Submitted:
14 June 2024
Posted:
17 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Theory
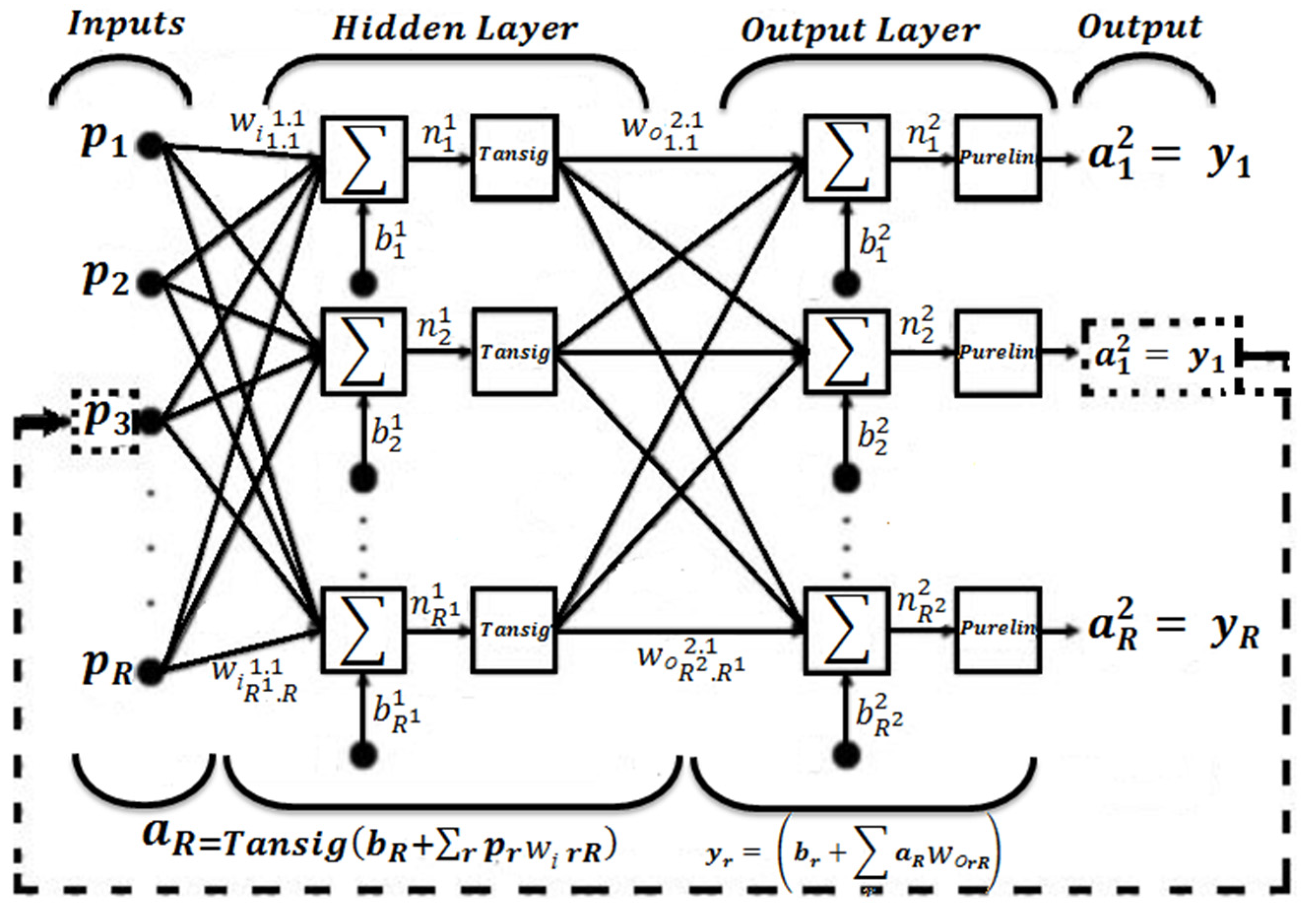
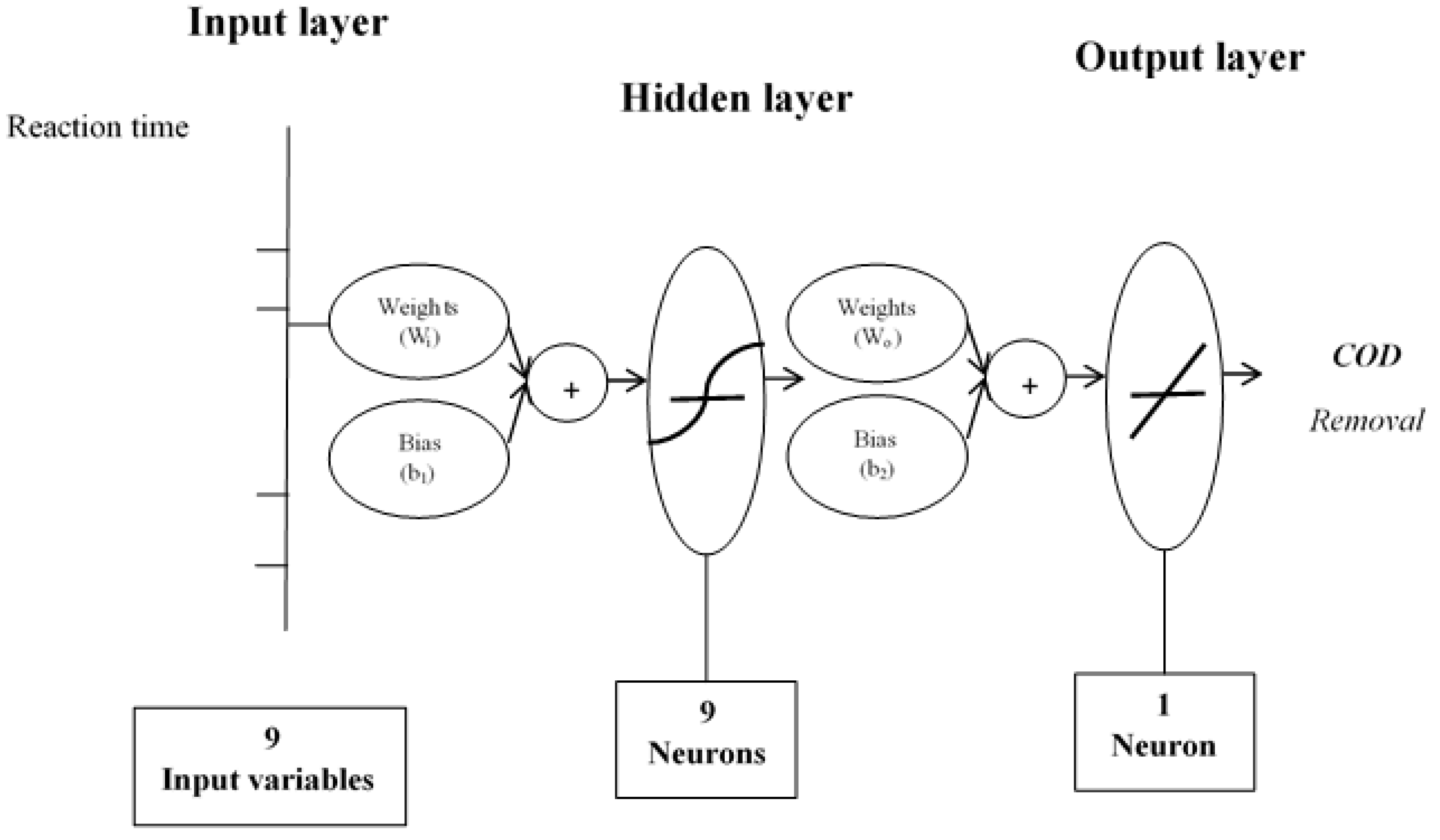
2.1. Artificial Neural Network Inverse
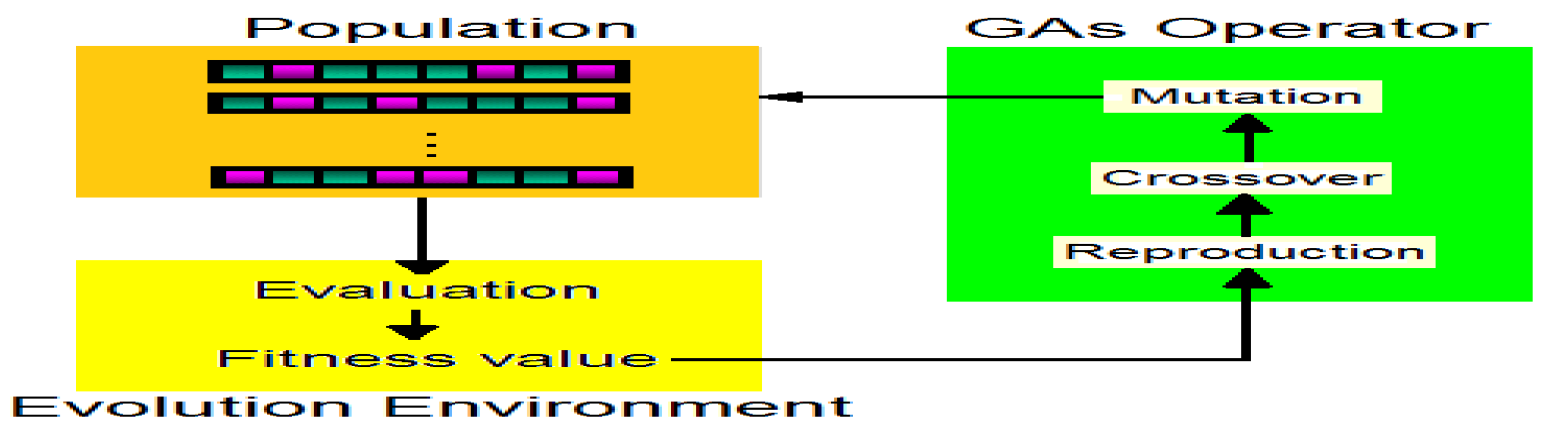
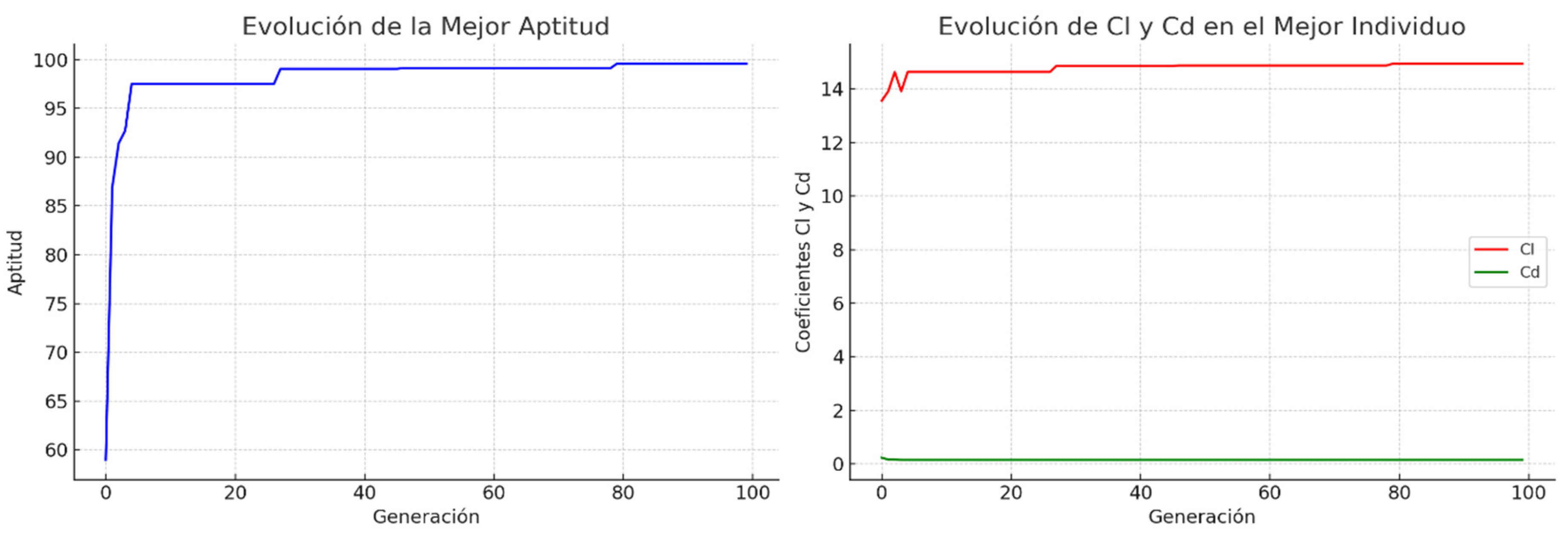
2.2. Genetic Algorithms with Crossed Binary Coding
- (1)
- The problem to be addressed is defined and captured by an objective function that indicates the fitness of any potential solution.
- (2)
- A population of candidate solutions was initialized subject to certain constraints. Typically, each trial solution is coded as a vector , termed a chromosome, with elements described as solutions represented by binary strings. The desired degree of precision indicates the appropriate length of the binary coding.
- (3)
- Each chromosome , in the population was decoded into an appropriate form for evaluation and was then assigned a fitness score, , according to the objective.
- (4)
- Selection in genetic algorithms is often accomplished via differential reproduction according to fitness. In a typical approach, each chromosome is assigned a probability of reproduction, Pi, i=1,2,..., P, so that its likelihood of being selected is proportional to its fitness relative to the other chromosomes in the population. If the fitness of each chromosome is a strictly positive number to be maximized, this is often accomplished using roulette-wheel selection [15]. Successive trials were conducted in which a chromosome was selected until all available positions were filled. Chromosomes with above-average fitness tended to generate more copies than those with below-average fitness.
- (5)
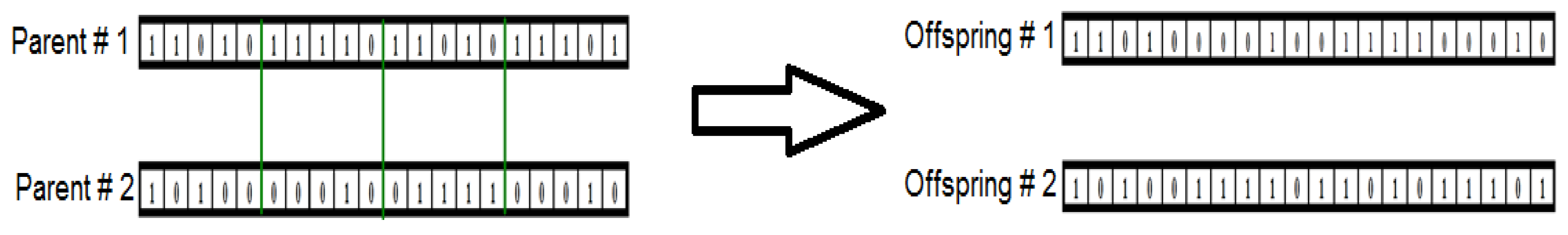
- According to the assigned probabilities of reproduction, ) , a new population of chromosomes is generated by probabilistically selecting strings from the current population. The selected chromosomes generate “offspring” using specific genetic operators, such as crossover and bit mutation. Crossover was applied to two chromosomes (parents) and creates two new chromosomes (offspring) by selecting a random position along the coding and splicing the section that appears before the selected position in the first string with the section that appears after the selected position in the second string and vice versa (see Figure 2). A bit mutation offers the chance to flip each bit in the coding of a new solution (see Figure 3).
- We start with a randomly generated population of n chromosomes (candidate solutions to a problem).
- Calculate the fitness of each chromosome in the population.
-
Repeat the following steps until n offspring have been created:
- A pair of parent chromosomes is selected from the current population, and the probability of selection is an increasing function of fitness. Selection is performed with replacement, meaning that the same chromosome can be selected more than once to become a parent.
- With probability (crossover rate), crossover is performed on the pair at a randomly chosen point to form two offspring.
- Mutate the two offspring at each locus with probability (mutation rate), and place the resulting chromosomes in the new population.
- Replace the current population with the new population.
- Go to step 2.
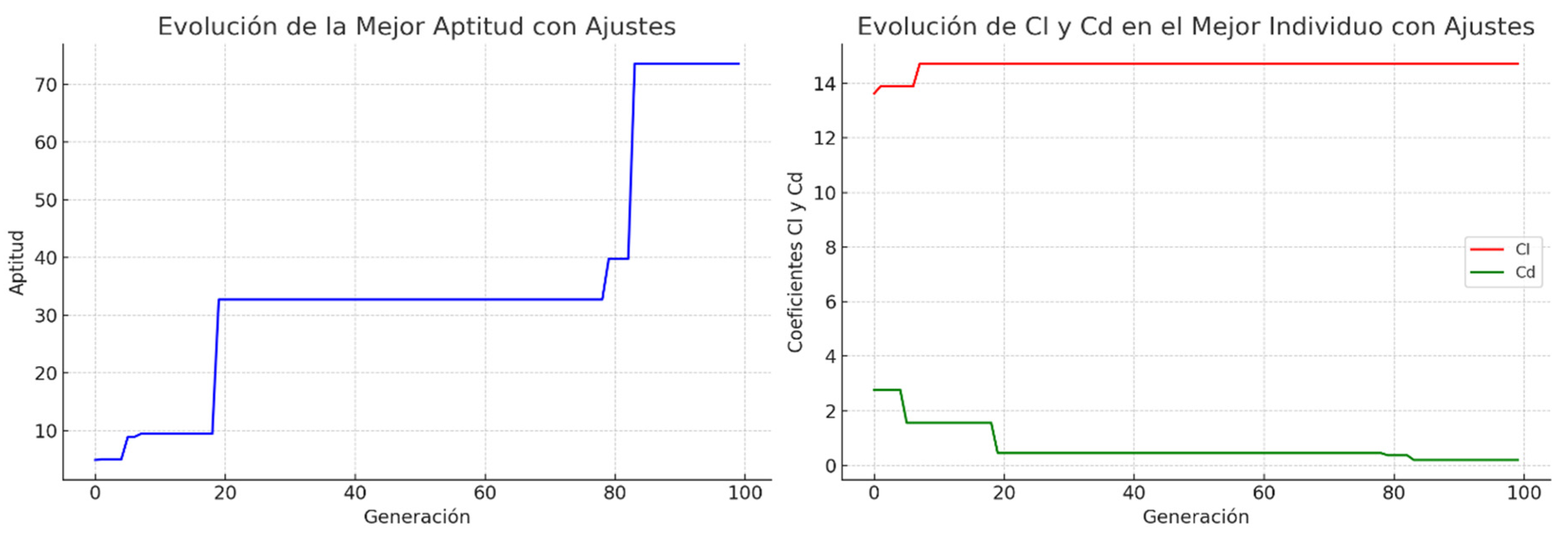
2.3. Implementation and Empirical Tuning Methods for GAs
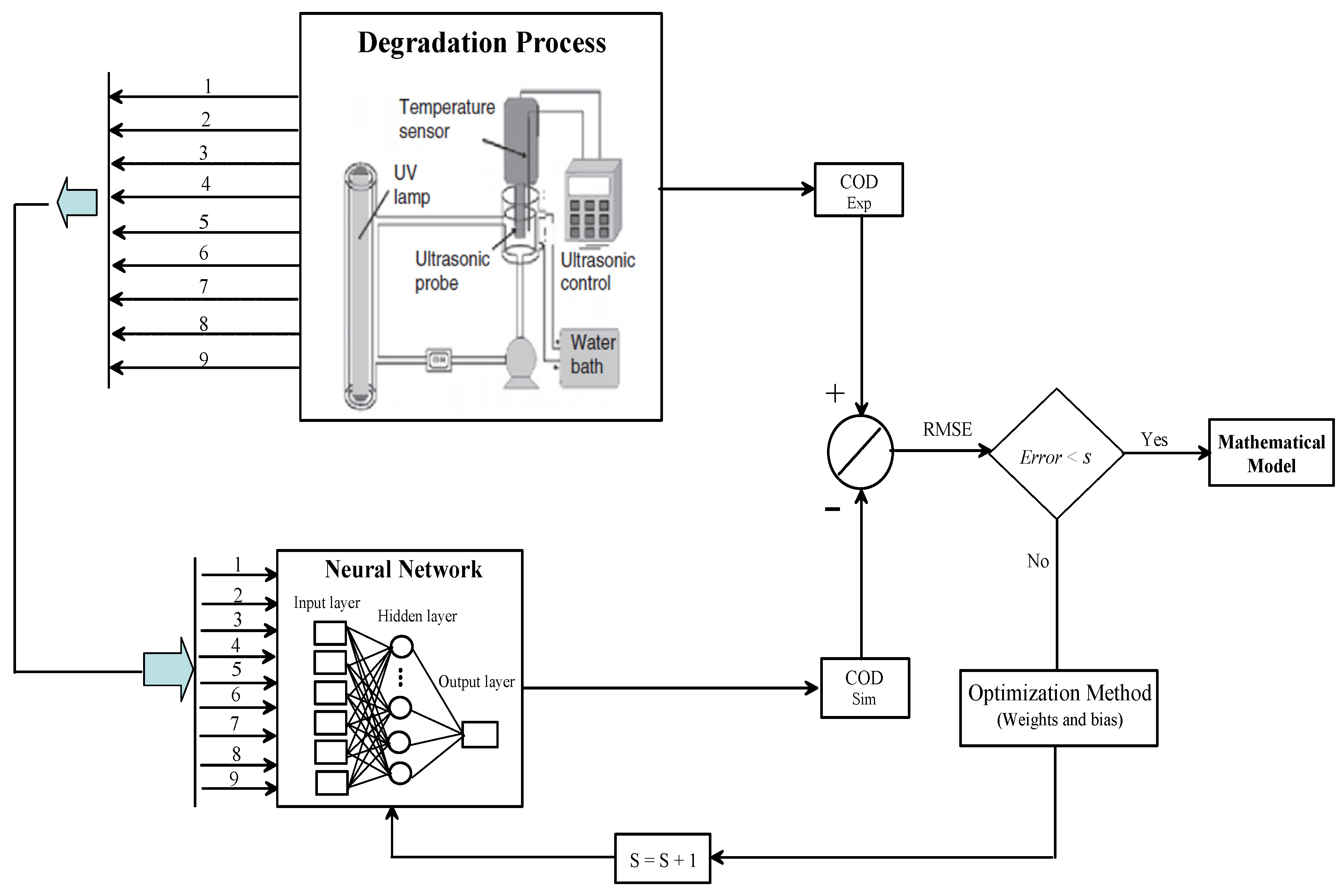
2.4. Sonophotocatalytic Process
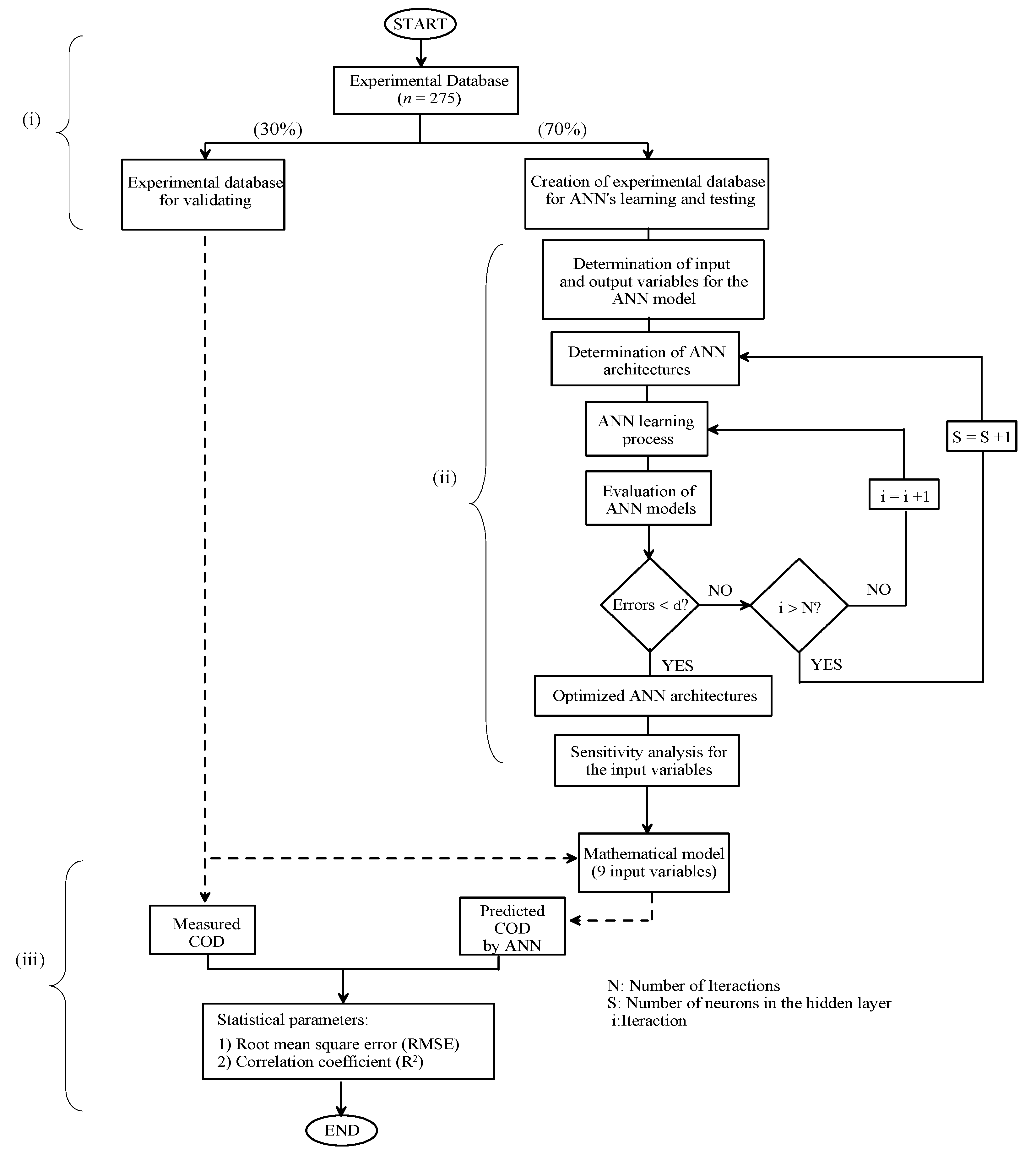
3. Experimental
4. Optimization Approach
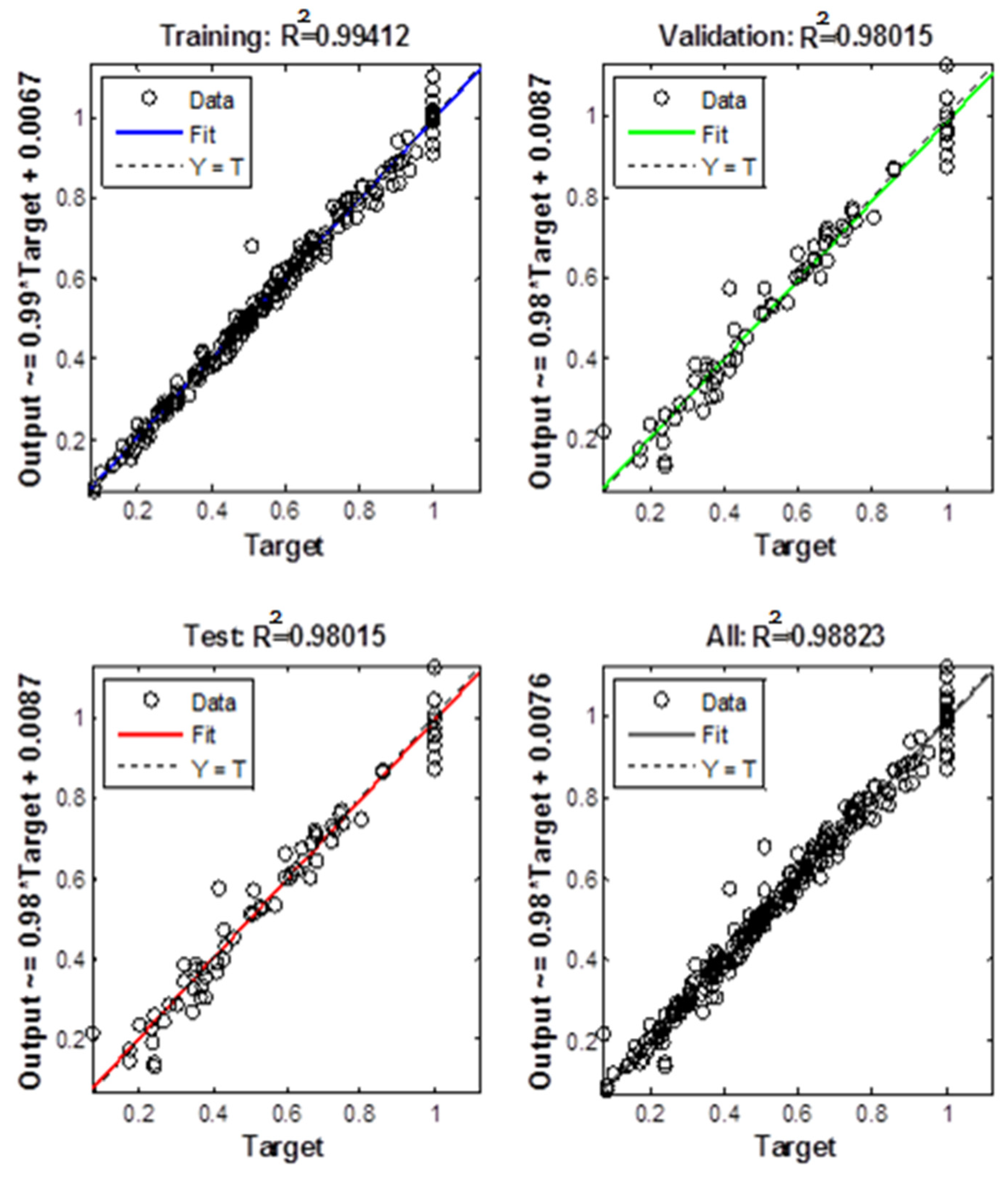
4.1. Neural Network Learning
4.2. Statistical Data Analysis
4.3. Inverse Artificial Neural Network
- ✓
- Population size = 50
- ✓
- Number of generations = 500;
- ✓
- Selection function = standard roulette selection mechanism is used.
- ✓
- Crossover rate = 0.6, 0.8 and 1.0 respectively;
- ✓
- Mutation rate = 0.6;
- ✓
- Elitism = 1.
5. Results and Discussion
| Test number | 25 | 55 | 102 | 130 | 163 | 180 | 207 | 228 | 245 | 275 |
|---|---|---|---|---|---|---|---|---|---|---|
| Input Variables | ||||||||||
| pH | 1 | 1.4 | 1.8 | 2.3 | 2.7 | 3.2 | 3.6 | 4.1 | 4.5 | 5 |
| Herbicide concentration | 0.1540 | 0.1712 | 0.1884 | 0.2057 | 0.2229 | 0.2401 | 0.2573 | 0.2746 | 0.2918 | 0.3090 |
| US ultrasound | 0 | 2.22 | 4.44 | 6.66 | 8.88 | 11.11 | 13.33 | 15.55 | 17.77 | 20 |
| UV light intensity | 0 | 39.11 | 78.22 | 117.33 | 156.44 | 195.55 | 234.66 | 273.77 | 312.88 | 352 |
| [TiO2]o | 0 | 33.33 | 66.66 | 100 | 133.33 | 166.66 | 200 | 233.33 | 266.66 | 300 |
| [K2S2O8]o | 0 | 1.44 | 2.88 | 4.33 | 5.77 | 7.22 | 8.66 | 10.11 | 11.56 | 13 |
| Solar radiation | 0 | 91.11 | 182.22 | 273.33 | 364.44 | 455.55 | 546.66 | 637.77 | 728.88 | 820 |
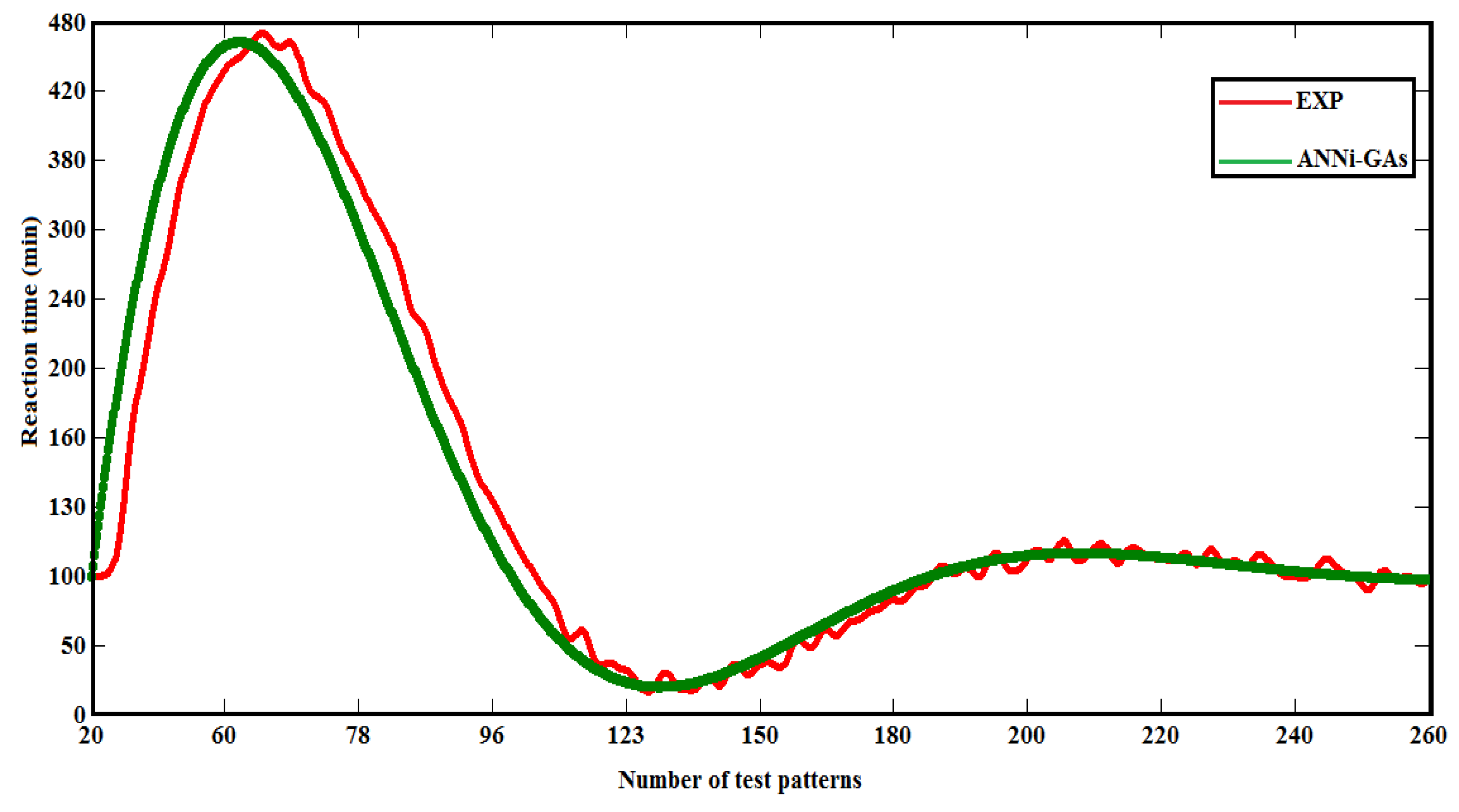
| Reaction time | 102 | 418 | 121 | 27 | 58 | 88(*) | 111 | 109 | 106 | 96 |
| Output | ||||||||||
| CODEXP | 0.73 | 0.57 | 0.44 | 0.35 | 0.32 | 0.3 | 0.26 | 0.23 | 0.21 | 0.17 |
| CODANN | 0.72 | 0.55 | 0.43 | 0.37 | 0.33 | 0.28 | 0.07 | 0.06 | 0.05 | 0.04 |
| (*) t exp= Reaction time that would be estimated by ANNi-GAs | ||||||||||
5.1. Comparative Results
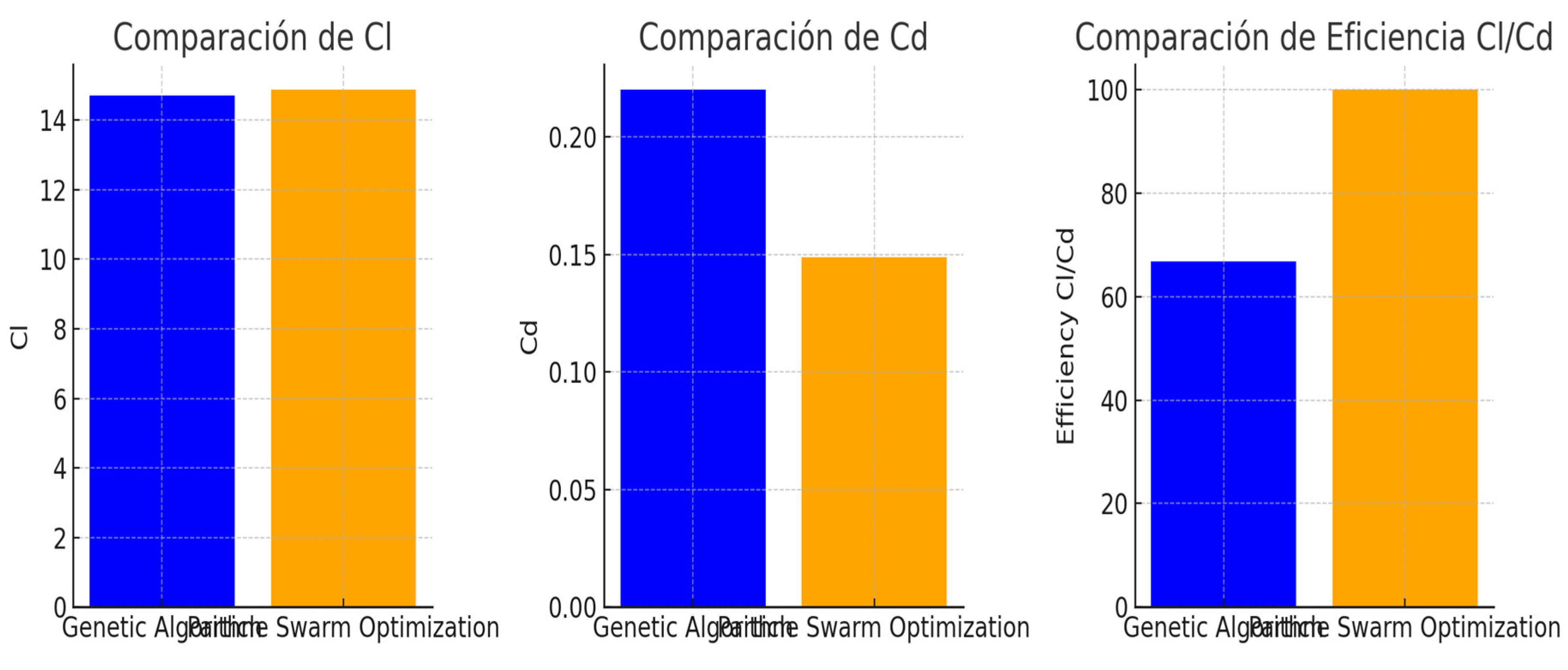
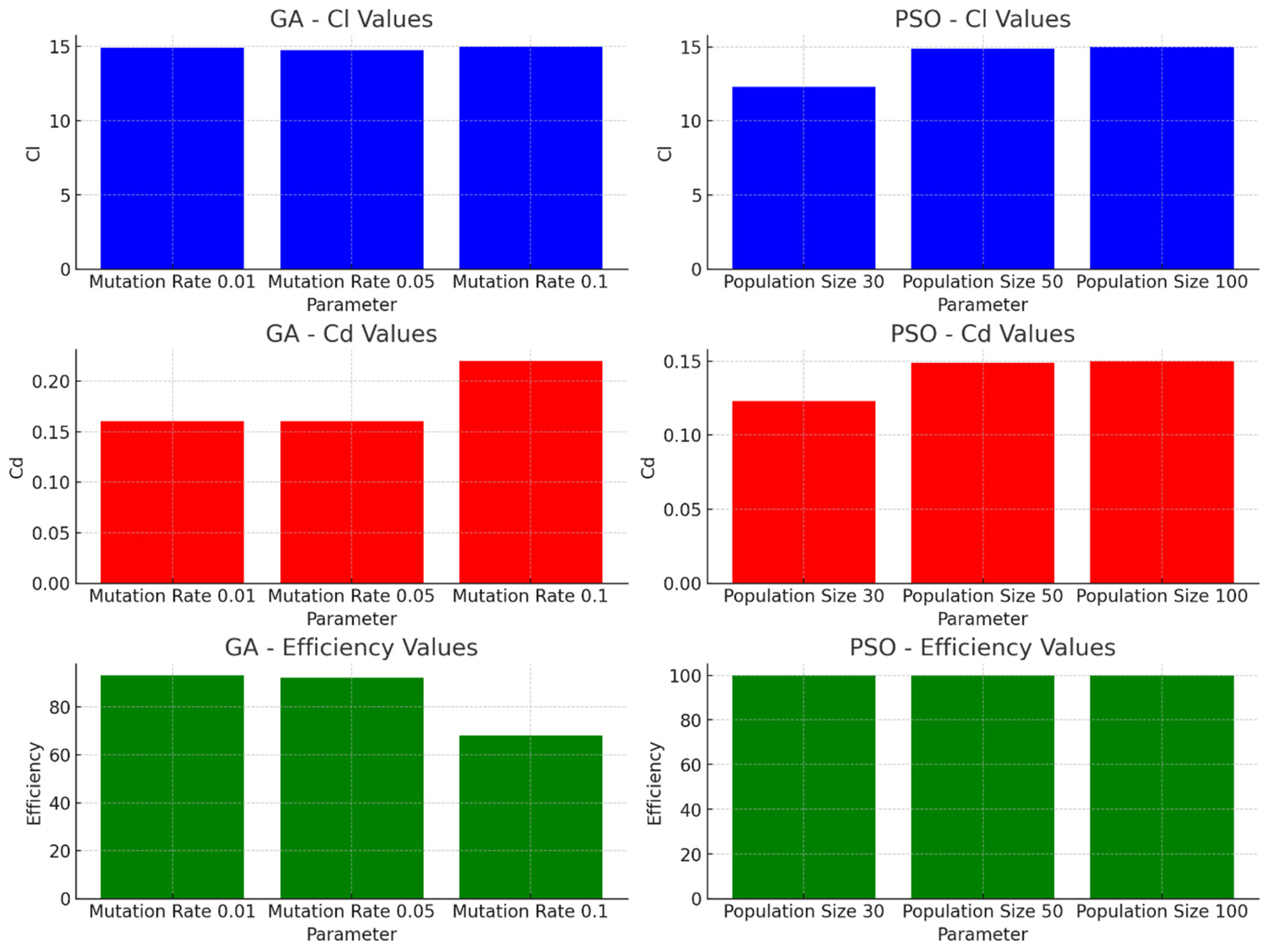
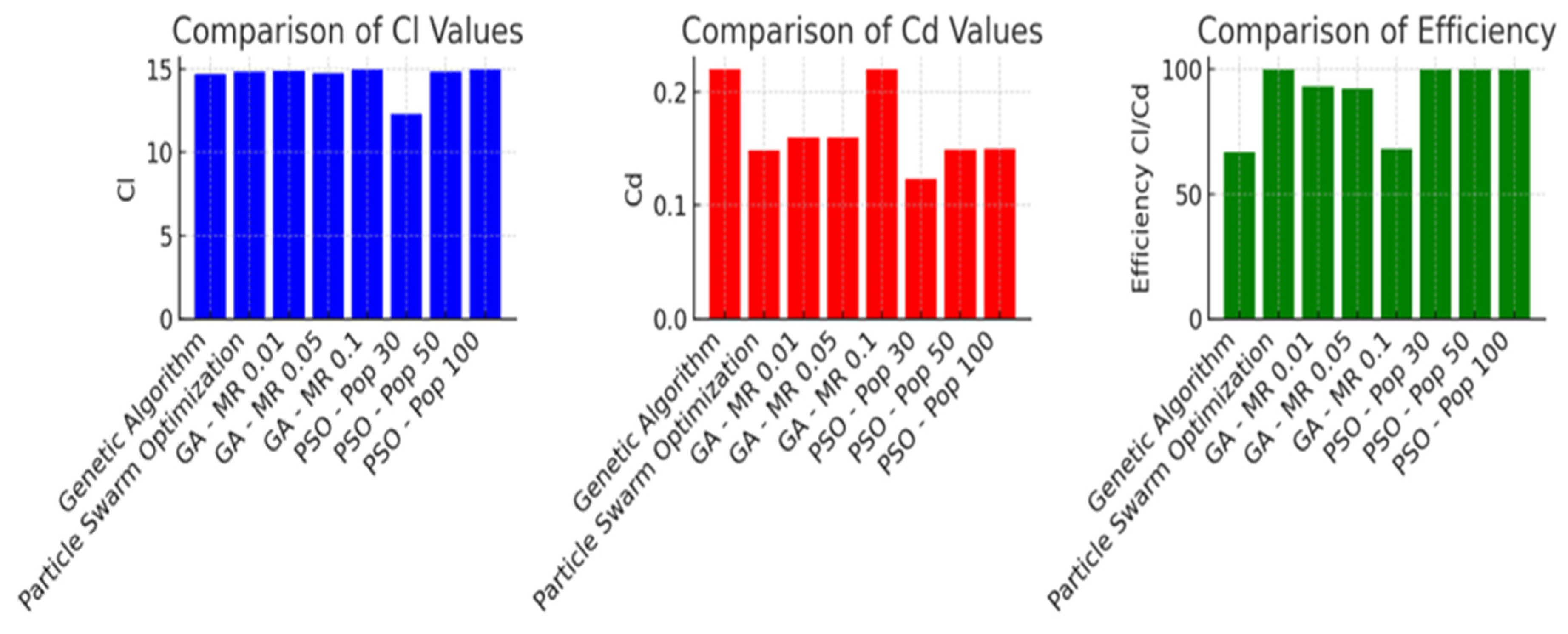
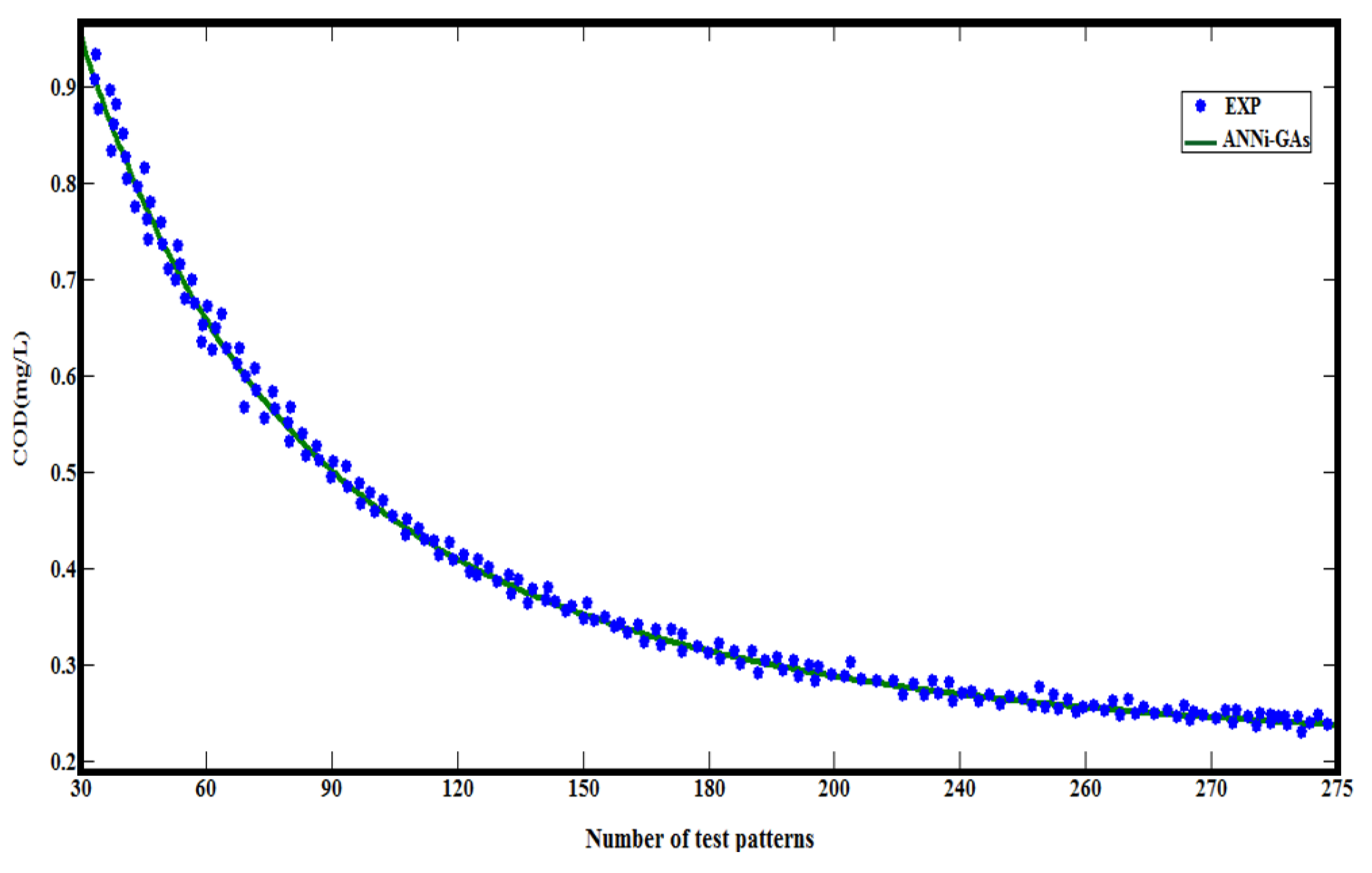
6. Conclusions
References
- H. Köhler and R. Triebskorn, "Wildlife Ecotoxicology of Pesticides: Can We Track Effects to the Population Level and Beyond?".
- P. G. Dennis, T. Kukulies, C. Forstner, T. G. Orton and A. B. Pattison, "The effects of glyphosate, glufosinate, paraquat and paraquat-diquat on soil microbial activity and bacterial, archaeal and nematode diversity".
- M. Greaves, H. A. Davies, J. A. P. Marsh, G. I. Wingfield and S. J. L. Wright, "Herbicides and Soil Microorganisms".
- G. Rammohan and M. N. Nadagouda, "Green Photocatalysis for Degradation of Organic Contaminants: A Review".
- Y. Liu et al., "Photoelectrocatalytic degradation of tetracycline by highly effective TiO2 nanopore arrays electrode".
- Goli et al., "Applications of artificial neural networks for water quality prediction and modeling: A review," Sci. Total Environ., vol. 711, 2020, Art no. 137191. [CrossRef]
- N. A. Khan and S. M. Ali, "Artificial neural networks and genetic algorithm based model for prediction of dissolved oxygen in river," Environ. Sci. Pollut. Res., vol. 29, no. 43, pp. 65992–66009, 2022. [CrossRef]
- S. Heddam and S. Belkacem, "Inverse artificial neural network for the prediction of optimal operating conditions in a photovoltaic thermal collector," Mater. Today: Proc., 2021. [CrossRef]
- Khataee, A. R., Fathinia, M., Zarei, M., Izadkhah, B., Joo, S.W. 2014. Modeling and optimization of photocatalytic/photoassisted-electro-Fenton like degradation of phenol using a neural network coupled with genetic algorithm. Journal of Industrial and Engineering Chemistry. 20 (4). 1852-1860.
- Li, X., Maier, H. R., Zecchin, A. C. 2015. Improved PMI-based input variable selection approach for artificial neural network and other data driven environmental and water resource models. Environmental Modelling & Software. 65. 15-29.
- Hoffmann, M.R.; Martin, S.T.; Choi, W.; Bahnemannt, D.W. Environmental applications of semiconductor photocatalysis. Chem. Rev., 1995, 95(1), 6996.
- Herrmann, J.M. Heterogeneous photocatalysis: Fundamentals and applications to the removal of various types of aqueous pollutants. Catal. Today., 1999, 53(1), 115-129.
- Gaya, U.I.; Abdullah, A.H. Heterogeneous photocatalytic degradation of organic contaminants over titanium dioxide: A review of fundamentals, progress and problems. J. Photoch. Photobio. C., 2008, 9(1), 1-12.
- Wang, C., Quan, H., Xu, X., Optimal design of multiproduct batch chemical process using genetic algorithm. Industrial engineering and chemistry research,1996, 35, 10, 3560-6.
- Goldberg, D. E. (1989). Genetic algorithms in search optimization, and machine learning. Reading, MA: Addison Wesley.
- Baker, J.E. Adaptive selection methods for Genetic Algoriyhms. Pages 101-111 of proceedings of an international conference on Genetic Algorithms, Grefenstette, J.J. (ed), Lawrence Earlbaum. 1985.
- Frantz, D. R. Non-linearities in genetic adaptive search. Diss. Abstr. Int. B 1972, 33 (11), 5240.
- Jaimes-Ramírez R., Pineda-Arellano C.A., Varia J.C., Silva-Martínez S. Visible light-induced photocatalytic elimination of organic pollutants by TiO2: A Review. Current Organic Chemistry, 2015, 19, 540-555.
- Suslick, K.S., the chemical effects of ultrasound, Sci. Am., 1989, 260, 80-86.
- Hach Company, 1992. Water Analysis Handbook, Loveland, Colorado.
- David E. Goldberg. Genetic Algorithms in Search, Optimization, and Machine Learning.
- El Hamzaoui, Y., J.A. Hernandez, J. A., Silva-Martínez, S., Bassam, A., Alvarez, A., Lizama-Bahena, C., 2011. Optimal performance of COD removal during aqueous treatment of alazine and gesaprim commercial herbicides by direct and inverse neural network. Desalination. 37, 325-337.
- Hernández, J. A., Colorado, D., Cortés-Aburto, O., El Hamzaoui, Y., Velazquez, V., Alonso, B., 2013. Inverse neural network for optimal performance in polygeneration systems. Applied Thermal Engineering. 50. 1399–1406.
- Mas, D.M.L., & Ahlfeld, D.P. (2007). Comparing artificial neural networks and regression models for predicting faecal coliform concentrations. Hydrological Sciences.
- Journal, 52, 713-731.
- Crowther, J., Kay, D.,& Wyer, M.D. (2001). Relationships between microbial water quality and environmental conditions in coastal recreational waters: the fylde coast, UK. Water Research, 35, 4029-4038.
- Motamarri, S., & Boccelli, D.L. (2012). Development of a neural-based forecasting tool to classify recreational water quality using fecal indicator organisms. Water Research, 46, 4508-4520.
- A. J. F. Van Rooij, L. C. Jain, R. P. Johnson. Neural Network Training Using Genetic Algorithms. Series in Machine Perception and Artificial Intelligence. World Scientific Pub Co Inc (March 1997).


| Active components | Oxidation reaction (mineralization) | Concentration (mg/l) | COD (mg/l) |
|---|---|---|---|
| Alachlor | C14H20ClNO2 + 17O2 → 4CO2 + NH3 + HCl + H2O | 83.3 | 170.4 |
| Atrazine | C8H14ClN5 + 7.5O2 + H2O → CO2 + 5NH3 + HCl | 62.5 | 68.0 |
| Variable | Range | Unit |
|---|---|---|
| Input layer | ||
| Reaction time | 0 – 480 | min |
| pH | 1 – 5 | |
| Concentration of herbicides | 0.1540 – 0.3090 | mM |
| Contaminant | 0.1 – 0.9 | |
| US Ultrasound | 0 – 20 | kHz |
| UV Ultraviolet | 0 – 352 | nm |
| [TiO2]o | 0 – 300 | mg/l |
| [K2S2O8]o | 0 – 13 | mM |
| Solar radiation | 0 – 820 | W/m2 |
| Output layer | ||
| COD | 0.07 – 1 | mg/l |
| Number of neurons |
Weights | Bias | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hidden layer Wi (s, k) |
Output layer Wo (s, l) |
|||||||||||
| (s)* | (k=1) | (k=2) | (k=3) | (k=4) | (k=5) | (k=6) | (k=7) | (k=8) | (k=9) |
COD (l=1) |
b(1,s) | b(2,l) |
| 1 | -0.3166 | 0.2236 | 7.2842 | 2.0881 | 1.811 | 3.3471 | -1.0447 | 1.7596 | 2.0459 | 0.6031 | -5.25 | 12.9598 |
| 2 | -0.5515 | -0.956 | -3.8864 | 2.0139 | -2.867 | -1.294 | -0.45 | -6.4122 | 2.1782 | -5.9361 | 5.6355 | |
| 3 | 1.4092 | -8.029 | -12.034 | 15.7374 | -3.313 | -0.242 | -3.6828 | 11.6702 | -3.655 | 0.6256 | -11.9996 | |
| 4 | -0.7383 | -0.169 | -3.2518 | 1.2309 | -0.327 | -1.031 | -0.541 | -2.8458 | 1.668 | 7.0453 | 2.6748 | |
| 5 | 50.505 | -0.110 | 6.7242 | 1.7719 | -1.248 | -3.151 | 1.6575 | -1.4106 | -3.0969 | -6.087 | -2.4115 | |
| 6 | -1.3825 | -0.466 | -5.9643 | 5.8058 | 0.9334 | -2.855 | -1.6653 | -3.5912 | 2.8908 | -0.6846 | 1.2615 | |
| 7 | -29.020 | 0.7569 | 0.4108 | 0.2687 | -0.086 | -0.630 | 1.0245 | -0.7546 | -7.7876 | 5.127 | 1.4984 | |
| 8 | -3.7356 | 0.0012 | 0.9109 | -0.667 | -0.786 | -1.226 | 1.2131 | -1.3882 | -0.1462 | 1.7649 | -0.0617 | |
| 9 | -5.5039 | 1.1252 | 3.23 | -2.7131 | -0.735 | 0.2545 | 1.9287 | 3.0638 | -1.139 | -0.6662 | -0.6785 | |
| *s is the number of neurons in the hidden layer, k is the number of neurons in the input layer, l is the number of neurons in output layer (l=1). | ||||||||||||
| Crossover type | Average of the runs2 | Geometric Mean of the runs3 | ||||
|---|---|---|---|---|---|---|
| 1-point | 0.515 | 0.583 | 0.379 | 0.401 | 6.81E-2 | 2.33E-2 |
| 2-point | 0.216 | 0.753 | 0.474 | 1.28E-2 | 5.02E-3 | 3.13E-3 |
| uniform | 0.113 | 0.183 | 0.203 | 9.52E-2 | 6.14E-3 | 3.17E-2 |
| NN-2-point | 0.553 | 0.277 | 0.927 | 3.35E-2 | 0.79E-3 | 7.01E-4 |
| NN-uniform | 0.376 | 0.305 | 0.229 | 3.47E-4 | 8.50E-5 | 9.12E-6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





