Submitted:
19 June 2024
Posted:
21 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Sediment and Water Sampling
2.2. Lagoon Status of Contamination - Analytical Development
2.2.1. Chemicals and Materials
- For water sample extraction:
- For sediment sample extraction:
- For UHPLC-MS/MS analyses:
2.2.2. Extraction Protocols
- Water samples
- Sediment samples
2.2.3. UHPLC-MS(/MS) Analyses
2.2.4. Data Processing
2.3. Omics Approaches Set Up
2.3.1. Non Targeted Metabolomics
- Chemicals and Materials
- Extraction protocols
- UHPLC-HRMS analyses
- Data processing
2.3.2. DNA Extraction and Metabarcoding Analyses
- Protocols for extracting DNA from sediments and assessing DNA quantity and quality
- 16S rRNAgene metabarcoding
- Bioinformatic analysis of amplicons sequences
3. Results and Discussion
3.1. Lagoon Status of Chemical Contamination
3.2. Omics Approaches Set Up
3.2.1. Non Targeted Metabolomics
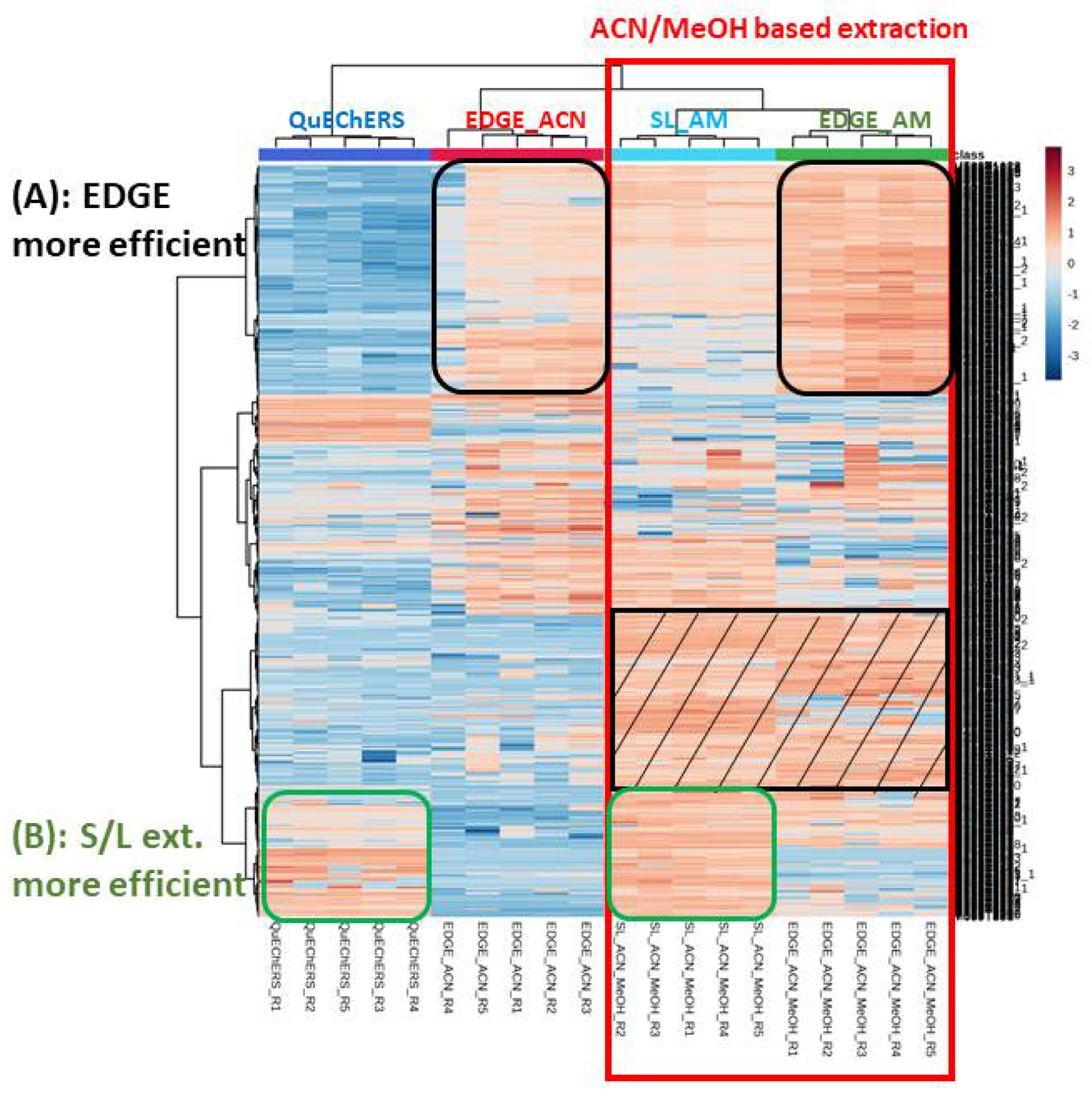
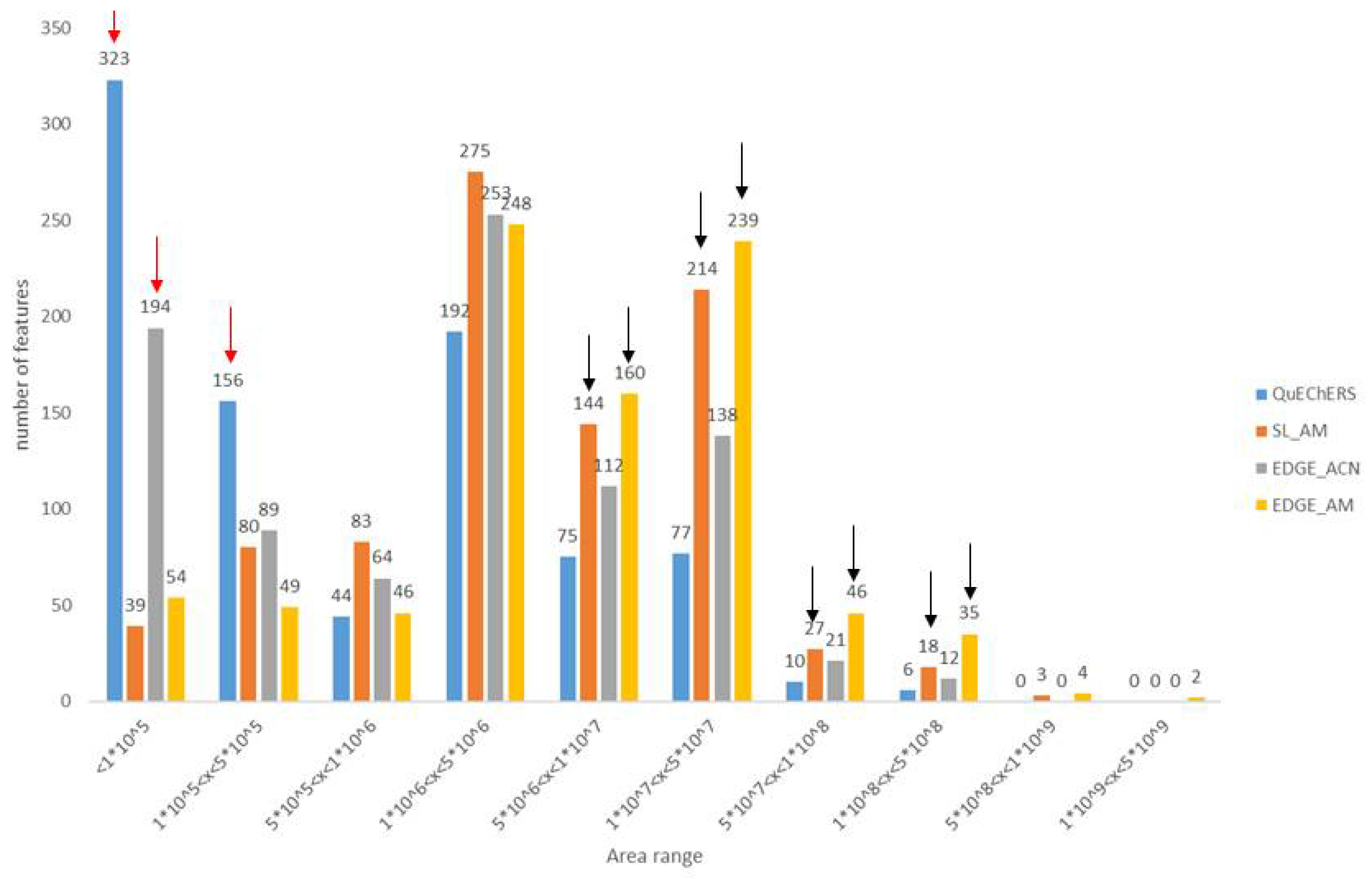
- Sediment meta-metabolome coverage and method efficiency
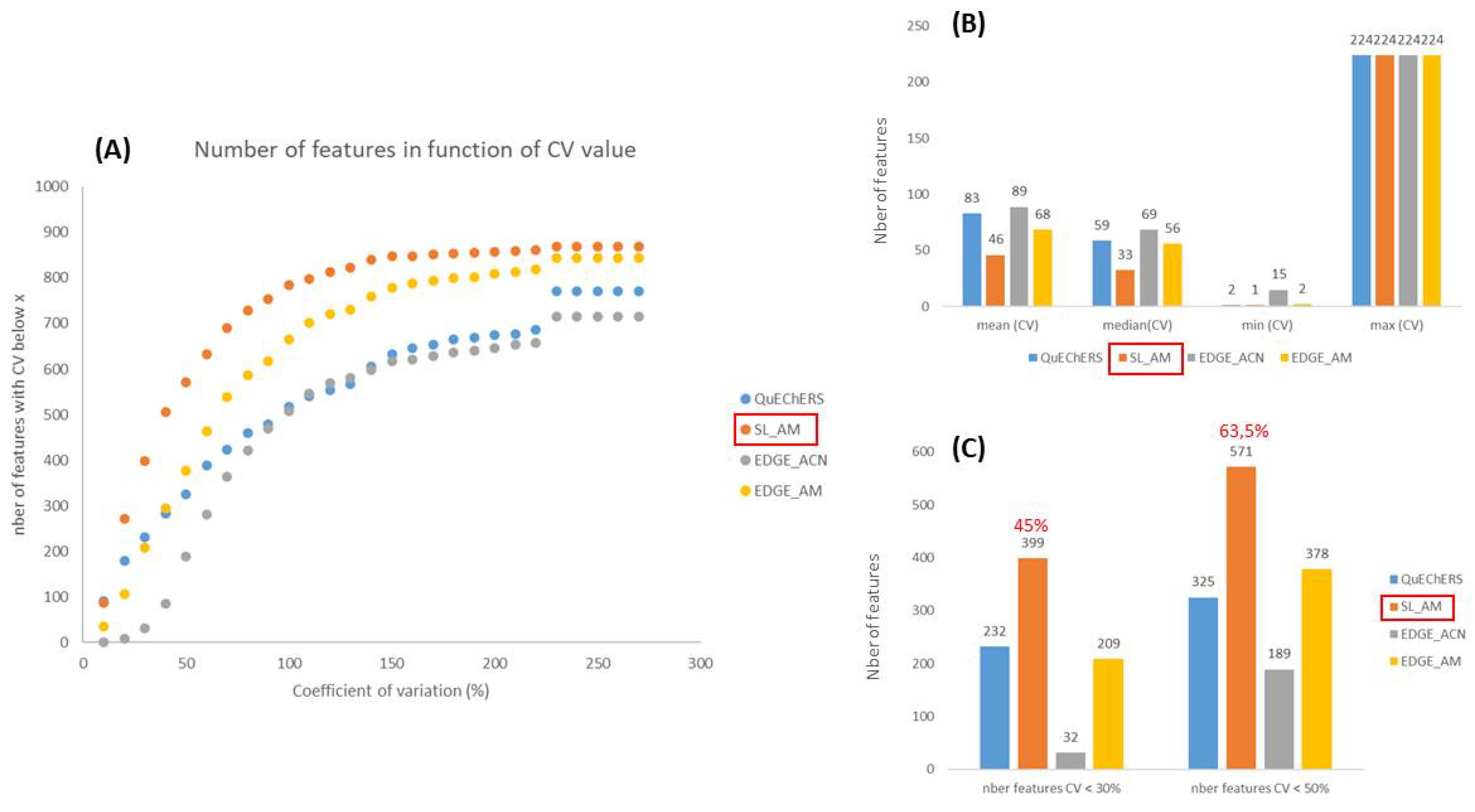
- Method reproducibility
- Putative family/metabolites identification
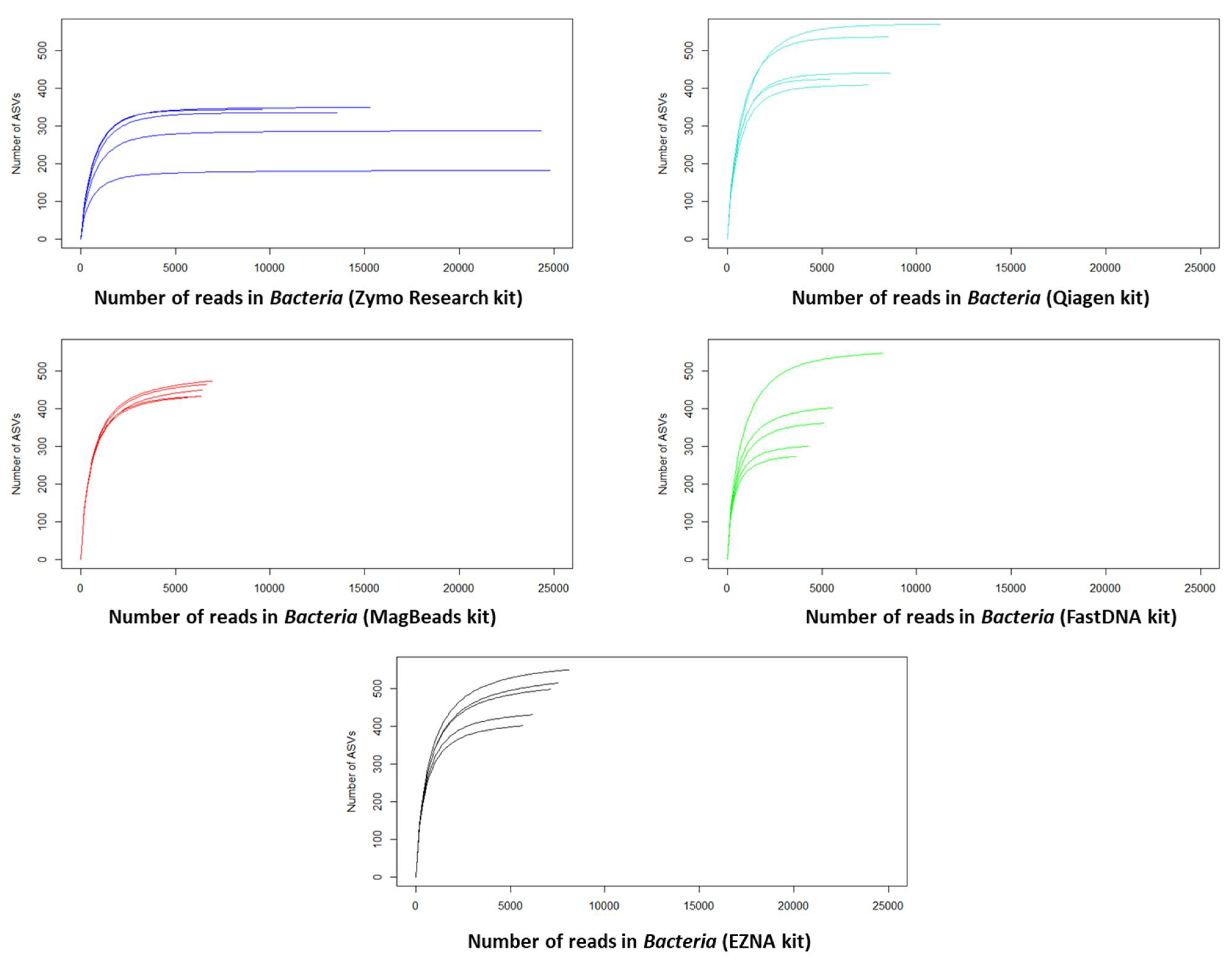
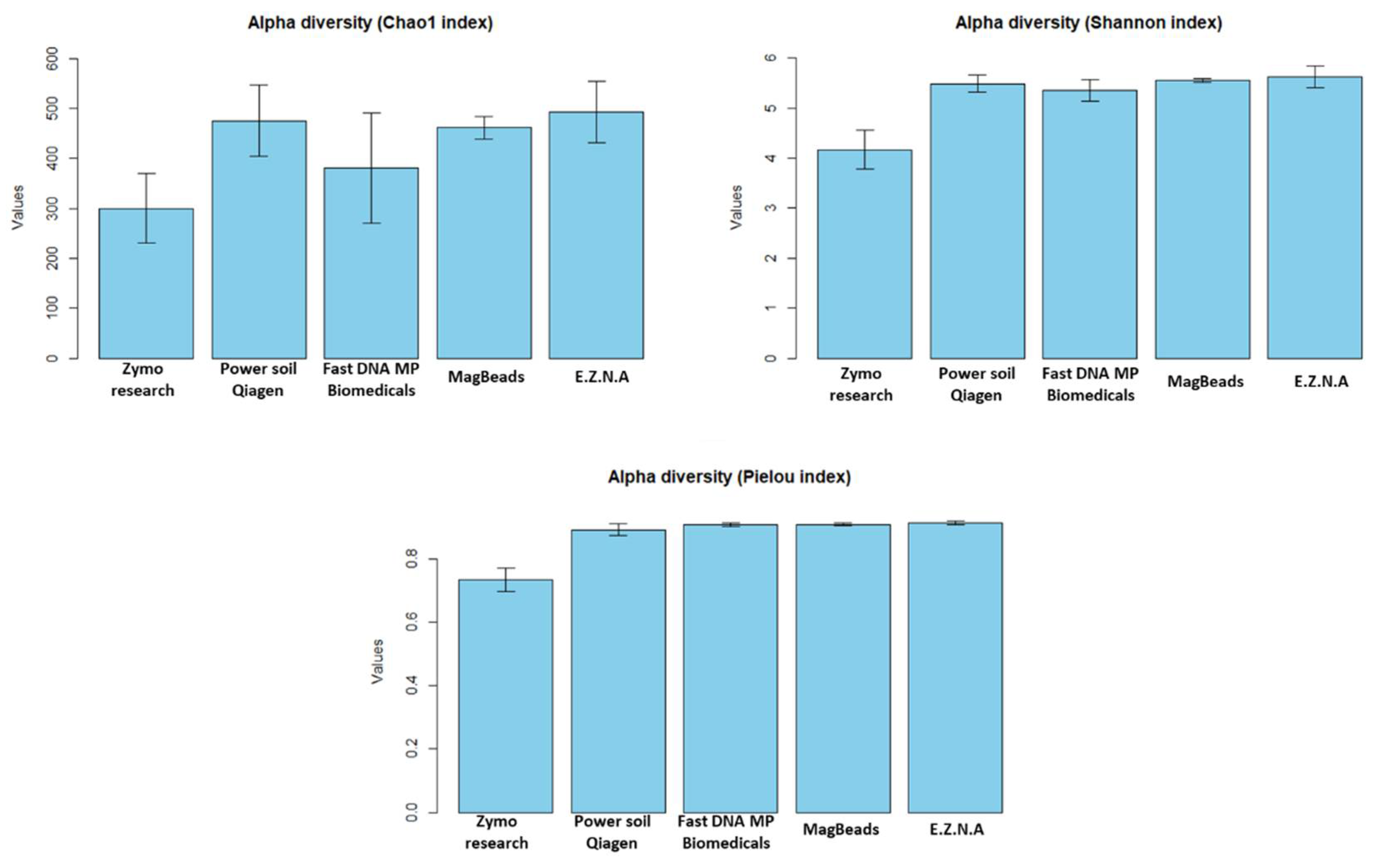
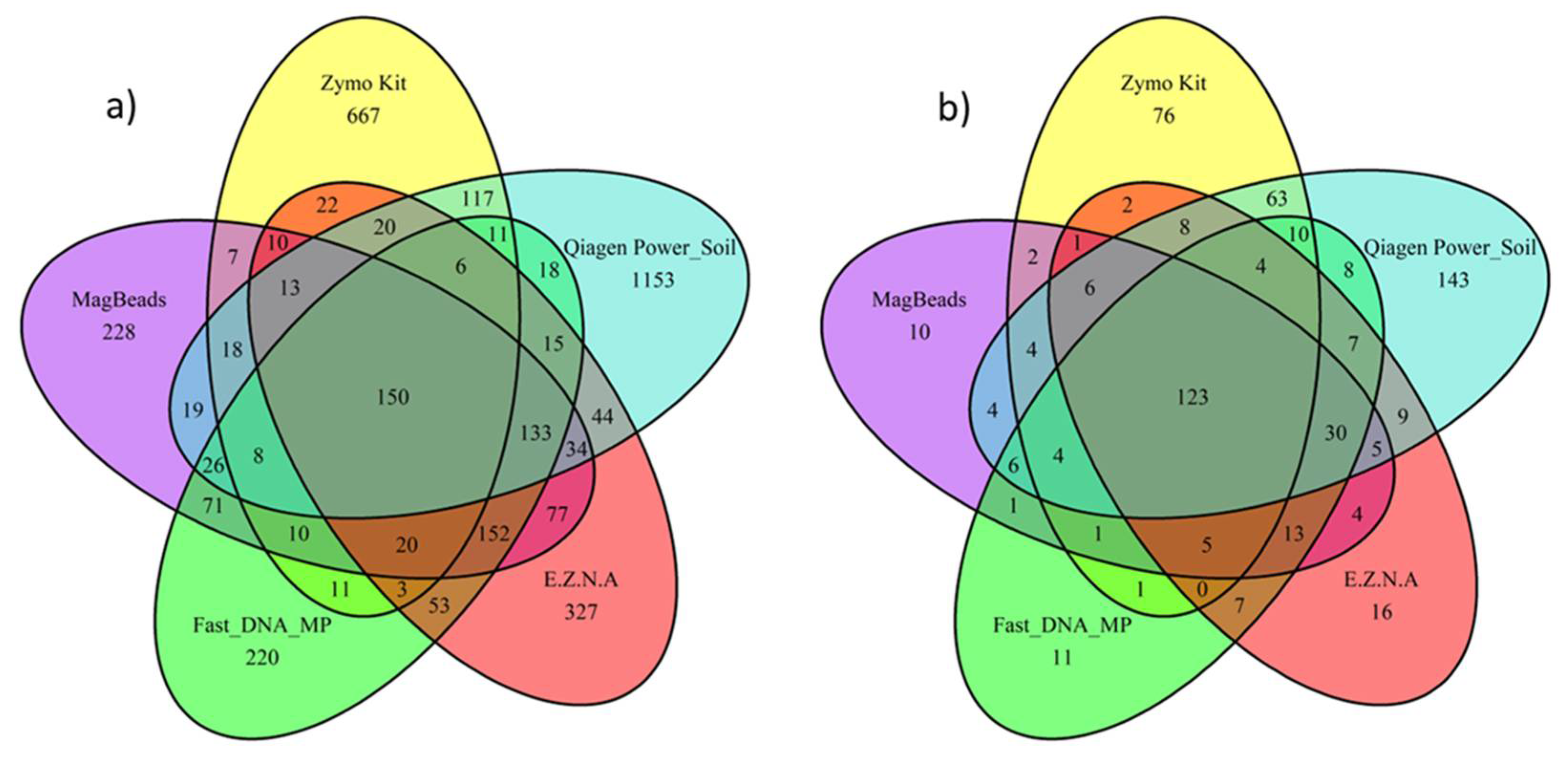
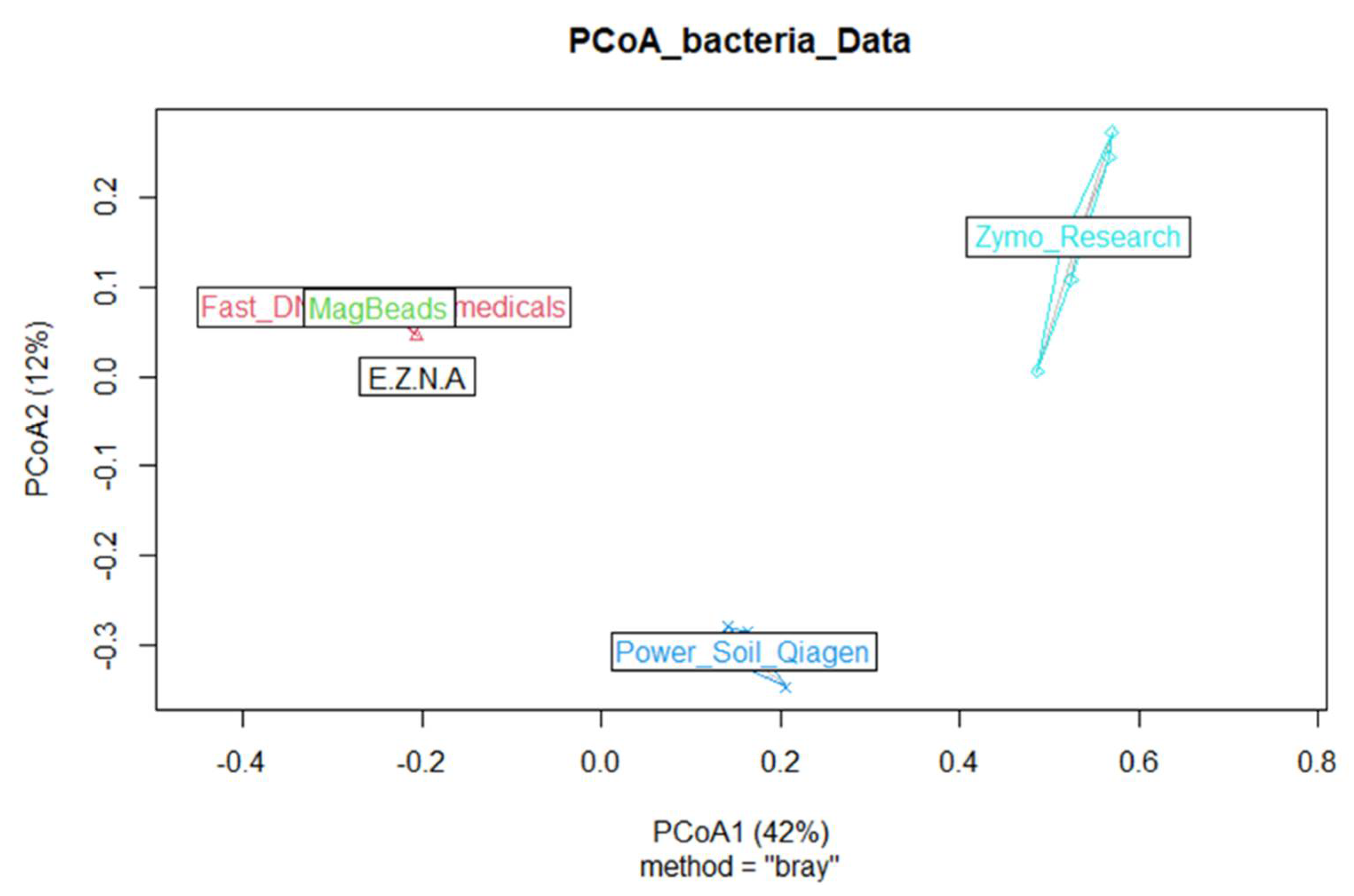
3.2.2. Effects of DNA Extraction Methods on the Determination of Soil Microbial Diversity in sediment
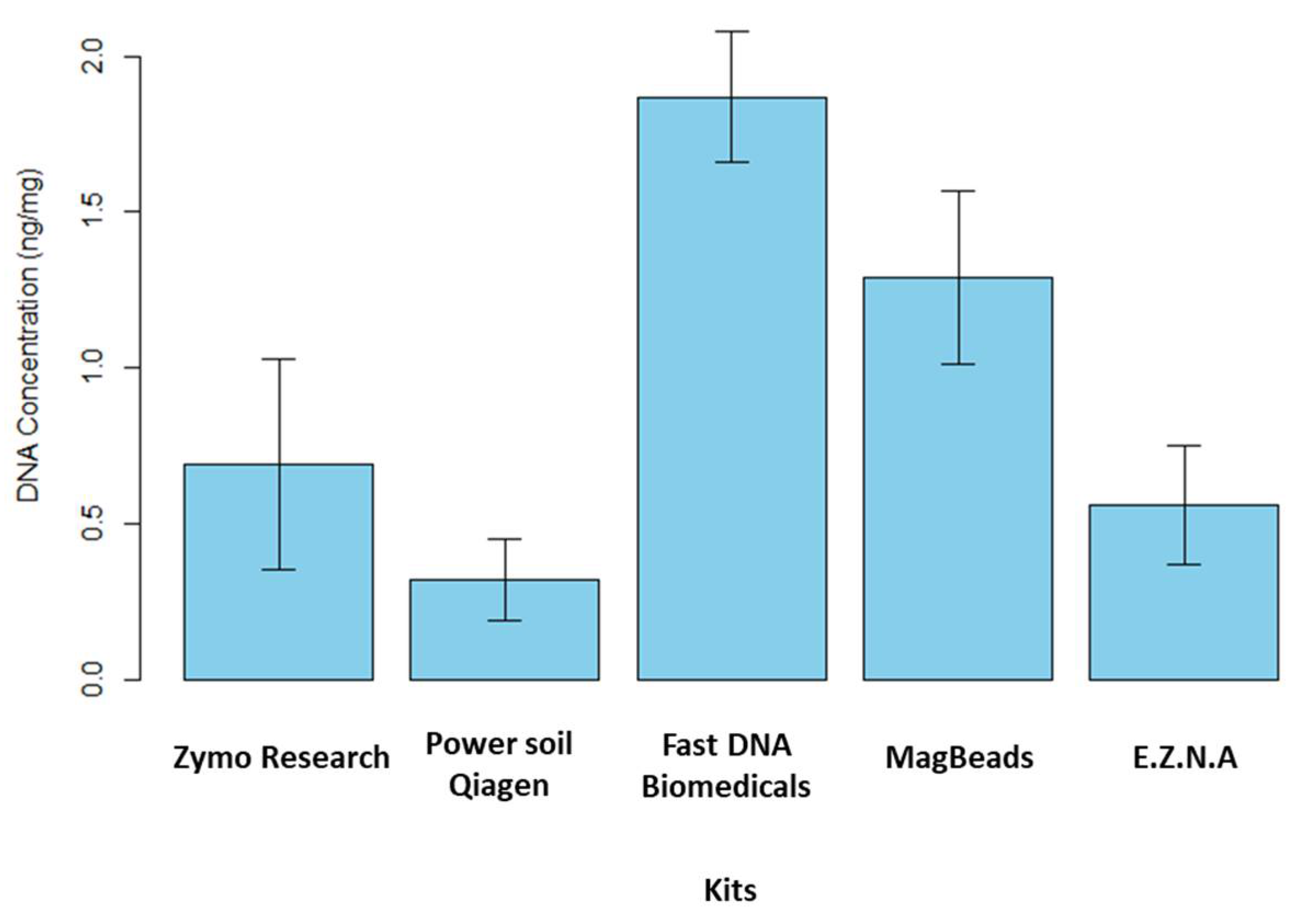
- DNA extraction and PCR amplification differences between DNA extraction kits
- DNA extraction kit influenced alpha-diversity and composition of the sediment microbiome
4. Conclusion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Munaron, D.; Hubert, M.; Gonzalez, J.-L.; Tapie, N.; Budzinski, H.; Guyomarch, J.; Andral, B. Projet échantillonneurs passifs pour la surveillance de la contamination chimique des lagunes méditerranéennes. Rapport Ifremer 2013.
- Munaron, D.; Derolez, V.; Foucault, E.; Cimiterra, N.; Tapie, N.; Budzinski, H.; Giraud, A. OBSLAG - Volet Pesticides. Bilan 2017-2019 du suivi des lagunes méditerranéennes. Rapport Ifremer 2020.
- Lin, C.Y.; Viant, M.R.; Tjeerdema, R.S. Metabolomics: Methodologies and applications in the environmental sciences. J. Pestic. Sci. 2006, 31, 245–251. [Google Scholar] [CrossRef]
- Lankadurai, B.-P.; Nagato, E.-G.; Simpson, M.-J. Environmental metabolomics: an emerging approach to study organism responses to environmental stressors. Envi Rev. 2013, 21, 180+. [Google Scholar] [CrossRef]
- Bundy, J.-G.; Davey, M.-P.; Viant, M. Environmental metabolomics: A critical review and future perspectives. Metabolomics 2009, 5, 3–21. [Google Scholar] [CrossRef]
- Longnecker, K.; Futrelle, J.; Coburn, E.; Soule, Melissa C.-K.; Kujawinski, E.-B. Environmental metabolomics: Databases and tools for data analysis. Marine Chem. 2015, 177, 366–373. [Google Scholar] [CrossRef]
- Bedia, C.; Cardoso, P.; Dalmau, N.; Garreta-Lara, E.; Gómez-Canela, C.; Gorrochategui, E.; Navarro-Reig, M.; Ortiz-Villanueva, E.; Puig-Castellví, F.; Tauler, R. Applications of Metabolomics Analysis in Environmental Research. Comprehensive Analy Chemi. 2018, 82, 533–582. [Google Scholar]
- Romdhane, S.; Devers-Lamrani, M.; Beguet, J.; Bertrand, C.; Calvayrac, C.; Salvia, M.-V.; Ben Jrad, B.; Dayan, F.E.; Spor, A.; Barthelmebs, L.; Martin-Laurent, F. Assessment of the ecotoxicological impact of natural and synthetic β-triketone herbicides on the diversity and activity of the soil bacterial community using omic approaches. Sci. Total Environ. 2019, 651, 1–11. [Google Scholar] [CrossRef]
- Patil, C.; Calvayrac, C.; Zhou, Y.; Romdhane, S.; Salvia, M.-V.; Cooper, J.-F.; Dayan, F.; Bertrand, C. Environmental Metabolic Footprinting: A novel application to study the impact of a natural and a synthetic β-triketone herbicide in soil. Sci Total Environ, 2016; 566–567, 552–558. [Google Scholar]
- Salvia, M.-V.; Ben Jrad, A.; Raviglione, D.; Zhou, Y.; Bertrand, C. Environmental Metabolic Footprinting (EMF) vs. half-life: a new and integrative proxy for the discrimination between control and pesticides exposed sediments in order to further characterise pesticides’ environmental impact. Environ Sci Pollut Res. 2017, 25, 29841–29847. [Google Scholar] [CrossRef]
- Ghosson, H.; Raviglione, D.; Salvia, M.-V.; Bertrand, C. Online Headspace-Solid Phase Microextraction-Gas Chromatography-Mass Spectrometry-based untargeted volatile metabolomics for studying emerging complex biopesticides: A proof of concept. Anal Chim Acta. 2020, 1134, 58–74. [Google Scholar] [CrossRef]
- Ghosson, H.; Guitton, Y.; Ben Jrad, A.; Patil, C.; Raviglione, D.; Salvia, M.-V.; Bertrand, C. Electrospray ionization and heterogeneous matrix effects in liquid chromatography/mass spectrometry based meta-metabolomics: A biomarker or a suppressed ion? Rapid Commun. Mass Spectrom. 2020, 35, e8977. [Google Scholar] [CrossRef]
- Ramos, M.; Ghosson, H.; Raviglione, D.; Bertrand, C.; Salvia, M.-V. Untargeted metabolomics as a tool to monitor biocontrol product residues' fate on field-treated Prunus persica. Sci. Total Environ. 2022, 807, 150717. [Google Scholar] [CrossRef]
- Ghosson, H.; Raviglione, D.; Salvia, M.-V.; Bertrand, C. Characteristic response of formulation ingredients revealed by ultra high-performance liquid chromatography-electrospray ionization-high resolution mass spectrometry-based untargeted screening of pesticides in soil. J. Mass Spectrom. 2023, 58, e4962. [Google Scholar] [CrossRef]
- Ghosson, H.; Raviglione, D.; Bertrand, C.; Salvia, M.-V. LC-HRMS-Driven Computational Toolbox to Assess Extraction Protocols Dedicated to Untargeted Analysis: How to Ease Analyzing Pesticide-Contaminated Soils? . Anal. Chem. 2024, 96, 2810–2821. [Google Scholar] [CrossRef]
- Shi, Z.; Kong, Q.; Li, X.; Xu, W.; Mao, C.; Wang, Y.; Song, W.; Huang, J. The effects of DNA extraction kits and primers on prokaryotic and eukaryotic microbial community in freshwater sediments. Microorganisms. Microorganisms. 2022, 10, 1213. [Google Scholar] [CrossRef]
- Pearman, J.K.; Keeley, N.B.; Wood, S.A.; Laroche, O.; Zaiko, A.; Thomson-Laing, G.; Biessy, L.; Atalah, J.; Pochon, X. Comparing sediment DNA extraction methods for assessing organic enrichment associated with marine aquaculture. PeerJ. 2020, 8, 10231. [Google Scholar] [CrossRef] [PubMed]
- Yudha, D.S.; Priyono, D.S.; Izzati, R.; Nainggolan, A.P. Comparing DNA extraction from environmental DNA samples to reveal the diversity of freshwater metazoans. Biogenesis Jurnal Ilmiah Biologi. 2021, 9, 206–212. [Google Scholar] [CrossRef]
- Iturbe-Espinoza, P.; Brandt, B.W.; Braster, M.; Bonte, M.; Brown, D.M.; van Spanning, R.J.M. Effects of DNA preservation solution and DNA extraction methods on microbial community profiling of soil. Folia Microbiol. 2021, 66(4), 597–606. [Google Scholar]
- Gűrtler, V.; Stanisich, V.A. New approaches to typing and identification of bacteria using the 16S-23S rDNA spacer region. Microbiology 1996, 142, 3–16. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Research, 2013, 41, e1. [Google Scholar] [PubMed]
- Callahan, B.; McMurdie, P.; Rosen, M.; et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O'Hara, R.B.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Szoecs, E.; Wagner, H. Vegan: Community Ecology Package. R package Version 2.4-3, 2017.
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag New York, 2016.
- Chen, H.; Boutros, P.C. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics. 2011.
- Wiest, L.; Gosset, A.; Fildier, A.; Libert, C.; Hervé, M.; Sibeud, E.; Giroud, B.; Vulliet, E.; Bastide, T.; Polomé, P.; Perrodin, Y. Occurrence and removal of emerging pollutants in urban sewage treatment plants using LC-QToF-MS suspect screening and quantification. Sci. Total Environ. 2021, 774, 145779. [Google Scholar] [CrossRef]
- Lafay, F.; Daniele, G.; Fieu, M.; Pelosi, C.; Fritsch, C.; Vulliet, E. Ultrasound-assisted QuEChERS-based extraction using EDTA for determination of currently-used pesticides at trace levels in soil. Environ Sci Pollut Res Int. 2022.
- Bonnard, N.; Jargot, D.; Falcy, M. Base de données fiches toxicologiques, Carbendazime, fiche toxicologique n°214, INRS 2009, France, https://www.inrs.fr/dms/ficheTox/FicheFicheTox/FICHETOX_214-3/FicheTox_214.pdf.
- Lesueur, C.; Gartner, M.; Mentler, A.; Fuerhacker, M. Comparison of four extraction methods for the analysis of 24 pesticides in soil samples with gas chromatography–mass spectrometry and liquid chromatography–ion trap–mass spectrometry. Talanta. 2008, 75, 284–293. [Google Scholar] [CrossRef] [PubMed]
- Drożdżyński, D.; Kowalska, J. Rapid analysis of organic farming insecticides in soil and produce using ultra-performance liquid chromatography/tandem mass spectrometry. Anal Bioanal Chem. 2009, 394, 2241–2247. [Google Scholar] [CrossRef] [PubMed]
- Asensio-Ramos, M.; Hernández-Borges, J.; Ravelo-Pérez, L.M.; Rodríguez-Delgado, M.A. Evaluation of a modified QuEChERS method for the extraction of pesticides from agricultural, ornamental and forestal soils. Anal Bioanal Chem. 2010, 396, 2307–2319. [Google Scholar] [CrossRef] [PubMed]
- Rashid, A.; Nawaz, S.; Barker, H.; Ahmad, I.; Ashraf, M. Development of a simple extraction and clean-up procedure for determination of organochlorine pesticides in soil using gas chromatography–tandem mass spectrometry. ABC. 2010, 1217, 2933–2939. [Google Scholar] [CrossRef] [PubMed]
- Salvia, M.-V.; Vulliet, E.; Wiest, L.; Baudot, R.; Cren-Olivé, C. Development of a multi-residue method using acetonitrile-based extraction followed by liquid chromatography–tandem mass spectrometry for the analysis of steroids and veterinary and human drugs at trace levels in soil. Journal of Chromatography A. 2012, 1245, 122–133. [Google Scholar] [CrossRef]
- Berlioz-Barbier, A.; Buleté, A.; Faburé, J.; Garric, J.; Cren-Olivé, C.; Vulliet, E. Multi-residue analysis of emerging pollutants in benthic invertebrates by modified micro-quick-easy-cheap-efficient-rugged-safe extraction and nanoliquid chromatography–nanospray–tandem mass spectrometry analysis. Journal of Chromatography A. 2014, 1367, 16–32. [Google Scholar] [CrossRef] [PubMed]
- Salvia, M.-V.; Cren-Olivé, C.; Wiest, L.; Baudot, R.; Vulliet, E. Comparison of Two Analytical Methods for the Determination of Traces of Veterinary Antibiotics and Steroid Hormones in Soil Based on Pressurised Liquid Extraction (PLE) and Quick, Easy, Cheap, Effective, Rugged, Safe (Modified-QuEChERS) Extraction. Pharm. Anal. Acta. 2014, 5, 1–10. [Google Scholar]
- Daniele, G.; Fieu, M.; Joachim, S.; Bado-Nilles, A.; Beaudouin, R.; Baudoin, P.; James-Casas, A.; Andres, S.; Bonnard, M.; Bonnard, I.; Geffard, A.; Vulliet, E. Determination of carbamazepine and 12 degradation products in various compartments of an outdoor aquatic mesocosm by reliable analytical methods based on liquid chromatography-tandem mass spectrometry. ESPR. 2017, 24, 16893–16904. [Google Scholar] [CrossRef]
- Guironnet, A.; Wiest, L.; Vulliet, E. Advantages of MS/MS/MS (MRM3) vs classic MRM quantification for complex environmental matrices: Analysis of beta-lactams in WWTP sludge. Analytica Chimica Acta 2022, 1205, 339–773. [Google Scholar]
- Kinross, A.D.; Hageman, K.J.; Doucette, W.J.; Foster, A.L. Comparison of Accelerated Solvent Extraction (ASE) and Energized Dispersive Guided Extraction (EDGE) for the analysis of pesticides in leaves. J. Chromatogr A. 2020, 1627, 461414. [Google Scholar] [CrossRef] [PubMed]
- Olivier, C.; Allen, B.; Luies, L. Optimising a urinary extraction method for non-targeted GC–MS metabolomics. Sci. Rep. 2023, 13, 17591. [Google Scholar] [CrossRef] [PubMed]
- van Bruggen, A.H.C.; Goss, E.M.; Havelaar, A.; van Diepeningen, A.D.; Finckh, M.R.; Morris Jr, J.G. One Health - Cycling of diverse microbial communities as a connecting force for soil, plant, animal, human and ecosystem health. Sci. Total Environ. 2019, 664, 927–937. [Google Scholar] [CrossRef] [PubMed]
- Lim, M.Y.; Song, E.-J.; Kim, S.H.; Lee, J.; Nam, Y.-D. Comparison of DNA extraction methods for human gut microbial community profiling. Syst Appl Microbiol. 2018, 41, 151–157. [Google Scholar] [CrossRef]
- Gand, M.; Bloemen, B.; Vanneste, K.; Roosens, N.H.C.; De Keersmaecker, S.C.J. Comparison of 6 DNA extraction methods for isolation of high yield of high molecular weight DNA suitable for shotgun metagenomics Nanopore sequencing to detect bacteria. BMC Genomics 2023, 24, 438. [Google Scholar] [CrossRef] [PubMed]
- Dopheide, A.; Xie, D.; Buckley, T.R.; Drummond, A.J.; Newcomb, R.D. Impacts of DNA extraction and PCR on DNA metabarcoding estimates of soil biodiversity. Meth Ecol Evol. 2018, 10, 120–133. [Google Scholar] [CrossRef]
- Wasimuddin, K.S.; Schlaeppi, K.; Ronchi, F.; Leib, S.L.; Erb, M.; Ramette, A. Evaluation of primer pairs for microbiome profiling from soils to humans within the One Health framework. Mol Ecol Resour 2020, 20, 1558–1571. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.-R.; Shin, J.; Guevarra, R.; Lee, J.H.; Kim, D.W.; Seol, K.-H.; Lee, J.-H.; Kim, H.B.; Isaacson, R. Deciphering Diversity Indices for a Better Understanding of Microbial Communities. J. Microbiol Biotechnol. 2017, 27, 2089–2093. [Google Scholar] [CrossRef] [PubMed]
- Willis, A.D. Rarefaction, Alpha Diversity, and Statistics. Front Microbiol 2019, 10, 2407. [Google Scholar] [CrossRef]
- Yang, N.; Tian, C.; Lv, Y.; Hou, J.; Yang, Z.; Xiao, X.; Zhang, Y. Novel primers for 16S rRNA gene-based archaeal and bacterial community analysis in oceanic trench sediments. Appl Microbiol Biotechnol. 2022, 106, 2795–2809. [Google Scholar] [CrossRef]
| Substances | MLQ Water (ng/L) |
Mean Conc Water (ng/L) |
CV Water (%) | MLQ Sed. (ng/g) |
Mean Conc Sed. (ng/g) |
CV Sed. (%) |
|---|---|---|---|---|---|---|
| PHARMACEUTICALS | ||||||
| Flecainide (Antiarrhythmic) | 0.04 | 35.6 | 6.15 | 0.001 | 0.180 | 46.5 |
| Oxazepam (Psycholeptic) | 0.06 | 30.6 | 5.05 | 0.007 | 0.088 | 14.8 |
| Carbamazepine (Psycholeptic) | 0.03 | 22.6 | 3.30 | 0.007 | 0.076 | 7.21 |
| Fluconazole* (Antimycotic) | 0.06 | 12.1 | 7.55 | 0.001 | 0.030 | 0 |
| Paracetamol (Analgesic) | 1.2 | 9.15 | 8.50 | 0.04 | 0.092 | 14.2 |
| Telmisartan* (Hypertensive) | 0.01 | 8.44 | 20.4 | 0.02 | 0.240 | 47.5 |
| Tiapride (Neuroleptic) | 0.002 | 7.72 | 6.94 | 0.001 | 0.072 | 22.8 |
| Sulfamethoxazole (Antibiotic) | 0.02 | 7.54 | 7.36 | 0.008 | nd | - |
| Valsartan (Hyperstensive) | 0.06 | 7.50 | 5.16 | 0.1 | nd | - |
| Cetirizine* (Antihistamine/ Antiallergic) | 0.01 | 5.34 | 3.88 | 0.007 | 0.042 | 10.6 |
| Venlafaxine (Antidepressant) | 0.05 | 2.86 | 24.8 | 0.001 | 0.050 | 0 |
| Disopyramide*(Antiarrhythmic) | 0.05 | 2.76 | 8.34 | 0.001 | 0.054 | 10.1 |
| Diclofenac (Analgesic) | 0.05 | 1.58 | 12.1 | 0.004 | 0.120 | 37.3 |
| Amisulpride (Antiarrhythmic) | 0.01 | 1.14 | 4.80 | 0.003 | 0.036 | 15.2 |
| Citalopram*(Antidepressant) | 0.06 | 0.34 | 16.1 | 0.005 | 0.040 | 0 |
| PESTICIDES | ||||||
| Terbuthylazine (Herbicide) | 0.02 | 0.200 | 0 | 0.001 | 0.030 | 0 |
| Terbuthylazine-2-OH*(by-product) | 0.06 | 47.4 | 2.78 | 0.001 | 0.340 | 82.1 |
| Terbuthylazine Desethyl hydroxy* (by-product) | 0.04 | 4.40 | 14.9 | 0.001 | 0.028 | 16.0 |
| Carbendazim (Fungicide) | 0.02 | 12.8 | 7.85 | 0.001 | 26.3 | 9.55 |
| Atrazine (Herbicide) | 0.01 | 0.660 | 81.3 | 0.001 | 0.660 | 8.30 |
| Atrazine-2-Hydroxy* (by-product) | 0.02 | 2.98 | 7.65 | 0.001 | 0.0078 | 44.8 |
| Diuron (Herbicide) | 0.05 | 2.52 | 3.32 | 0.01 | 0.048 | 17.4 |
| Boscalid (Fungicide) | 0.08 | 2.00 | 10.0 | 0.01 | 0.046 | 68.0 |
| Propazine-2-Hydroxy*(by-product) | 0.02 | 1.48 | 3.02 | 0.001 | 0.058 | 39.3 |
| Tebuconazole* (Fungicide) | 0.05 | 1.14 | 4.80 | 0.001 | 0.072 | 6.21 |
| Imidaclopride (Insecticide) | 0.06 | 0.880 | 5.08 | 0.008 | nd | - |
| HORMONAL STEROIDS | ||||||
| Progesterone* (Progestogen) | 0.06 | 0.0900 | 11.1 | 0.005 | 0.064 | 34.2 |
| Testosterone* (Androgen) | 0.06 | nd | nd | 0.005 | 0.042 | 10.6 |
| Exp. m/z | RT (min) | Adduct | Elemental composition |
Putative compound class / family |
Putative Compound identity |
|---|---|---|---|---|---|
| 96.9599 | 0.6 | [M-H]- | H2SO4 | acid | sulfuric acid |
| 121.0295 | 6.8 | [M-H]- | C7H6O2 | acid | benzoic acid |
| 268.1043 | 0.9 | [M-H]+ | C10H13N5O4 | nucleoside | adenosine |
| 287.0890 | 0.8 | [M+FA-H]- | C10H14N2O5 | nucleoside | thymidine |
| 327.2913 | 15.6 | [M-H]- | C20H40O3 | fatty acyl (fatty acid / fatty ester) | 20-hydroxyeicosanoic acid |
| 355.3226 | 16.5 | [M-H]- | C22H44O3 | fatty acyl (fatty acid / fatty ester) | 2-hydroxydocosanoic acid |
| 369.3378 | 16.9 | [M-H]- | C23H46O3 | fatty acyl (fatty acid / fatty ester) | 2-hydroxytricosanoic acid |
| 379.2613 | 11.2 / 11.3 | [M-H]+ | C19H39O5P | glycerophospholipid | (isomers) |
| 383.3540 | 17.4 | [M-H]- | C24H48O3 | fatty acid | 2-hydroxy Lignoceric acid |
| 393.2769 | 11.8 | [M-H]+ | C20H41O5P | glycerophospholipid | |
| 496.3394 | 12.8 | [M-H]+ | C24H50NO7P | glycerophospholipid (glycerophosphocholine) | 1-O-palmitoyl-sn-glycero-3-phosphocholine |
| 497.2898 | 12.6 | [M-H]- | C23H47O9P | glycerophospholipid | [3-[2,3-dihydroxypropoxy(hydroxy)phosphoryl]oxy-2-hydroxypropyl] heptadecanoate |
| 552.4642 | 17.8 | [M-H]- | C33H63NO5 | sphingolipid | |
| 566.4282 | 7.71 | [M-H]+ | C30H55N5O5 | natural cyclopeptide | clavatustide C |
| 566.4799 | 18.2 | [M-H]- | C34H65NO5 | lipid / sphingolipid | |
| 653.5118 | 17.6 | [M-H]- | C37H70N2O7 | lipopeptide (containing serine) | |
| 753.5292 | 16.1 | [M-H]- | C42H70N6O6 | peptide |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





