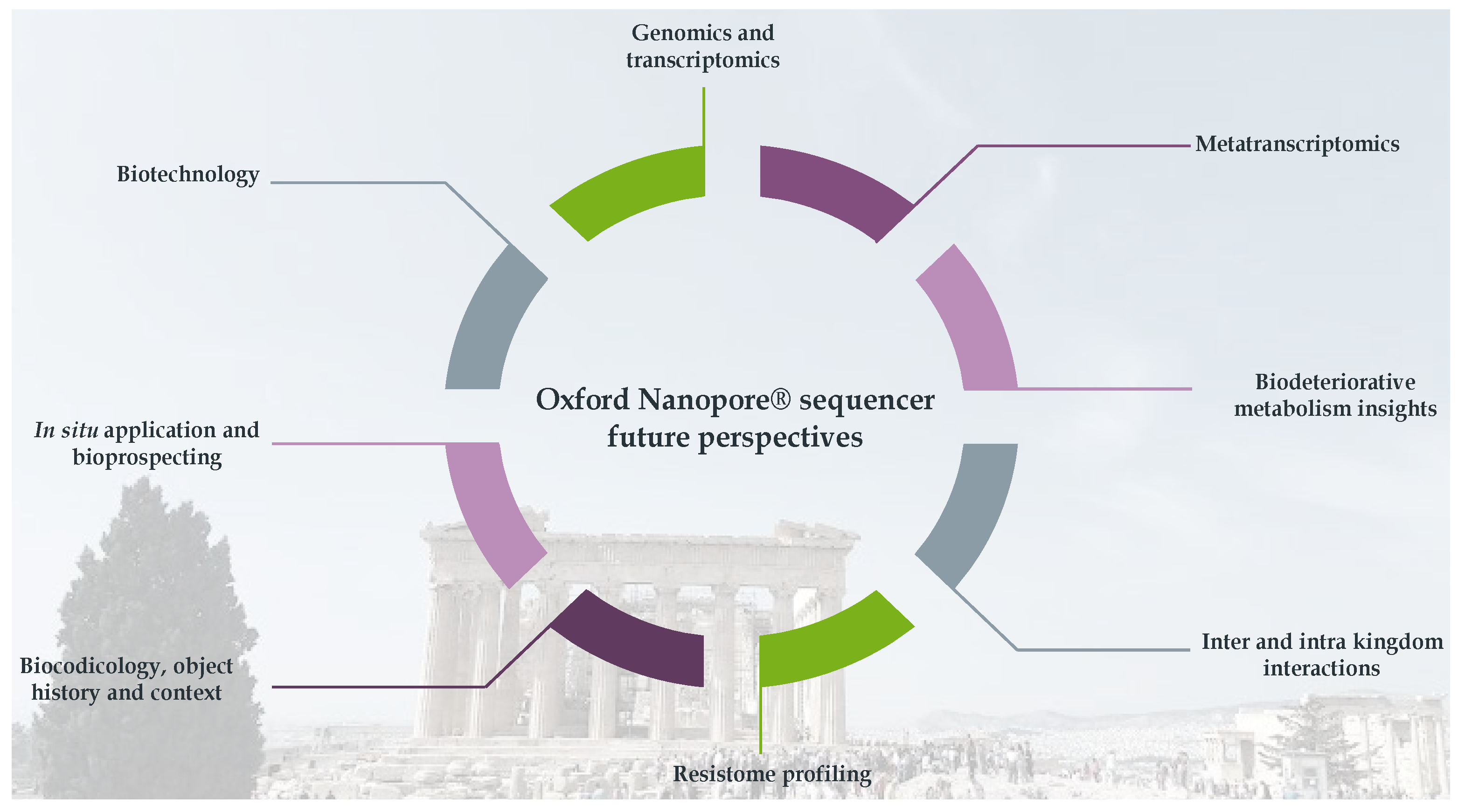
3. Future Directions for Oxford Nanopore® Applications in Biodeterioration Studies
3.1. Biodeteriogens Genomics and Transcriptomics
3.2. Metranscriptomics
3.3. Biodeteriorative Metabolic Insights
3.4. Ecological Multi-Kingdom Interactions
3.5. Cultural Heritage Resistome
3.6. Cultural Heritage Object History
3.7. On-Site Application
3.8. Biotechnology and Restoration Efforts
4. Challenges in the Oxford Nanopore® Application in Biodeterioration Studies
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hueck, H.J. The biodeterioration of materials as part of hylobiology. Mater Org 1965, 1, 1. 5– 34. [Google Scholar]
- Hueck, H.J. The biodeterioration of materials – an appraisal. In Biodeterioration of Materials. Walters, A.H., Elphick, J.S., Eds, Elsevier, London, UK, 1968, 6–12.
- Urzì, C., Krumbein. W.E., Microbiological impacts on the cultural heritage. In Durability and change: the science, responsability and cost of sustaining cultural heritage. Krumbein, W.E., Brimblecombe, P., Cosgrove D.E., Stainforth, S., Eds, John Wiley & Sons Ltd., London, UK, 1994, 107–135.
- Sterflinger, K.; Piñar, G. Microbial Deterioration of Cultural Heritage and Works of Art--Tilting at Windmills? Appl Microbiol Biotechnol 2013, 97, 9637–9646. [Google Scholar] [CrossRef]
- Liu, X.; Qian, Y.; Wu, F.; Wang, Y.; Wang, W.; Gu, J.-D. Biofilms on Stone Monuments: Biodeterioration or Bioprotection? Trends Microbiol 2022, 30, 816–819. [Google Scholar] [CrossRef]
- Beata, G. The Use of -Omics Tools for Assessing Biodeterioration of Cultural Heritage: A Review. J Cult Herit 2020, 45, 351–361. [Google Scholar] [CrossRef]
- Sterflinger, K.; Piñar, G. Molecular-Based Techniques for the Study of Microbial Communities in Artworks. In Microorganisms in the Deterioration and Preservation of Cultural Heritage; Joseph, E., Ed.; Springer International Publishing: Cham, 2021; pp. 59–77. ISBN 978-3-030-69411-1. [Google Scholar]
- Piñar, G.; Sterflinger, K. Natural Sciences at the Service of Art and Cultural Heritage: An Interdisciplinary Area in Development and Important Challenges. Microb Biotechnol 2021, 14, 806–809. [Google Scholar] [CrossRef]
- Quail, M.A.; Smith, M.; Coupland, P.; Otto, T.D.; Harris, S.R.; Connor, T.R.; Bertoni, A.; Swerdlow, H.P.; Gu, Y. A tale of three next generation sequencing platforms: Comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genom 2012, 13, 341. [Google Scholar] [CrossRef]
- Shokralla, S.; Spall, J.L.; Gibson, J.F.; Hajibabaei, M. Next-generation sequencing technologies for environmental DNA research. Mol Ecol 2012, 21, 1794–1805. [Google Scholar] [CrossRef]
- Buermans, H.P.J.; den Dunnen, J.T. Next generation sequencing technology: Advances and applications. Biochim Biophys Acta 2014, 1842, 1932–1941. [Google Scholar] [CrossRef]
- Goodwin, S.; McPherson, J.D.; McCombie, W.R. Coming of age: Ten years of next-generation sequencing technologies. Nat Rev Genet 2016, 17, 333–351. [Google Scholar] [CrossRef]
- Amarasinghe, S.L.; Su, S.; Dong, X.; Zappia, L.; Ritchie, M.E.; Gouil, Q. Opportunities and challenges in long-read sequencing data analysis. Genome Biol 2020, 21, 30. [Google Scholar] [CrossRef]
- Mohammed, A.A.; Senbeta, B.; Worku, T.; Mohammed, A.A.; Senbeta, B.; Worku, T. Pacific bioscience sequence technology: Review. Int J Vet Sci Res 2022, 8, 27–33. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, Y.; Bollas, A.; Wang, Y.; Au, K.F. Nanopore sequencing technology, bioinformatics and applications. Nat Biotechnol 2021, 39, 1348–1365. [Google Scholar] [CrossRef]
- Pavlovic, J.; Cavalieri, D.; Mastromei, G.; Pangallo, D.; Perito, B.; Marvasi, M. MinION Technology for Microbiome Sequencing Applications for the Conservation of Cultural Heritage. Microbiol Res 2021, 247, 126727. [Google Scholar] [CrossRef]
- Grottoli, A.; Beccaccioli, M.; Zoppis, E.; Fratini, R.S.; Schifano, E.; Santarelli, M.L.; Uccelletti, D.; Reverberi, M. Nanopore Sequencing and Bioinformatics for Rapidly Identifying Cultural Heritage Spoilage Microorganisms. Front Mater 2020, 7. [Google Scholar] [CrossRef]
- Kisová, Z.; Planý, M.; Pavlović, J.; Bučková, M.; Puškárová, A.; Kraková, L.; Kapustová, M.; Pangallo, D.; Šoltys, K. Biodeteriogens Characterization and Molecular Analyses of Diverse Funeral Accessories from XVII Century. Appl Sci 2020, 10, 5451. [Google Scholar] [CrossRef]
- Šoltys, K.; Planý, M.; Biocca, P.; Vianello, V.; Bučková, M.; Puškárová, A.; Sclocchi, M.C.; Colaizzi, P.; Bicchieri, M.; Pangallo, D.; et al. Lead Soaps Formation and Biodiversity in a XVIII Century Wax Seal Coloured with Minium. Environ Microbiol 2020, 22, 1517–1534. [Google Scholar] [CrossRef]
- Pinar, G.; Poyntner, C.; Lopandic, K.; Tafer, H.; Sterflinger, K. Rapid Diagnosis of Biological Colonization in Cultural Artefacts Using the MinION Nanopore Sequencing Technology. Int Biodeterior Biodegradation 2020, 148. [Google Scholar] [CrossRef]
- Piñar, G.; Sclocchi, M.C.; Pinzari, F.; Colaizzi, P.; Graf, A.; Sebastiani, M.L.; Sterflinger, K. The Microbiome of Leonardo Da Vinci’s Drawings: A Bio-Archive of Their History. Front Microbiol 2020, 11. [Google Scholar] [CrossRef]
- Planý, M.; Pinzari, F.; Šoltys, K.; Kraková, L.; Cornish, L.; Pangallo, D.; Jungblut, A.D.; Little, B. Fungal-Induced Atmospheric Iron Corrosion in an Indoor Environment. Int Biodeterior Biodegradation 2021, 159, 105204. [Google Scholar] [CrossRef]
- Brimblecombe, P.; Sterflinger, K.; Derksen, K.; Haltrich, M.; Querner, P. Thermohygrometric Climate, Insects and Fungi in the Klosterneuburg Monastic Library. Heritage 2022, 5, 4228–4244. [Google Scholar] [CrossRef]
- Delegou, E.; Karapiperis, C.; Hilioti, Z.; Chasapi, A.; Valasiadis, D.; Alexandridou, A.; Rihani, V.; Kroustalaki, M.; Bris, T.; Ouzounis, C.; et al. Metagenomics of the built cultural heritage: microbiota characterization of the building materials of the holy aedicule of the holy sepulchre in Jerusalem. CIENTIFIC CULTURE 2022, 8, 59–83. [Google Scholar] [CrossRef]
- Pavlović, J.; Bosch-Roig, P.; Rusková, M.; Planý, M.; Pangallo, D.; Sanmartín, P. Long-Amplicon MinION-Based Sequencing Study in a Salt-Contaminated Twelfth Century Granite-Built Chapel. Appl Microbiol Biotechnol 2022, 106, 4297–4314. [Google Scholar] [CrossRef] [PubMed]
- Pavlović, J.; Sclocchi, M.C.; Planý, M.; Ruggiero, D.; Puškárová, A.; Bučková, M.; Šoltys, K.; Colaizzi, P.; Riccardi, M.L.; Pangallo, D.; et al. The Microbiome of Candle Beeswax Drops on Ancient Manuscripts. Int Biodeterior Biodegradation 2022, 174, 105482. [Google Scholar] [CrossRef]
- Rabbachin, L.; Piñar, G.; Nir, I.; Kushmaro, A.; Pavan, M.J.; Eitenberger, E.; Waldherr, M.; Graf, A.; Sterflinger, K. A Multi-Analytical Approach to Infer Mineral–Microbial Interactions Applied to Petroglyph Sites in the Negev Desert of Israel. Appl Sci 2022, 12, 6936. [Google Scholar] [CrossRef]
- Timoncini, A.; Costantini, F.; Bernardi, E.; Martini, C.; Mugnai, F.; Mancuso, F.; Sassoni, E.; Ospitali, F.; Chiavari, C. Insight on Bacteria Communities in Outdoor Bronze and Marble Artefacts in a Changing Environment. Sci Total Environ 2022, 850, 157804. [Google Scholar] [CrossRef]
- Beccaccioli, M.; Moricca, C.; Faino, L.; Reale, R.; Mineo, M.; Reverberi, M. The Neolithic Site “La Marmotta”: DNA Metabarcoding to Identify the Microbial Deterioration of Waterlogged Archeological Wood. Front Microbiol 2023, 14. [Google Scholar] [CrossRef]
- Li, Q.; Wu, C.; He, J.; Zhang, B. Unraveling the Microbiotas and Key Genetic Contexts Identified on Stone Heritage Using Illumina and Nanopore Sequencing Platforms. Int Biodeterior Biodegradation 2023, 185. [Google Scholar] [CrossRef]
- Nir, I.; Barak, H.; Rabbachin, L.; Arielle, K.; Pavan, M.; Winter, E.; Pinar, G.; Sterflinger, K.; Ariel, K. Trichocoleus Desertorum Isolated from Negev Desert Petroglyphs: Characterization, Adaptation and Bioerosion Potential. The Sci Total Environ 2023, 904, 166739. [Google Scholar] [CrossRef]
- Pavlović, J.; Puškárová, A.; Planý, M.; Farkas, Z.; Rusková, M.; Kvalová, K.; Kraková, L.; Bučková, M.; Pangallo, D. Colored Stains: Microbial Survey of Cellulose-Based and Lignin Rich Papers. Int J Biol Macromol 2023, 241, 124456. [Google Scholar] [CrossRef]
- Rabbachin, L.; Pinar, G.; Nir, I.; Kushmaro, A.; Eitenberger, E.; Waldherr, M.; Graf, A.; Sterflinger, K. Natural Biopatina on Historical Petroglyphs in the Austrian Alps: To Clean or Not to Clean? Int Biodeterior Biodegradation 2023, 183, 105632. [Google Scholar] [CrossRef]
- Tichy, J.; Waldherr, M.; Ortbauer, M.; Graf, A.; Sipek, B.; Jembrih-Simbuerger, D.; Sterflinger, K.; Piñar, G. Pretty in Pink? Complementary Strategies for Analysing Pink Biofilms on Historical Buildings. Sci Total Environ 2023, 904, 166737. [Google Scholar] [CrossRef]
- Bastholm, C.J.; Andersen, B.; Frisvad, J.C.; Oestergaard, S.K.; Nielsen, J.L.; Madsen, A.M.; Richter, J. A Novel Contaminant in Museums? A Cross-Sectional Study on Xerophilic Aspergillus Growth in Climate-Controlled Repositories. Sci Total Environ 2024, 173880. [Google Scholar] [CrossRef]
- Haedar, N.; Iqram, M.; Priosambodo, D.; Lebe, R. Bacterial Communities on Degraded Prehistoric Rock Paintings in Maros-Pangkep Global Geopark. Philippine Journal of Science 2024, 153, 391–402. [Google Scholar]
- Marvasi, M.; Pangallo, D.; Cavalieri, D.; Poyatos-Jiménez, F. Editorial: Multi-Omics Revolution in Microbial Cultural Heritage Conservation. Front Microbiol 2021, 12. [Google Scholar] [CrossRef]
- De Leo, F.; Marchetta, A.; Urzì, C. Black Fungi on Stone-Built Heritage: Current Knowledge and Future Outlook. Appl Sci 2022, 12, 3969. [Google Scholar] [CrossRef]
- Trovão, J.; Tiago, I.; Soares, F.; Paiva, D.S.; Mesquita, N.; Coelho, C.; Catarino, L.; Gil, F.; Portugal, A. High-Quality Draft Genome Sequence of the Microcolonial Black Fungus Aeminium Ludgeri DSM 106916. Microbiol Resour Announc 2019, 8, e00202–19. [Google Scholar] [CrossRef]
- Quach, N.T.; Ngo, C.C.; Nguyen, T.H.; Nguyen, P.L.; Vu, T.H.N.; Phan, T.H.T.; Nguyen, Q.H.; Le, T.T.M.; Chu, H.H.; Phi, Q.-T. Genome-Wide Comparison Deciphers Lifestyle Adaptation and Glass Biodeterioration Property of Curvularia Eragrostidis C52. Sci Rep 2022, 12, 11411. [Google Scholar] [CrossRef]
- Paiva, D.S.; Fernandes, L.; Portugal, A.; Trovão, J. First Genome Sequence of the Microcolonial Black Fungus Saxispiralis Lemnorum MUM 23.14: Insights into the Unique Genomic Traits of the Aeminiaceae Family. Microorganisms 2024, 12, 104. [Google Scholar] [CrossRef]
- Pei, S.; Wu, F.; Chen, Y.; Ma, W.; He, D.; Zhang, Q.; Gu, J.-D.; Wang, W.; Tian, T.; Feng, H. Mechanisms of Lead-Containing Pigment Discoloration Caused by Naumannella Cuiyingiana AFT2T Isolated from 1500 Years Tomb Wall Painting of China. Int Biodeterior Biodegradation 2023, 185, 105689. [Google Scholar] [CrossRef]
- Wang, Y.; Han, Y.; Li, N.; Wang, C.; Ma, K.; Huang, X.; Du, J.; Guo, H.; Pan, J. Study on Biodegradation Mechanism of Fusarium Solani NK-NH1 on the Hull Wood of the Nanhai No. 1 Shipwreck. Front Microbiol 2024, 15, 1382653. [Google Scholar] [CrossRef]
- Zhao, W.; Zeng, W.; Pang, B.; Luo, M.; Peng, Y.; Xu, J.; Kan, B.; Li, Z.; Lu, X. Oxford Nanopore Long-Read Sequencing Enables the Generation of Complete Bacterial and Plasmid Genomes without Short-Read Sequencing. Front Microbiol 2023, 14. [Google Scholar] [CrossRef] [PubMed]
- Salazar, A.N.; Gorter de Vries, A.R.; van den Broek, M.; Wijsman, M.; de la Torre Cortés, P.; Brickwedde, A.; Brouwers, N.; Daran, J.-M.G.; Abeel, T. Nanopore Sequencing Enables Near-Complete de Novo Assembly of Saccharomyces Cerevisiae Reference Strain CEN.PK113-7D. FEMS Yeast Res 2017, 17, fox074. [Google Scholar] [CrossRef] [PubMed]
- McGinnis, J.L.; Giguere, D.J. High-Quality Genome Assembly of a Pestalotiopsis Fungus Using DIY-Friendly Methods 2022. [version 1; peer review: 3 approved with reservations]. F1000Research 2022, 11, 442. [Google Scholar] [CrossRef]
- Witte, T.E.; Hicks, C.; Shoukouhi, P.; Dadej, K.; Findlay, W.; Liu, M.; Overy, D.P. Chromosome-Level Draft Genome Sequences of Three Isolates of the Toxigenic Fungus Claviceps Purpurea Showing Structural Rearrangements. Microbiol Resour Announc 2023, 12, e00234-23. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wu, F.; Gu, J.-D.; He, K.; Fang, Z.; Liu, X.; He, D.; Ding, X.; Li, J.; Han, Z.; et al. Dominance by Cyanobacteria in the Newly Formed Biofilms on Stone Monuments under a Protective Shade at the Beishiku Temple in China. Env Res 2024, 251, 118576. [Google Scholar] [CrossRef] [PubMed]
- Haveman, N.J.; Khodadad, C.L.M.; Dixit, A.R.; Louyakis, A.S.; Massa, G.D.; Venkateswaran, K.; Foster, J.S. Evaluating the Lettuce Metatranscriptome with MinION Sequencing for Future Spaceflight Food Production Applications. npj Microgravity 2021, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Koestler, R.J.; Warscheid, T.; Katayama, Y.; Gu, J.-D. Microbial Deterioration and Sustainable Conservation of Stone Monuments and Buildings. Nat Sustain 2020, 3, 991–1004. [Google Scholar] [CrossRef]
- Wu, F.; Ding, X.; Zhang, Y.; Gu, J.-D.; Liu, X.; Guo, Q.; Li, J.; Feng, H. Metagenomic and Metaproteomic Insights into the Microbiome and the Key Geobiochemical Potentials on the Sandstone of Rock-Hewn Beishiku Temple in Northwest China. Sci Total Environ 2023, 893, 164616. [Google Scholar] [CrossRef] [PubMed]
- Qian, Z.; Li, Y.; Pratush, A.; Kan, J.; Gu, J.-D.; Peng, T.; Huang, T.; Hu, Z. A Comparative Analysis of the Microbial Communities and Functional Genes of the Nitrogen Cycling in Mangroves of China, Indian and Malaysia. Int Biodeterior Biodegradation 2024, 190, 105767. [Google Scholar] [CrossRef]
- Liu, W.; Zhou, X.; Jin, T.; Li, Y.; Wu, B.; Yu, D.; Yu, Z.; Su, B.; Chen, R.; Feng, Y.; et al. Multikingdom Interactions Govern the Microbiome in Subterranean Cultural Heritage Sites. PNAS 2022, 119, e2121141119. [Google Scholar] [CrossRef]
- Yu, Y.; Zhang, J.; Chen, R.; Coleine, C.; Liu, W.; Delgado-Baquerizo, M.; Feng, Y. Unearthing the Global Patterns of Cultural Heritage Microbiome for Conservation. Int Biodeterior Biodegradation 2024, 190, 105784. [Google Scholar] [CrossRef]
- Trovão, J.; Portugal, A. The Impact of Stone Position and Location on the Microbiome of a Marble Statue. The Microbe 2024, 2, 100040. [Google Scholar] [CrossRef]
- He, J.; Zhang, N.; Shen, X.; Muhammad, A.; Shao, Y. Deciphering Environmental Resistome and Mobilome Risks on the Stone Monument: A Reservoir of Antimicrobial Resistance Genes. Sci Total Environ 2022, 838, 156443. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Lan, W.; Li, J.; Deng, M.; Li, Y.; Katayama, Y.; Gu, J.-D. Metagenomic Insight into the Pathogenic-Related Characteristics and Resistome Profiles within Microbiome Residing on the Angkor Sandstone Monuments in Cambodia. Sci Total Environ 2024, 918, 170402. [Google Scholar] [CrossRef] [PubMed]
- Solcova, M.; Demnerova, K.; Purkrtova, S. Application of Nanopore Sequencing (MinION) for the Analysis of Bacteriome and Resistome of Bean Sprouts. Microorganisms 2021, 9, 937. [Google Scholar] [CrossRef] [PubMed]
- Piñar, G.; Poyntner, C.; Tafer, H.; Sterflinger, K. A Time Travel Story: Metagenomic Analyses Decipher the Unknown Geographical Shift and the Storage History of Possibly Smuggled Antique Marble Statues. Ann Microbiol 2019, 69, 1001–1021. [Google Scholar] [CrossRef]
- Vassallo, Y.; Waldherr, M.; Lehner, E.; Graf, A.; Cappa, F.; Hartl, A.; Schober, R.; Beccaccioli, M.; Sterflinger, K.; Piñar, G.; et al. Oxford Nanopore Technologies for Biocodicology: A Case Study on a 15th-Century Parchment - Vassallo Y., Waldherr M., Lehner E., Graf A., Cappa F., Hartl A., Schober R., Beccaccioli M., Sterflinger K., Piñar G., Reverberi M. In; DTC Lazio: Tecnologie e patrimonio culturale: nuove competenze e professioni. 2023, https://iris.uniroma1.it/handle/11573/1695400?mode=complete.
- Simon, L.M.; Flocco, C.; Burkart, F.; Methner, A.; Henke, D.; Rauer, L.; Müller, C.L.; Vogel, J.; Quaisser, C.; Overmann, J.; et al. Microbial Fingerprints Reveal Interaction between Museum Objects, Curators, and Visitors. iScience 2023, 26, 107578. [Google Scholar] [CrossRef]
- Cao, Y.; Bowker, M.A.; Delgado-Baquerizo, M.; Xiao, B. Biocrusts Protect the Great Wall of China from Erosion. Sci Adv 2023, 9, eadk5892. [Google Scholar] [CrossRef] [PubMed]
- Castro-Wallace, S.L.; Chiu, C.Y.; John, K.K.; Stahl, S.E.; Rubins, K.H.; McIntyre, A.B.R.; Dworkin, J.P.; Lupisella, M.L.; Smith, D.J.; Botkin, D.J.; et al. Nanopore DNA Sequencing and Genome Assembly on the International Space Station. Sci Rep 2017, 7, 18022. [Google Scholar] [CrossRef]
- Goordial, J.; Altshuler, I.; Hindson, K.; Chan-Yam, K.; Marcolefas, E.; Whyte, L.G. In Situ Field Sequencing and Life Detection in Remote (79°26′N) Canadian High Arctic Permafrost Ice Wedge Microbial Communities. Front Microbiol 2017, 8. [Google Scholar] [CrossRef]
- Harcourt, J.; Tamin, A.; Lu, X.; Kamili, S.; Sakthivel, S.K.; Murray, J.; Queen, K.; Tao, Y.; Paden, C.R.; Zhang, J.; et al. Isolation and Characterization of SARS-CoV-2 from the First US COVID-19 Patient. bioRxiv 2020, 2020.03.02.972935. [Google Scholar]
- Latorre-Pérez, A.; Pascual, J.; Porcar, M.; Vilanova, C. A Lab in the Field: Applications of Real-Time, in Situ Metagenomic Sequencing. Biology Methods and Protocols 2020, 5, bpaa016. [Google Scholar] [CrossRef]
- Moore, S.C.; Penrice-Randal, R.; Alruwaili, M.; Dong, X.; Pullan, S.T.; Carter, D.P.; Bewley, K.; Zhao, Q.; Sun, Y.; Hartley, C.; et al. Amplicon Based MinION Sequencing of SARS-CoV-2 and Metagenomic Characterisation of Nasopharyngeal Swabs from Patients with COVID-19 bioRxiv 2020, 2020.03.05.20032011.
- Quick, J.; Loman, N.J.; Duraffour, S.; Simpson, J.T.; Severi, E.; Cowley, L.; Bore, J.A.; Koundouno, R.; Dudas, G.; Mikhail, A.; et al. Real-Time, Portable Genome Sequencing for Ebola Surveillance. Nature 2016, 530, 228–232. [Google Scholar] [CrossRef]
- Wasswa, F.B.; Kassaza, K.; Nielsen, K.; Bazira, J. MinION Whole-Genome Sequencing in Resource-Limited Settings: Challenges and Opportunities. Curr Clin Micro Rpt 2022, 9, 52–59. [Google Scholar] [CrossRef]
- Werner, D.; Acharya, K.; Blackburn, A.; Zan, R.; Plaimart, J.; Allen, B.; Mgana, S.M.; Sabai, S.M.; Halla, F.F.; Massawa, S.M.; et al. MinION Nanopore Sequencing Accelerates Progress towards Ubiquitous Genetics in Water Research. Water 2022, 14, 2491. [Google Scholar] [CrossRef]
- Tamames, J.; Jiménez-Lalana, D.; Redondo, Á.; Martínez-García, S.; De Los Rios, A. In Situ Metagenomics: A Platform for Rapid Sequencing and Analysis of Metagenomes in Less than One Day. Mol Ecol Resour 2024, 24, e13909. [Google Scholar] [CrossRef]
- Bastholm, C.J.; Madsen, A.M.; Andersen, B.; Frisvad, J.C.; Richter, J. The Mysterious Mould Outbreak - A Comprehensive Fungal Colonisation in a Climate-Controlled Museum Repository Challenges the Environmental Guidelines for Heritage Collections. J Cult Herit 2022, 55, 78–87. [Google Scholar] [CrossRef]
- Martin-Pozas, T.; Nováková, A.; Jurado, V.; Cuezva, S.; Fernandez-Cortes, A.; Saiz-Jimenez, C.; Sanchez-Moral, S. A Second Fungal Outbreak in Castañar Cave, Spain, Discloses the Fragility of Subsurface Ecosystems. Microb Ecol 2024, 87, 53. [Google Scholar] [CrossRef]
- Trovão, J.; Portugal, A. Current Knowledge on the Fungal Degradation Abilities Profiled through Biodeteriorative Plate Essays. Appl Sci 2021, 11, 4196. [Google Scholar] [CrossRef]
- Pyzik, A.; Ciuchcinski, K.; Dziurzynski, M.; Dziewit, L. The Bad and the Good—Microorganisms in Cultural Heritage Environments—An Update on Biodeterioration and Biotreatment Approaches. Materials 2021, 14, 177. [Google Scholar] [CrossRef]
- Latorre-Pérez, A.; Gimeno-Valero, H.; Tanner, K.; Pascual, J.; Vilanova, C.; Porcar, M. A Round Trip to the Desert: In Situ Nanopore Sequencing Informs Targeted Bioprospecting. Front Microbiol 2021, 12. [Google Scholar] [CrossRef]
- Andreolli, M.; Lampis, S.; Bernardi, P.; Calò, S.; Vallini, G. Bacteria from Black Crusts on Stone Monuments Can Precipitate CaCO3 Allowing the Development of a New Bio-Consolidation Protocol for Ornamental Stone. Int Biodeterior Biodegradation 2020, 153, 105031. [Google Scholar] [CrossRef]
- Dhami, N.K.; Reddy, M.S.; Mukherjee, A. Application of Calcifying Bacteria for Remediation of Stones and Cultural Heritages. Front Microbiol 2014, 5. [Google Scholar] [CrossRef]
- Reddy, M.S. Biomineralization of Calcium Carbonates and Their Engineered Applications: A Review. Front Microbiol 2013, 4. [Google Scholar] [CrossRef]
- Cappitelli, F. Biocleaning of Cultural Heritage Surfaces. Open Conf Proc J 2016, 7. [Google Scholar] [CrossRef]
- Ranalli, G.; Zanardini, E. Biocleaning on Cultural Heritage: New Frontiers of Microbial Biotechnologies. Journal of Applied Microbiology 2021, 131, 583–603. [Google Scholar] [CrossRef]
- Bosch-Roig, P.; Sanmartín, P. Bioremoval of Graffiti in the Context of Current Biocleaning Research. In Microorganisms in the Deterioration and Preservation of Cultural Heritage; Joseph, E., Ed.; Springer International Publishing: Cham, 2021; pp. 175–197. ISBN 978-3-030-69411-1. [Google Scholar]
- Cattò, C.; Sanmartín, P.; Gulotta, D.; Troiano, F.; Cappitelli, F. Bioremoval of Graffiti Using Novel Commercial Strains of Bacteria. Sci Total Environ 2021, 756, 144075. [Google Scholar] [CrossRef]
- Sanmartín, P.; Bosch-Roig, P.; Pangallo, D.; Kraková, L.; Serrano, M. Unraveling Disparate Roles of Organisms, from Plants to Bacteria, and Viruses on Built Cultural Heritage. Appl Microbiol Biotechnol 2023, 107, 2027–2037. [Google Scholar] [CrossRef]
- Kerkhof, L.J. Is Oxford Nanopore Sequencing Ready for Analyzing Complex Microbiomes? FEMS Microbiol. Ecol. 2021, 97, fiab001. [Google Scholar] [CrossRef]
- Ni, Y.; Liu, X.; Simeneh, Z.M.; Yang, M.; Li, R. Benchmarking of Nanopore R10.4 and R9.4.1 Flow Cells in Single-Cell Whole-Genome Amplification and Whole-Genome Shotgun Sequencing. Comput Struct Biotechnol J 2023, 21, 2352–2364. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Li, H.; Ma, S.; Cao, J.; Liao, H.; Huang, Q.; Chen, W. The Newest Oxford Nanopore R10.4.1 Full-Length 16S rRNA Sequencing Enables the Accurate Resolution of Species-Level Microbial Community Profiling. Appl Environ Microbiol 2023, 89, e0060523. [Google Scholar] [CrossRef] [PubMed]
- Sereika, M.; Kirkegaard, R.H.; Karst, S.M.; Michaelsen, T.Y.; Sørensen, E.A.; Wollenberg, R.D.; Albertsen, M. Oxford Nanopore R10.4 Long-Read Sequencing Enables the Generation of near-Finished Bacterial Genomes from Pure Cultures and Metagenomes without Short-Read or Reference Polishing. Nat Methods 2022, 19, 823–826. [Google Scholar] [CrossRef] [PubMed]
- Maghini, D.G.; Moss, E.L.; Vance, S.E.; Bhatt, A.S. Improved High-Molecular-Weight DNA Extraction, Nanopore Sequencing and Metagenomic Assembly from the Human Gut Microbiome. Nat Protoc 2021, 16, 458–471. [Google Scholar] [CrossRef]
- Zorz, J.; Li, C.; Chakraborty, A.; Gittins, D.A.; Surcon, T.; Morrison, N.; Bennett, R.; MacDonald, A.; Hubert, C.R.J. SituSeq: An Offline Protocol for Rapid and Remote Nanopore 16S rRNA Amplicon Sequence Analysis. ISME Commun 2023, 3, 1–11. [Google Scholar] [CrossRef]
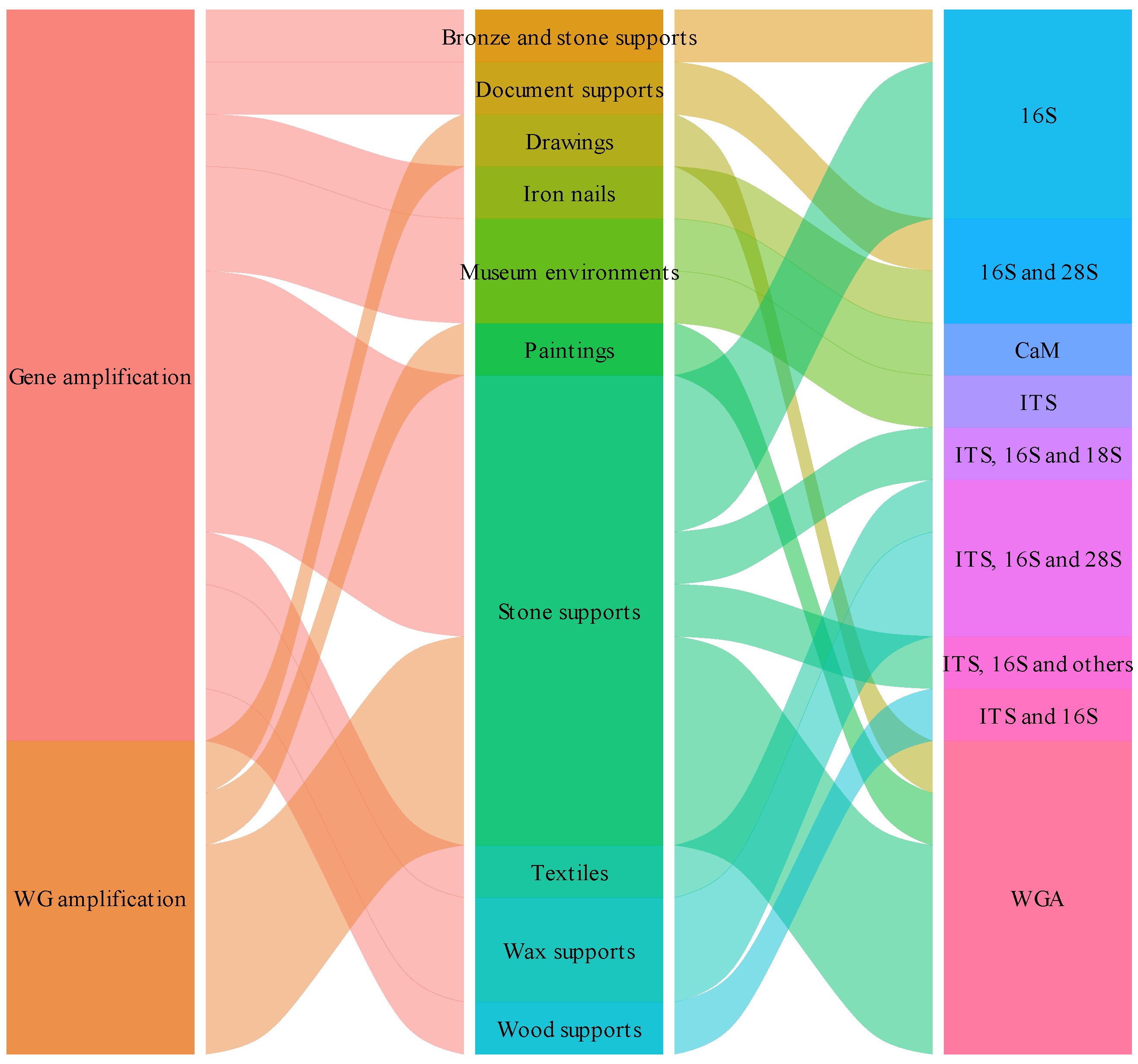
| Approach | Material | Reference |
|---|---|---|
| Gene amplification | Stone monument | [17] |
| Gene amplification | Funeral accessories textiles | [18] |
| Gene amplification | Wax seal | [19] |
| Whole genome amplification | Oil painting | [20] |
| Whole genome amplification | Drawings | [21] |
| Gene amplification | Iron nails from whale skeleton | [22] |
| Gene amplification | Museum surfaces | [23] |
| Gene amplification | Stone monument | [24] |
| Gene amplification | Granite chapel | [25] |
| Gene amplification | Beeswax drops | [26] |
| Whole genome amplification | Petroglyph sites | [27] |
| Gene amplification | Bronze and marble statues | [28] |
| Gene amplification | Waterlogged archeological wood | [29] |
| Whole genome amplification | Stone monument | [30] |
| Whole genome amplification | Petroglyph sites | [31] |
| Gene amplification | Documents | [32] |
| Whole genome amplification | Petroglyph sites | [33] |
| Gene amplification | Salt-weathered buildings | [34] |
| Gene amplification | Air and museum surfaces | [35] |
| Gene amplification | Rock Paintings | [36] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





