Submitted:
08 June 2024
Posted:
10 June 2024
You are already at the latest version
Abstract
Keywords:
1. Background
2. Results
2.1. Chloroplast Genome of Habenaria aitchisonii and Habenaria tibetica
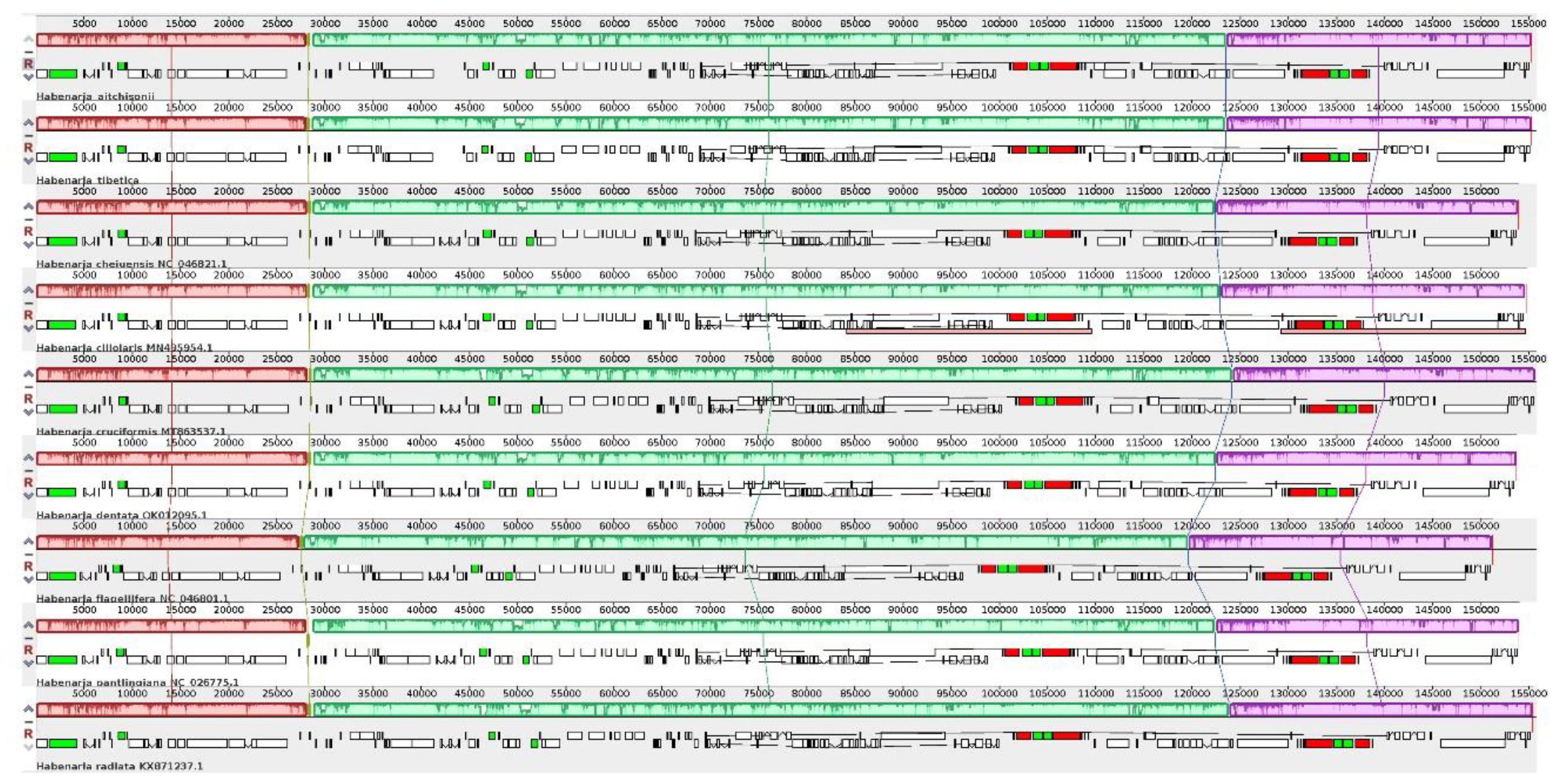
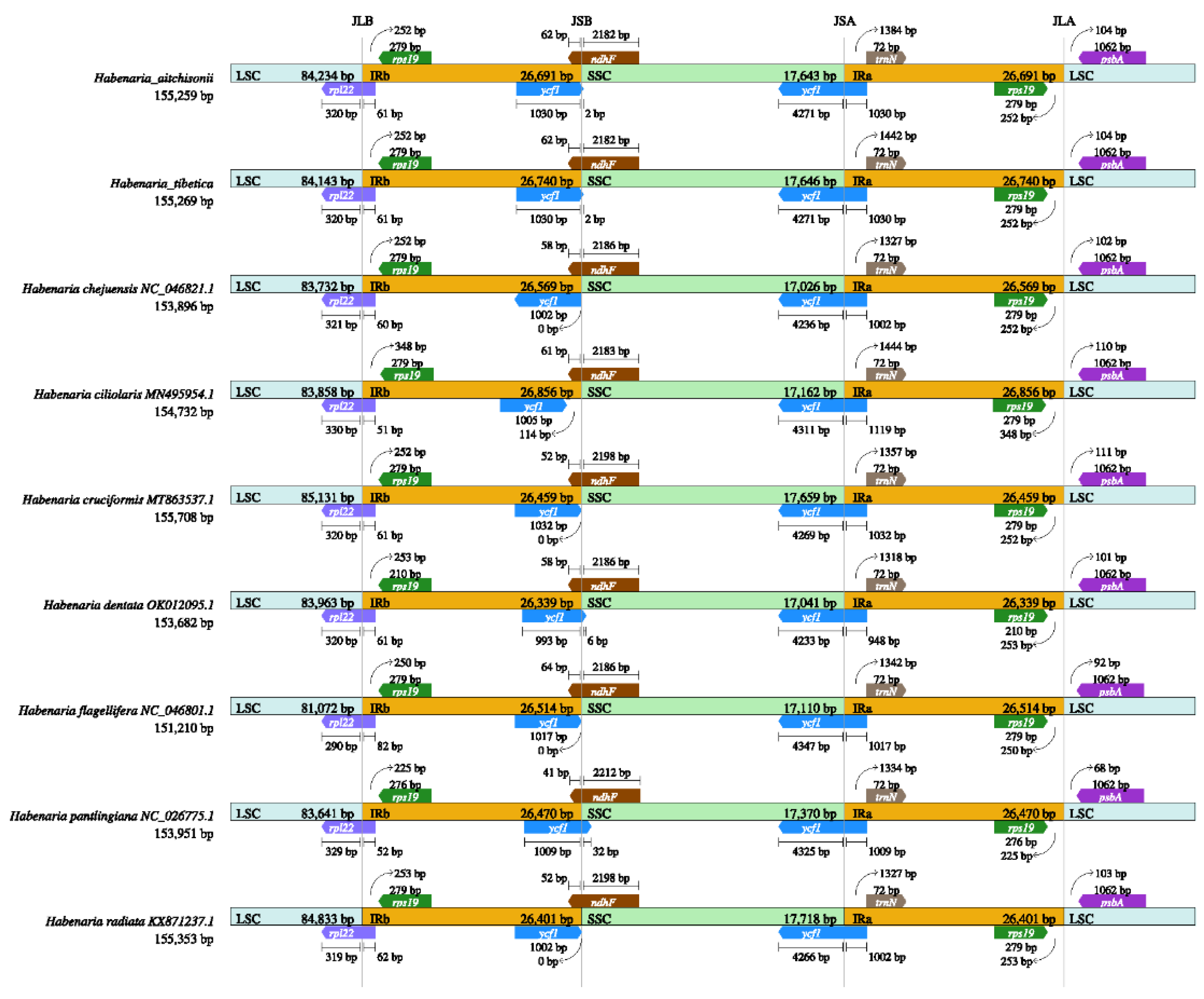
2.2. Comparation of Plastome Features of the Habenaria Genus
2.3. Relative Synonymous Codon usage and Amino Acid Frequency
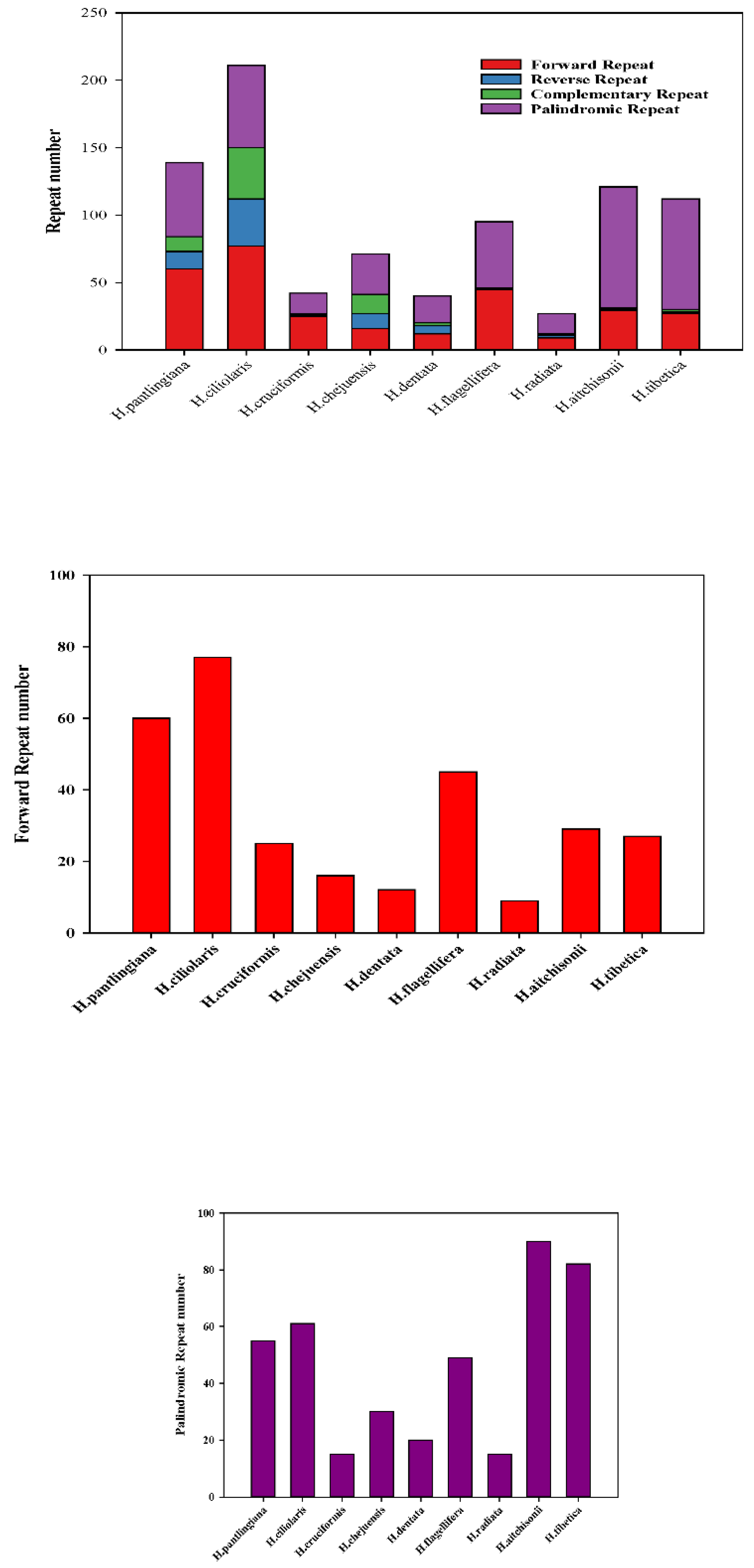
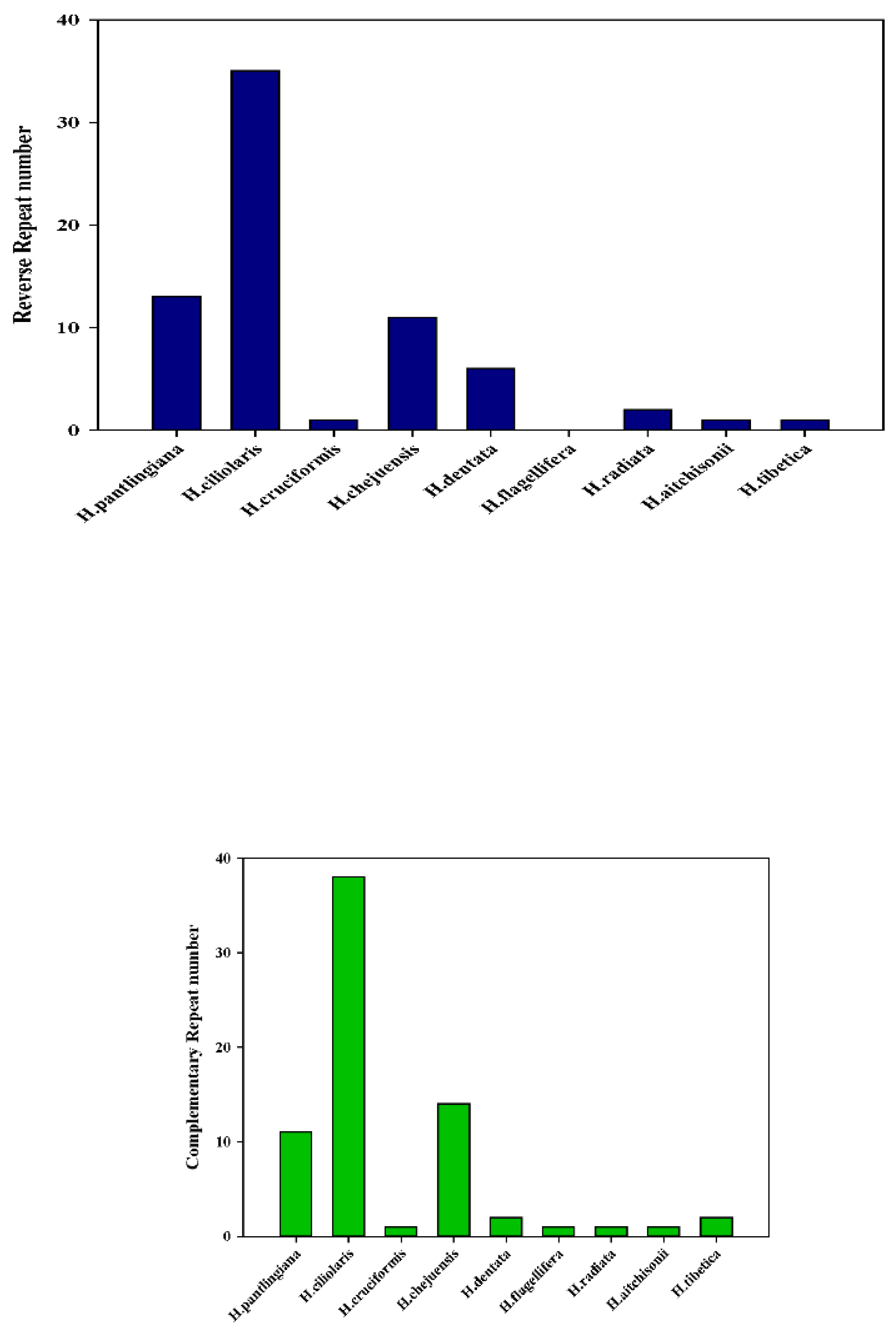
2.4. Analysis of Microsatellites and Oligonucleotide Repeats
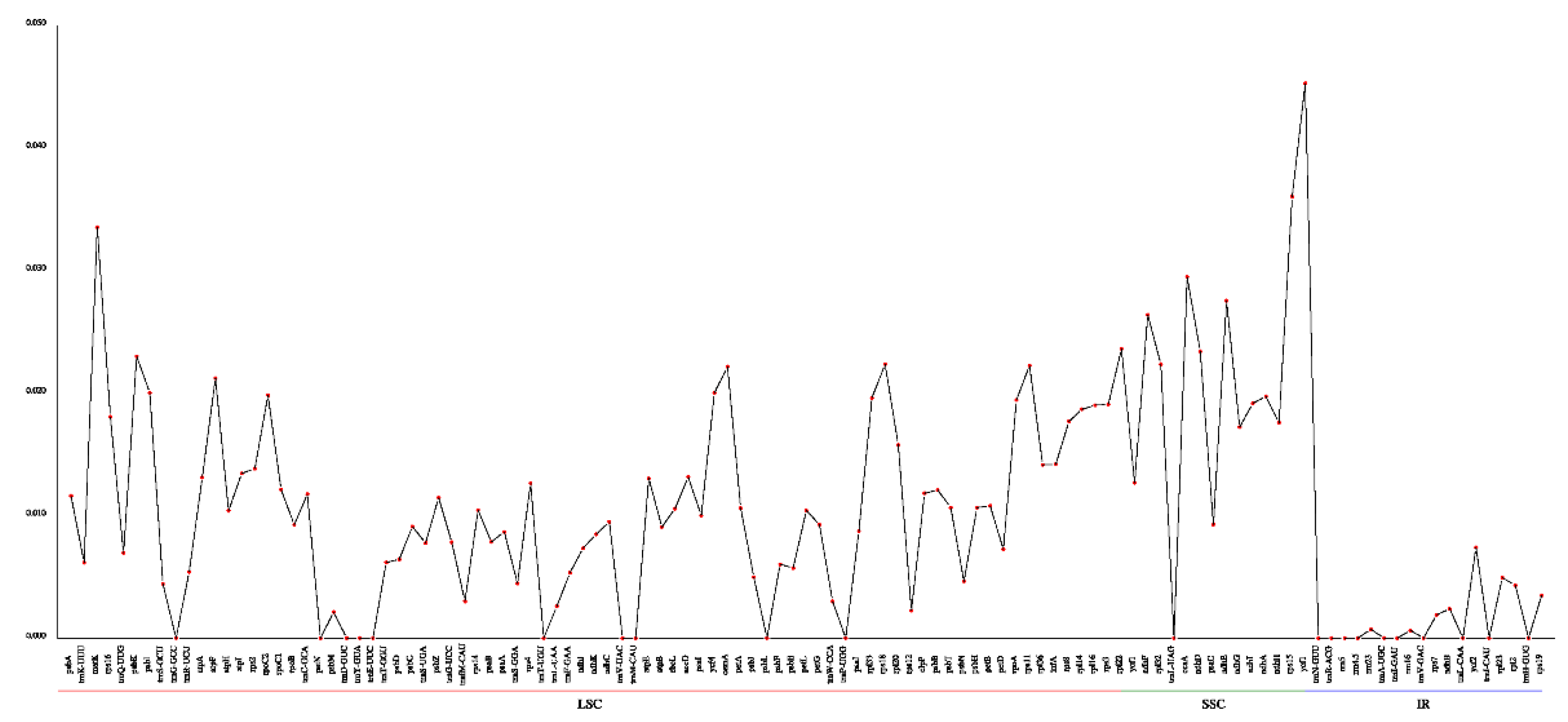
2.5. Identification of Polymorphic Loci
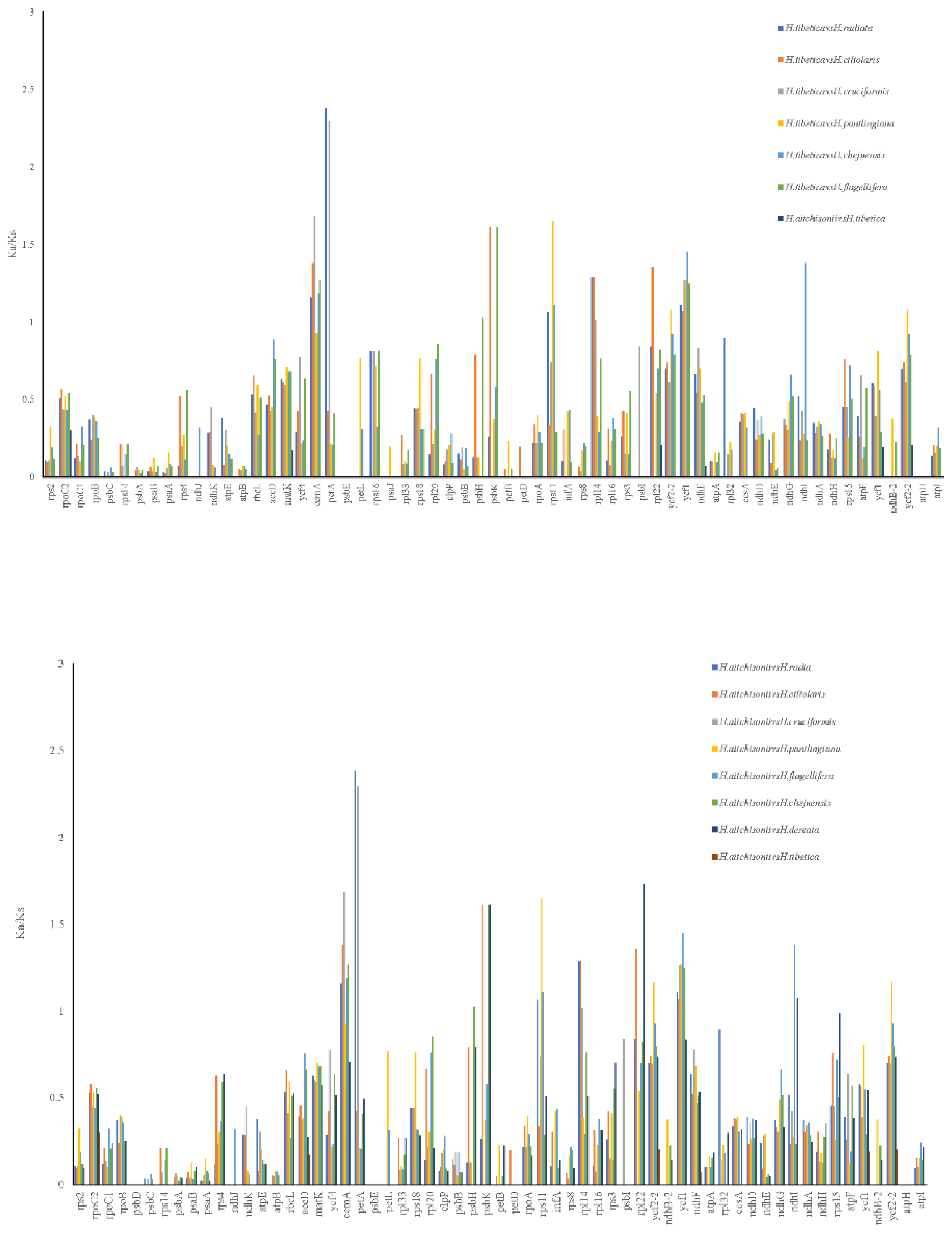
2.6. Molecular Evolution Analysis
2.7. Phylogenetic Relationship Analysis
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Sample Collection and DNA Extraction
5.2. Plastome Genome Sequencing, Assembling and Annotation
5.3. SSRs, Codon Usages and Nucleotide Diversity Analysis
5.4. Comparative Analysis of cp Genomes
5.5. Molecular Evolution Analysis
5.6. Phylogenetic Analysis
Availability of data and materials
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Govaerts, R.; Bernet, P.; Kratochvil, K.; Gerlach, G.; Carr, G.; Alrich, P.; et al. World Checklist of Orchidaceae. The board of Trustees of the Royal Botanic Gardens, Richmond, U.K, 2019.
- Pridgeon, A.M.; Cribb, P.J.; Chase, M.W.; Rasmussen, F.N. Genera Orchidacearum, Orchidoideae (part 1), Oxford: Oxford University Press, U.K, 2001.
- Chen, X.; Cribb, P.J. “Habenaria”, In Flora of China, eds Wu ZG, Raven PH, Hong DY. Beijing: Science Press, and St.Louis : Missouri Botanical Garden, 2009, 144-160.
- Tang, H. DNA barcoding identification and ecological suitability of important medicinal plants of Orchidaceae. Ph.D. thesis, Sichuan Agricultural University, Ya’an (China), 2016.
- Zhang, J.; Yang, Y.S.; Tang, J.M.; Luo, Y.J.; Lv, H.Q.; Chai, S.F. The complete chloroplast genome sequence of Habenaria dentata (Orchidaceae). Mitochondrial DNA B. Resour. 2022, 7, 969–970. [Google Scholar] [CrossRef] [PubMed]
- Mahnashi, M.H.; Alqahtani, Y.S.; Alyami, B.A.; Alqarni, A.O.; Alshrahili, M.A.; Abou-Salim, M.A.; Alqahtani, M.N.; Mushtaq, S.; Sadiq, A.; Jan, M.S. GC-MS analysis and various in vitro and in vivo pharmacological potential of Habenaria plantaginea Lindl. Evid-Based. Compl. Alt. Med. 2022, 7921408. [Google Scholar] [CrossRef] [PubMed]
- Challe, J.F.; Price, L.L. Endangered edible orchids and vulnerable gatherers in the context of HIV/AIDS in the Southern Highlands of Tanzania. J. Ethnobiol. Ethnomed. 2009, 5, 41. [Google Scholar] [CrossRef] [PubMed]
- Dressler, R.L. Phylogeny and classification of the orchid family. OR:Dioscorides Press, Portland, USA, 1993.
- Bateman, R.M.; Hollingsworth, P.M.; Preston, J.; Luo, Y.B.; Pridgeon, A.M.; Chase, M.W. Molecular phylogenetics and evolution of Orchidinae and selected Habenariinae (Orchidaceae). Bot. J. Linn. Soc. 2003, 142, 1–40. [Google Scholar] [CrossRef]
- Raskoti, B.B.; Ale, R. Molecular phylogeny and morphology reveal a new epiphytic species of Habenaria (Orchidaceae; Orchideae; Orchidinae) from Nepal. PLoS ONE. 2019, 14, e0223355. [Google Scholar] [CrossRef] [PubMed]
- Pandey, T.R.; Jin, X.H. Taxonomic revision of Habenaria josephi group (sect. Diphyllae s.l.) in the Pan Himalaya. PhytoKeys 2021, 175, 109–135. [Google Scholar] [CrossRef] [PubMed]
- Besi, E.E.; Hooi, W.K.; Sylvester. ; Pungga, R.; Yong, C.S.Y.; Mustafa, M.; Go, R. Rare orchid species in Malaysia: New records, recollections and amended descriptions. PLoS ONE 2022, 17, e0267485. [Google Scholar] [CrossRef] [PubMed]
- Batista, J.A.; Borges, K.S.; de Faria, M.W.; Proite, K.; Ramalho, A.J.; Salazar, G.A.; van den Berg, C. Molecular phylogenetics of the species-rich genus Habenaria (Orchidaceae) in the new world based on nuclear and plastid DNA sequences. Mol. Phylogenet. Evol. 2013, 67, 95–109. [Google Scholar] [CrossRef]
- Pedron, M.; Buzatto, C.R.; Ramalho, A.J.; Carvalho, B.M.; Radins, J.A.; Singer, R.B.; Batista, J.A. Molecular phylogenetics and taxonomic revision of Habenaria section Pentadactylae (Orchidaceae, Orchidinae). Bot. J. Linn. Soc. 2014, 175, 47–73. [Google Scholar] [CrossRef]
- Jin, W.T.; Jin, X.H.; Schuiteman, A.; Li, D.Z.; Xiang, X.G.; Huang, W.C.; Li, J.W.; Huang, L.Q. Molecular systematics of subtribe Orchidinae and Asian taxa of Habenariinae (Orchideae, Orchidaceae) based on plastid matK, rbcL and nuclear ITS. Mol. Phylogenet. Evol. 2014, 77, 41–53. [Google Scholar] [CrossRef]
- Smith, D.R. Mutation rates in plastid genomes: They are lower than you might think. Genome Biol. Evol. 2015, 7, 1227–1234. [Google Scholar] [CrossRef] [PubMed]
- Daniell, H.; Lin, C.S.; Yu, M.; Chang, W.J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef] [PubMed]
- Abdullah. ; Mehmood, F.; Rahim, A.; Heidari, P.; Ahmed, I.; Poczai, P. Comparative plastome analysis of Blumea, with implications for genome evolution and phylogeny of Asteroideae. Ecol. Evol. 2021, 11, 7810–7826. [Google Scholar] [CrossRef] [PubMed]
- Mehmood, F.; Abdullah. ; Ubaid, Z.; Shahzadi, I.; Ahmed, I.; Waheed, M.T.; Poczai, P.; Mirza, B. Plastid genomics of Nicotiana (Solanaceae): insights into molecular evolution, positive selection and the origin of the maternal genome of Aztec tobacco (Nicotiana rustica). Peer J 2020, 8, e9552. [Google Scholar] [CrossRef] [PubMed]
- Palmer, J.D. Comparative organization of chloroplast genomes. Ann. Rev. Genet. 1985, 19, 325–354. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Y.; Liao, M.; Cheng, Y.H.; Feng, Y.; Ju, W.B.; Deng, H.N.; Li, X.; Plenkovi ´c-Moraj, A.; Xu, B. Comparative chloroplast genomics of seven endangered Cypripedium species and phylogenetic relationships of Orchidaceae. Front. Plant Sci. 2022, 13, 911702. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.K.; Jo, S.; Cheon, S.H.; Joo, M.J.; Hong, J.R.; Kwak, M.; Kim, K.J. Plastome Evolution and Phylogeny of Orchidaceae, With 24 New Sequences. Front. Plant Sci. 2020, 11, 22. [Google Scholar] [CrossRef]
- Zhou, D.W.; Mehmood, F.; Lin, P.C.; Cheng, T.F.; Wang, H.; Shi, S.B.; Zhang, J.K.; Meng, J.; Zheng, K.; Poczai, P. Characterization of the evolutionary pressure on Anisodus tanguticus Maxim. with complete chloroplast genome sequence. Genes (Basel) 2022, 13, 2125. [Google Scholar] [CrossRef]
- Yang, Z.; Bielawski, J.P. Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 2000, 15, 496–503. [Google Scholar] [CrossRef]
- Yang, Z.; Nielsen, R. Codon-substitution Models for detecting molecular adaptation at individual sites along specific lineages. Mol. Biol. Evol. 2002, 19, 908–917. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, Q.; Shi, S.H.; Huang, Y.L.; Hasegawa, M. Detecting evolutionary rate heterogeneity among mangroves and their close terrestrial relatives. Ecol. Lett. 2002, 5, 427–432. [Google Scholar] [CrossRef]
- Kapralov, M.V.; Kubien, D.S.; Andersson, I.; Filatov, D.A. Changes in Rubisco kinetics during the evolution of C4 photosynthesis in Flaveria (Asteraceae) are associated with positive selection on genes encoding the enzyme. Mol. Biol. Evol. 2011, 28, 1491–1503. [Google Scholar] [CrossRef] [PubMed]
- Erixon, P.; Oxelman, B. Whole-gene positive selection, elevated synonymous substitution rates, duplication, and indel evolution of the chloroplast clpP1 gene. PLoS One 2008, 3, e1386. [Google Scholar] [CrossRef] [PubMed]
- Fan, W.B.; Wu, Y.; Yang, J.; Shahzad, K.; Li, Z.H. Comparative chloroplast genomics of Dipsacales species: insights into sequence variation, adaptive evolution, and phylogenetic relationships. Front. Plant Sci. 2018, 9, 689. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.Y.; Yang, J.X.; Li, H.K.; Zhao, H.S. Chloroplast genomes of two species of Cypripedium: expanded genome size and proliferation of AT-biased repeat sequences. Front. Plant Sci. 2021, 12, 609–729. [Google Scholar] [CrossRef] [PubMed]
- Clément, Y.; Fustier, M.A.; Nabholz, B.; Glémin, S. The bimodal distribution of genic GC content is ancestral to monocot species. Genome Biol. Evol. 2014, 7, 336–348. [Google Scholar] [CrossRef] [PubMed]
- Muyle, A.; Serres-Giardi, L.; Ressayre, A.; Escobar, J.; Glémin, S. GC-biased gene conversion and selection affect GC content in the Oryza genus (rice). Mol. Biol. Evol. 2011, 28, 2695–2706. [Google Scholar] [CrossRef] [PubMed]
- Powell, W.; Morgante, M.; Mcdevitt, R.; Vendramin, G.G.; Rafalski, J.A. Polymorphic simple sequence repeat regions in chloroplast genomes:applications to the population genetics of pines. Proc. Natl. Acad. Sci. U. S. A. 1995, 92, 7759–7763. [Google Scholar] [CrossRef] [PubMed]
- Xie, D.F.; Yu, H.X.; Price, M.; Xie, C.; Deng, Y.Q.; Chen, J.P.; Yu, Y.; Zhou, S.D.; He, X.J. Phylogeny of Chinese Allium species in section Daghestanica and adaptive evolution of Allium (Amaryllidaceae, Allioideae) species revealed by the Chloroplast complete genome. Front. Plant Sci. 2019, 10, 460. [Google Scholar] [CrossRef]
- Myers, N. , Mittermeier, R.A.; Mittermeier, C.G.; da Fonseca, G.A.; Kent, J. Biodiversity hotspots for conservation priorities. Nature 2000, 403, 853–858. [Google Scholar] [CrossRef]
- Wang, J.; Li, J.; Lin, W.; Deng, B.; Lin, L.; Lv, X.; Hu, Q.; Liu, K.; Fatima, M.; He, B.; et al. Genome-wide identification and adaptive evolution of CesA/Csl superfamily among species with different life forms in Orchidaceae. Front. Plant Sci. 2022, 13, 994679. [Google Scholar] [CrossRef] [PubMed]
- Acharya, K.P.; Vetaas, O.R.; Birks, H.J.B. Orchid species richness along Himalayan elevational gradients. J. Biogeogr. 2011, 38, 1821–1833. [Google Scholar] [CrossRef]
- Hu, H.; Wei, Y.; Wang, W.; Suonan, J.; Wang, S.; Chen, Z.; Guan, J.; Deng, Y. Richness and distribution of endangered orchid species under different climate scenarios on the Qinghai-Tibetan Plateau. Front. Plant Sci. 2022, 13, 948189. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987, 19, 11–15. [Google Scholar]
- Luo, R.; Liu, B.; Xie, Y.; Li, Z.; Huang, W.; Yuan, J.; He, G.; Chen, Y.; Pan, Q.; Liu, Y.; et al. Erratum: SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 2015, 4, 30. [Google Scholar] [CrossRef] [PubMed]
- Boetzer, M.; Henkel, C.V.; Jansen, H.J.; Butler, D.; Pirovano, W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 2011, 27, 578–579. [Google Scholar] [CrossRef] [PubMed]
- Wyman, S.K.; Jansen, R.K.; Boore, J.L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 2004, 20, 3252–3255. [Google Scholar] [CrossRef] [PubMed]
- Lohse, M.; Drechsel, O.; Bock, R. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 2007, 52, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Thiel, T.; Michalek, W.; Varshney, R.K.; Graner, A. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor. Appl. Genet. 2003, 106, 411–422. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, SE.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Darling, A.C.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef] [PubMed]
- Amiryousefi, A.; Hyvönen, J.; Poczai, P. IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics 2018, 34, 3030–3031. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Liu, F.; Wang, L.; Huang, S.; Yu, J. Nonsynonymous substitution rate (Ka) is a relatively consistent parameter for defining fast-evolving and slow-evolving protein-coding genes. Biol. Direct 2011, 6, 13. [Google Scholar] [CrossRef]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
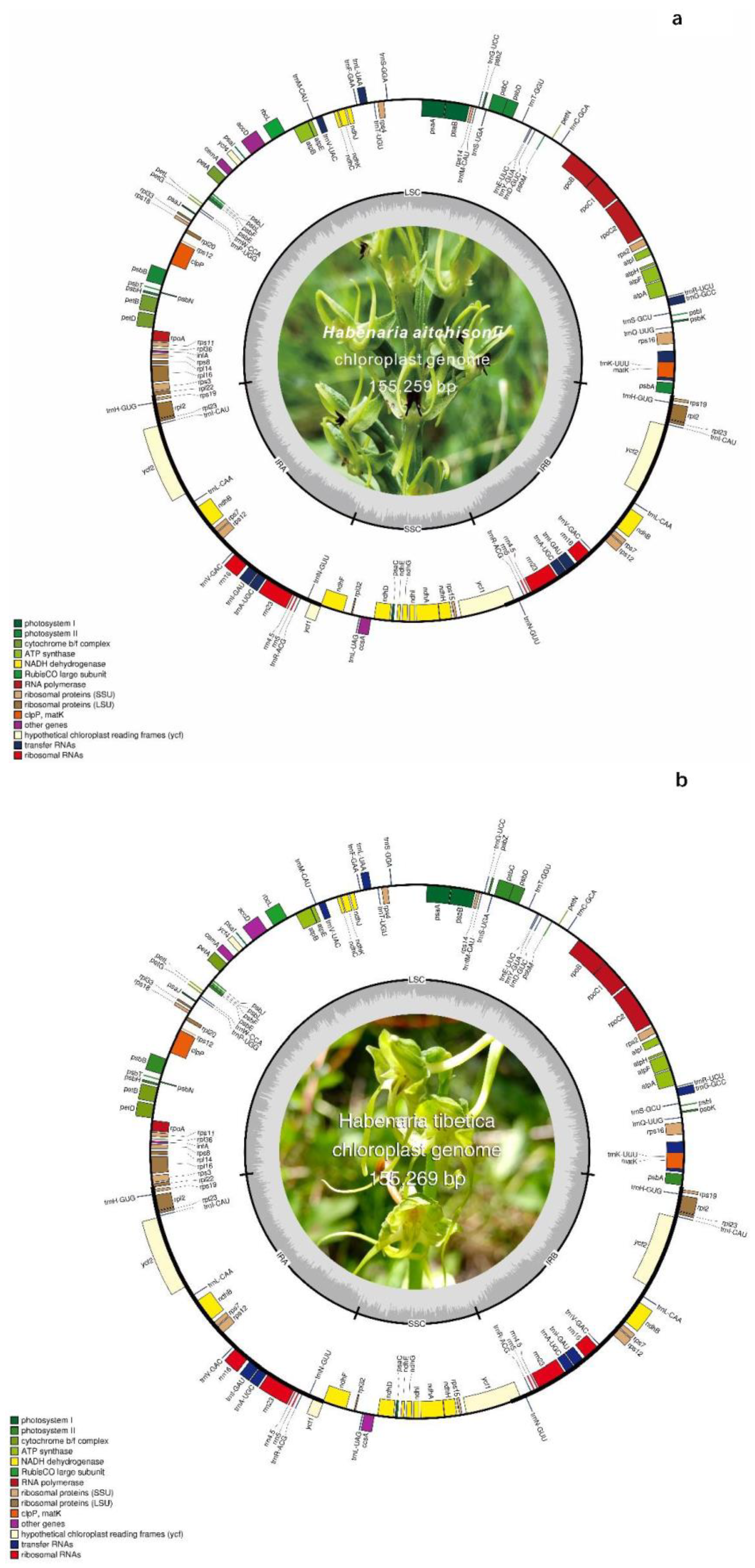



| Genome feature | H. aitchisonii | H.tibetica | H.dentata | H.cruciformis | H.ciliolaris | H.radiata | H.flagellifera | H.checiformis | H.pantlingiana |
| Genome Size(bp) | 155,259 | 155,269 | 153,682 | 155,708 | 154,544 | 155, 353 | 155,298 | 153,896 | 153,951 |
| LSC (bp) | 84,234 | 84,143 | 83,963 | 85,131 | 84,032 | 84, 833 | 85,749 | 83,732 | 83,641 |
| SSC (bp) | 17,643 | 17,646 | 17,041 | 17,659 | 19,602 | 17, 718 | 18,373 | 17,026 | 17,370 |
| IR (bp) | 26,691 | 26,740 | 26,339 | 26,459 | 25,455 | 26,401 | 25,595 | 26,569 | 26,470 |
| GC content (%) | 36.83 | 36.84 | 36.62 | 36.60 | 37.90 | 36.60 | 37.90 | 36.70 | 36.60 |
| Total numberof genes | 132 | 132 | 133 | 131 | 132 | 113 | 130 | 131 | 133 |
| Protein-coding gene | 86 | 86 | 87 | 79 | 86 | 79 | 81 | 85 | 87 |
| tRNA | 38 | 38 | 37 | 30 | 38 | 30 | 37 | 38 | 38 |
| rRNA | 8 | 8 | 8 | 4 | 8 | 4 | 8 | 8 | 8 |
| Gene name | Null hypothesis | Alternative hypothesis | Significant test | |||||
| InL | df | ω | InL | df | ω | BEB | p-value | |
| cemA | -1967.69 | 1 | 1 | -1971.76 | 3 | 0.547 | 104K | p<0.05 |
| petA | -2147.03 | 1 | 1 | -2147.03 | 3 | 1.000 | none | p>0.05 |
| rps11 | -1126.26 | 1 | 1 | -1124.08 | 3 | 999.0 | 38V,82A,88T | p<0.05 |
| ndhi | -1371.40 | 1 | 1 | -1369.59 | 3 | 147.6 | 38I,95F | p<0.05 |
| psbH | -493.70 | 1 | 1 | -492.37 | 3 | 999.0 | 10S | p<0.05 |
| psbK | -462.73 | 1 | 1 | -462.73 | 3 | 2.140 | none | p>0.05 |
| rpl14 | -851.21 | 1 | 1 | -850.77 | 3 | 52.192 | 17Q,119P | p>0.05 |
| rpl22 | -1130.19 | 1 | 1 | -1130.11 | 3 | 1.643 | 120V | p>0.05 |
| ycf1 | -23972.75 | 1 | 1 | -23972.75 | 3 | 1.000 | none | p<0.05 |
| ycf2 | -16608.99 | 1 | 1 | -16608.99 | 3 | 1.000 | None | p<0.05 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





