Submitted:
07 August 2024
Posted:
07 August 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results and Discussion
2.1. Chloroplast Genome Features of G. indica
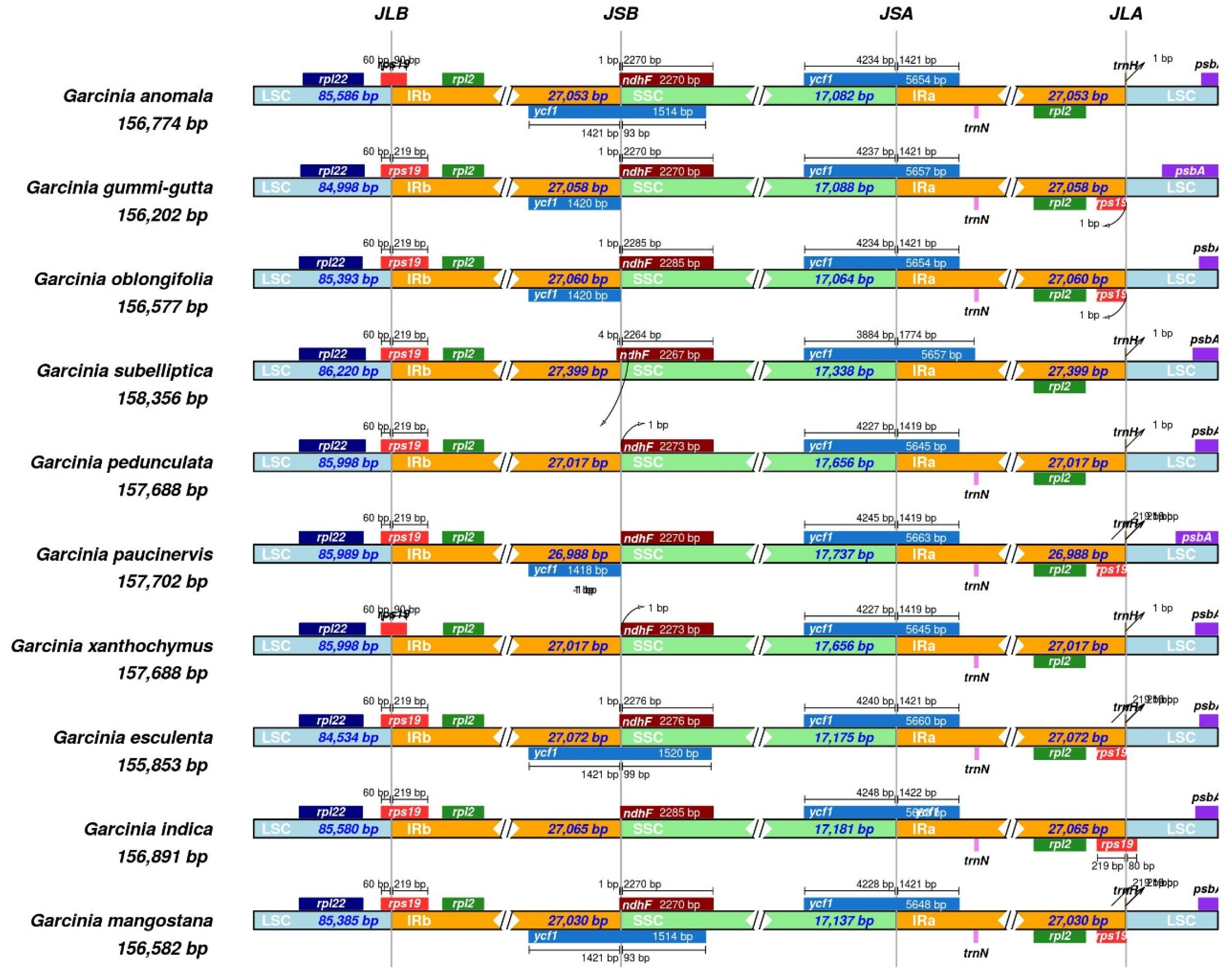
| S No | Garcinia species | GenBank Number | Chloroplast genome size (bp) | LSC (bp) | SSC (bp) | IRs (bp) | GC (%) | trnA genes | rrnA genes | Protein-coding genes | Total gene |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Garcinia anomala | MW582313.1 | 1,56,774 | 85,586 | 17,082 | 27,053 | 36 | 38 | 8 | 86 | 132 |
| 2 | Garcinia esculenta | OR834394.1 | 1,55,853 | 84,534 | 17,175 | 27,072 | 36 | 38 | 8 | 88 | 134 |
| 3 | Garcinia gummi-gutta | MN746309.1 | 1,56,202 | 84,996 | 17,088 | 27,059 | 36 | 36 | 8 | 86 | 130 |
| 4 | Garcinia mangostana | KX822787.1 | 1,58,179 | 86,458 | 17,703 | 27,009 | 36 | 38 | 8 | 86 | 132 |
| 5 | Garcinia oblongifolia | MT726019.1 | 1,56,577 | 85,393 | 17,064 | 27,060 | 36 | 36 | 8 | 86 | 130 |
| 6 | Garcinia paucinervis | MT501656.1 | 1,57,702 | 85,989 | 17,737 | 26,988 | 36 | 38 | 8 | 86 | 132 |
| 7 | Garcinia pedunculata | MN106251.1 | 1,57,688 | 85,998 | 17,656 | 27,017 | 36 | 36 | 8 | 86 | 130 |
| 8 | Garcinia subelliptica | MZ929421.1 | 1,58,356 | 86,220 | 17,338 | 27,399 | 36 | 38 | 8 | 85 | 131 |
| 9 | Garcinia xanthochymus | OP650213.1 | 1,57,688 | 85,998 | 17,656 | 27,017 | 36 | 38 | 8 | 85 | 131 |
| 10 | Garcinia indica | PP869627.1 | 1,56,891 | 85,580 | 17,181 | 27,065 | 36 | 32 | 8 | 86 | 126 |
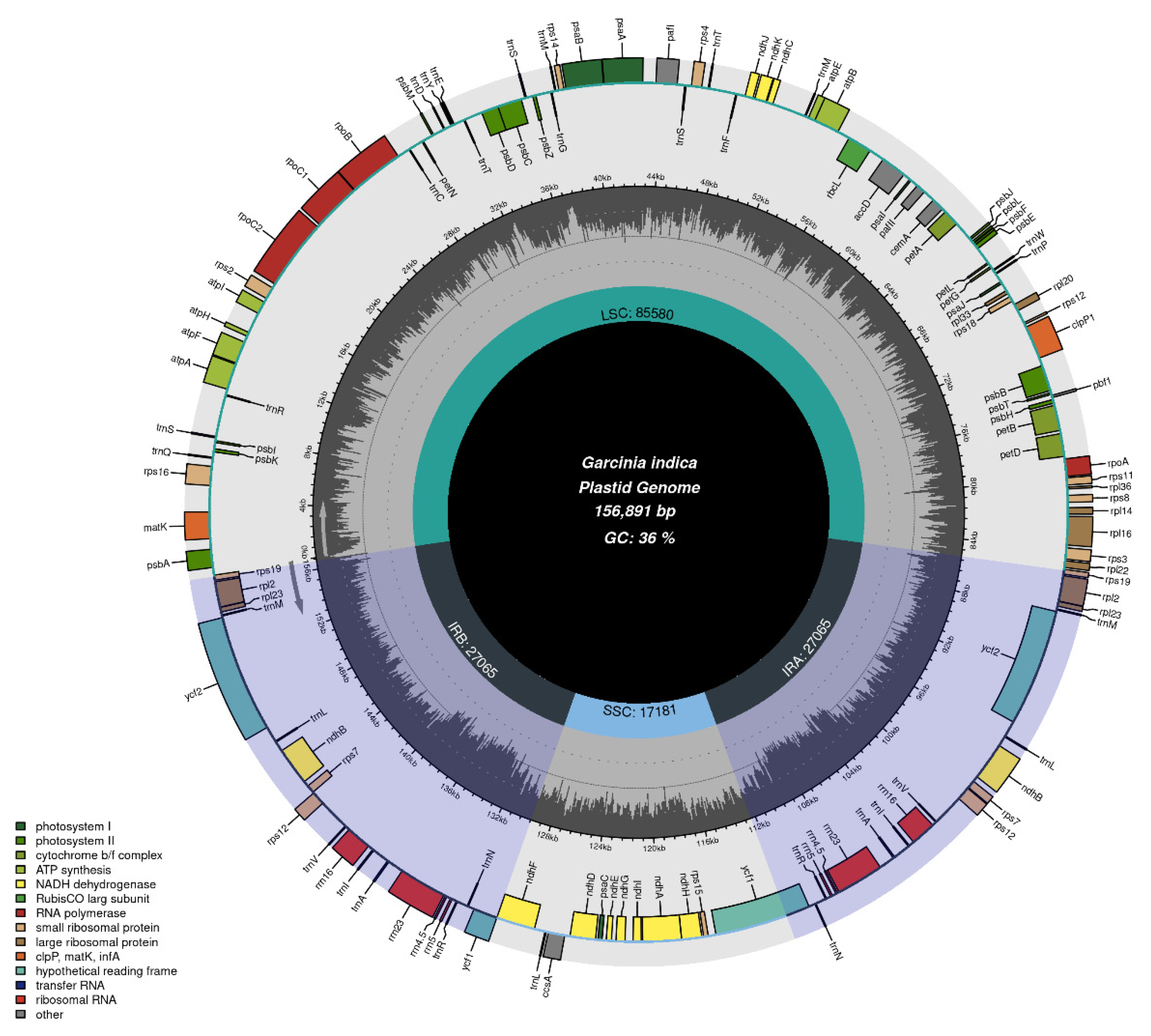
| Function of Genes | Category of Genes | Name of genes | ||||
|---|---|---|---|---|---|---|
| Self-replication | Large subunit of ribosome | rpl2a | rpl14 | rpl16b | rpl20 | rpl22 |
| rpl23a | rpl33 | rpl36 | ||||
| RNA polymerase | rpoA | rpoB | rpoC1b | rpoC2 | ||
| Ribosomal RNA genes | rrn4.5a | rrn5a | rrn16a | rrn23a | ||
| Small subunit of ribosome | rps2 | rps3 | rps4 | rps7a | rps8 | |
| rps11 | rps12abd | rps14 | rps15 | rps16b | ||
| rps18 | rps19a | |||||
| Transfer RNA genes | trnA-UGCab | trnC-GCA | trnD-GUC | trnE-UUC | trnF-GAA | |
| trnG-GCC | trnI-GAUab | trnL-CAAa | trnL-UAG | trnM-CAUa | ||
| trnN-GUUa | trnP-UGG | trnQ-UUG | trnR-ACGa | trnR-UCU | ||
| trnS-GCU | trnS-GGA | trnS-UGA | trnT-GGU | trnT-UGU | ||
| trnV-GACa | trnW-CCA | trnY-GUA | ||||
| Photosynthesis genes | ATP synthase | atpA | atpB | atpE | atpFb | atpH |
| atpI | ||||||
| NADH dehydrogenase | ndhAa | ndhBab | ndhC | ndhD | ndhE | |
| ndhF | ndhG | ndhH | ndhI | ndhJ | ||
| ndhK | ||||||
| ATPdependent protease subunit p gene | clpP1c | |||||
| Photosystem I | psaA | psaB | psaC | psaI | psaJ | |
| Photosystem II | psbA | psbB | psbC | psbD | psbE | |
| psbF | psbH | psbI | psbJ | psbK | ||
| psbL | psbM | psbT | psbZ | |||
| Cytochrome b/f complex | petA | petBb | petDb | petG | petL | |
| petN | ||||||
| Photosystem assembly factor | pafI | pafII | ||||
| Rubisco large subunit | rbcL | |||||
| Other genes | Subunit of acetyl-CoAcarboxylase | accD | ||||
| C-type cytpchrome synthesis gene | ccsA | |||||
| envelope membrane protein | cemA | |||||
| Maturase | matK | |||||
| Genes of unknown function | Conserved open reading frames | ycf1 | ycf2a | |||
2.2. Codon Usage Bias of G. indica Chloroplast Genome
2.3. Analysis of SSRs in G. indica Chloroplast
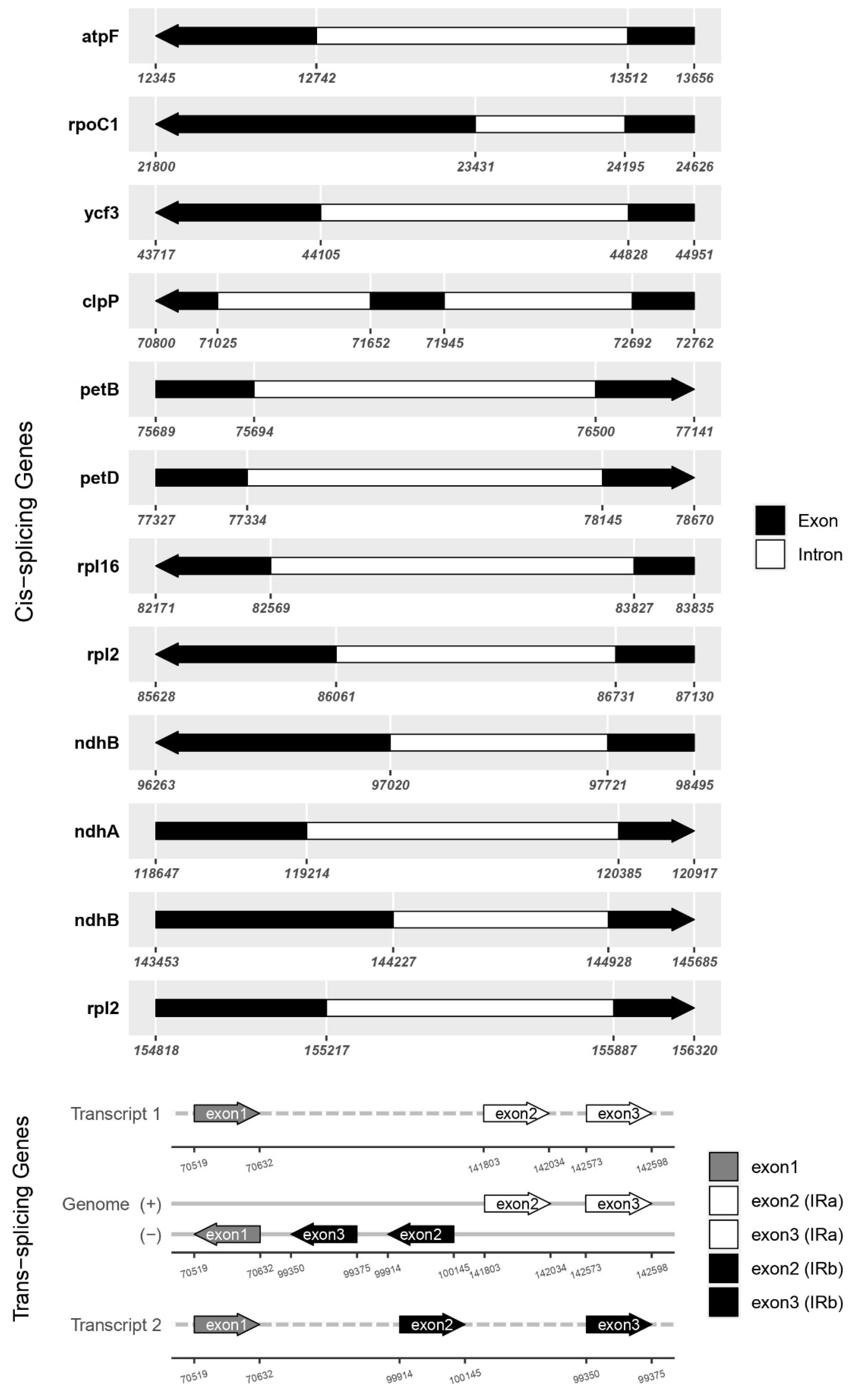
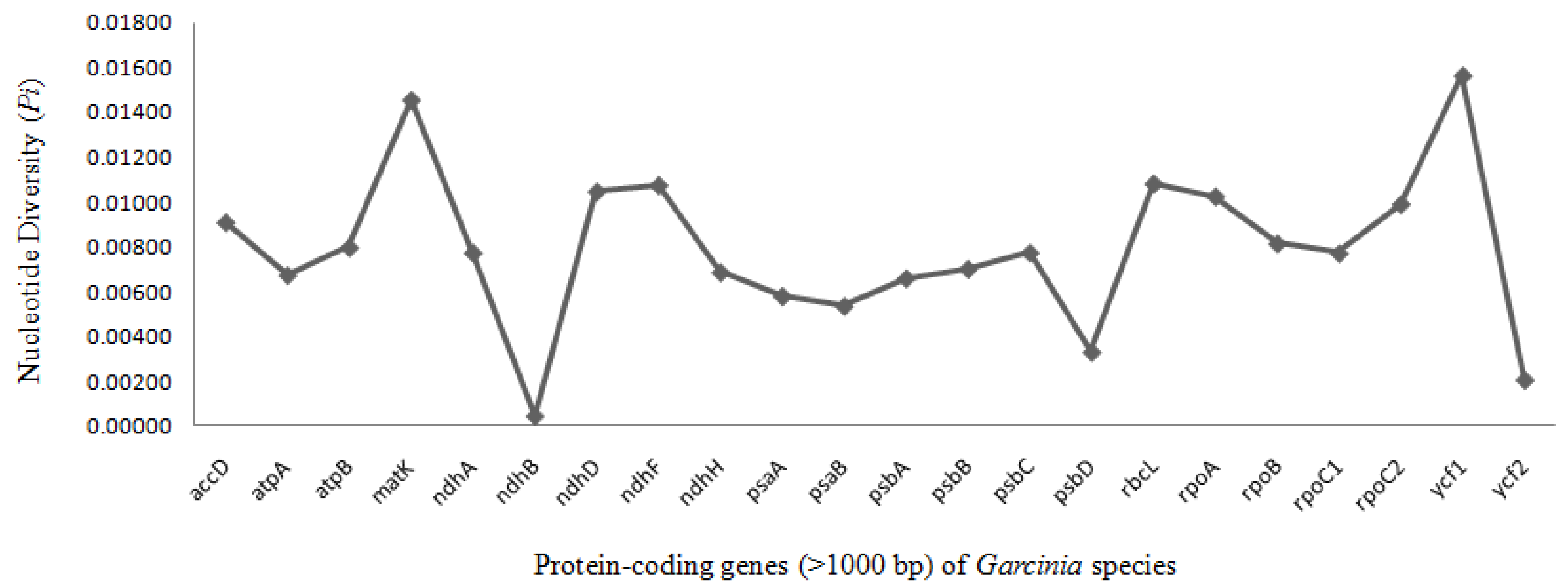
2.4. Comparative Chloroplast Genome and Divergence Hotspot Regions
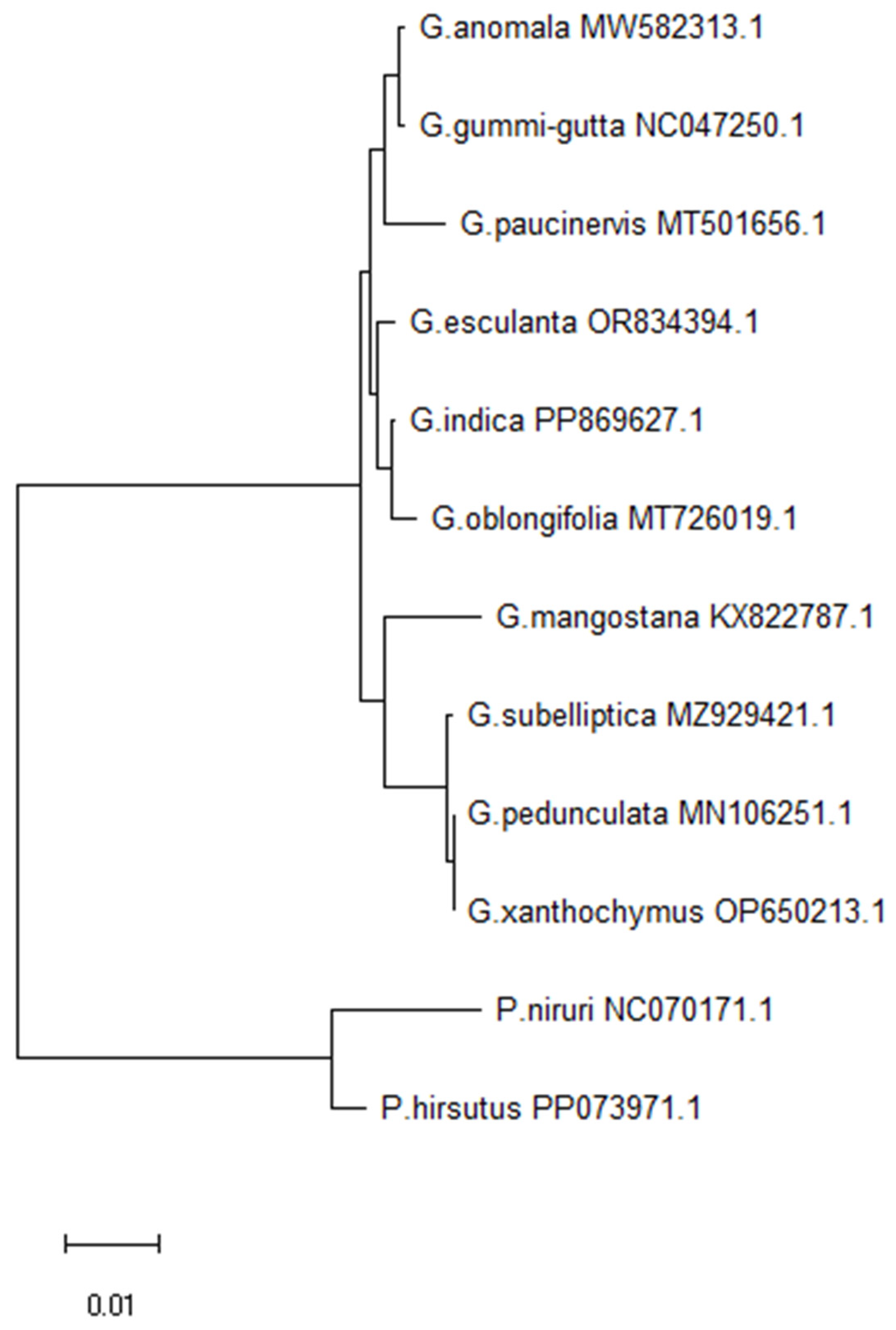
2.5. Phylogenetic Analysis
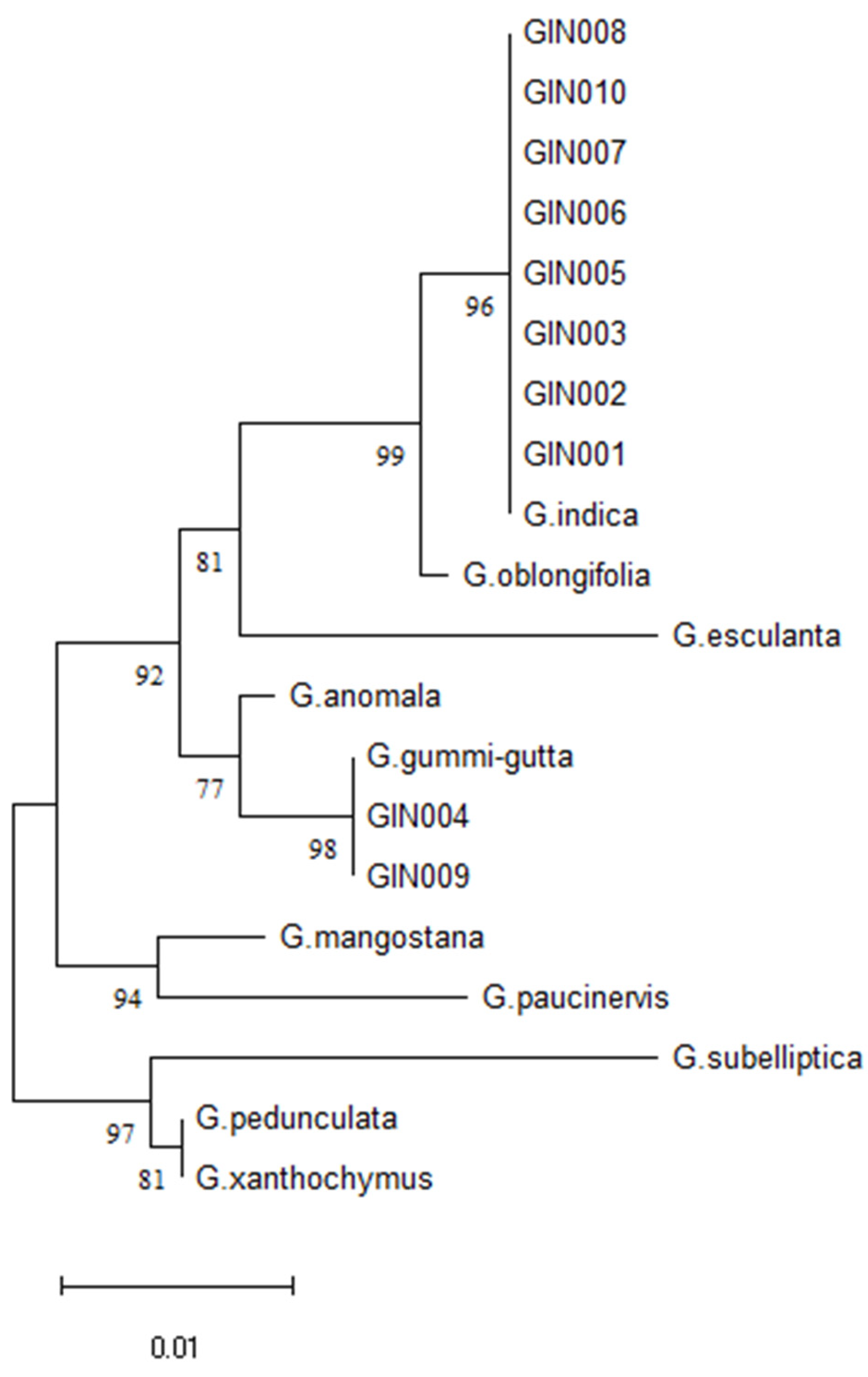
2.6. Authentication of Kokum (G. indica) Market Samples
3. Materials and Methods
3.1. Whole Genome Data
3.2. Chloroplast Genome Assembly and Annotation
3.3. SSR Identification and Codon Usuage Bias Analysis
3.4. Comparative Chloroplast Genomes and Nucleotide Diversity
3.5. Phylogenetic Analysis
3.6. Collection of Plants and Market Samples
3.7. Genomic DNA, DNA Barcoding and PCR Amplification
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mustardy, L.; Buttle, K.; Steinbach, G.; Garab, G. The three-dimensional network of the thylakoid membranes in plants: quasihelical model of the granum-stroma assembly. Plant Cell. 2008, 20, 2552–2557. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Wang, Z.; Cai, D.; Song, L.; Bai, J. The chloroplast genome sequence and phylogenetic analysis of Apocynum venetum L. PLoS ONE. 2022, 17, e0261710. [Google Scholar] [CrossRef] [PubMed]
- Howe, C.J.; Barbrook, A.C.; Koumandou, V.L.; Nisbet, R.E.R.; Symington, H.A; Wightman, T.F. Evolution of the chloroplast genome. Philos. Trans. R. Soc. Lond. Ser. B. Biol. Sci. 2003, 358, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Daniell, H.; Lin, C.S.; Yu, M.; Chang, W.J. Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 1–29. [Google Scholar] [CrossRef]
- Maheswari, P.; Kunhikannan, C.; Yasodha, R. Chloroplast genome analysis of Angiosperms and phylogenetic relationships among Lamiaceae members with particular reference to teak (Tectona grandis L.f). Journal of biosciences. 2021, 46, 43. [Google Scholar] [CrossRef]
- Oldenburg, D.J.; Bendich, A.J. The linear plastid chromosomes of maize: terminal sequences, structures, and implications for DNA replication. Current Genetics. 2016, 62, 431–442. [Google Scholar] [CrossRef] [PubMed]
- Vassou, S.L.; Nithaniyal, S.; Raju, B.; Parani, M. Creation of reference DNA barcode library and authentication of medicinal plant raw drugs used in Ayurvedic medicine. BMC Complement Altern Med. 2016, 18 (Suppl. S1), 186. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.V.; Patil, P.G.; Sowjanya, R.P.; Parashuram, S.; Natarajan, P.; Babu, K.D.; Pal, R.K.; Sharma, J.; Reddy, U.K. Chloroplast Genome Sequencing, Comparative Analysis, and Discovery of Unique Cytoplasmic Variants in Pomegranate (Punica granatum L.). Frontiers in genetics. 2021, 12, 704075. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.; Kim, H.W.; Kim, Y.K.; Sohn, J.Y.; Cheon, S.H.; Kim, K.J. The complete plastome of tropical fruit Garcinia mangostana (Clusiaceae). Mitochondrial DNA B Resour. 2017, 2, 722–724. [Google Scholar] [CrossRef]
- NCBI. 2024. Organelle Resources at NCBI. http://www.ncbi.nlm.nih.gov/genome/organelle/ Accessed 29 May 2024.
- Ravikumar, K.; Nooruunisa, S.B.; Ved, D.K.; Bhatt, J.R.; Goraya, G.S. 2018. Compendium of traded Indian Medicinal plants. Founda¬tion for Revitalization of Local Health Traditions (FRLHT), Ben¬galuru, Karnataka, India.
- Jagtap, P.; Bhise, K.; Prakya, V. A phytopharmacological review on Garcinia indica. Int J Herb Med 2015, 3, 2–7. [Google Scholar]
- Lim, S.H.; Lee, H.S.; Lee, C.H.; Choi, C.I. Pharmacological Activity of Garcinia indica (Kokum): An Updated Review. Pharmaceuticals. 2021, 14, 1338. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Ho, P.C.; Wong, F.C.; Sethi, G.; Wang, L.Z.; Goh, B.C. Garcinol: Current status of its anti-oxidative, anti-inflammatory and anti-cancer effects. Cancer letters. 2015, 362, 8–14. [Google Scholar] [CrossRef]
- Aggarwal, V.; Tuli, H.S.; Kaur, J.; Aggarwal, D.; Parashar, G.; Chaturvedi Parashar, N.; Kulkarni, S.; Kaur, G.; Sak, K.; Kumar, M.; Ahn, K.S. Garcinol Exhibits Anti-Neoplastic Effects by Targeting Diverse Oncogenic Factors in Tumor Cells. Biomedicines. 2020, 8, 103. [Google Scholar] [CrossRef] [PubMed]
- Padhye, S.; Ahmad, A.; Oswal, N.; Sarkar, F.H. Emerging role of Garcinol, the antioxidant chalcone from Garcinia indica Choisy and its synthetic analogs. J Hematol Oncol. 2009, 2, 38. [Google Scholar] [CrossRef]
- Ciochina, R.; Grossman, R.B. Polycyclic Polyprenylated Acylphloroglucinols. Chem. Rev. 2006, 106, 3963–3986. [Google Scholar] [CrossRef]
- Antala, B.V.; Patel, M.S.; Bhuva, S.V.; Gupta, S.; Rabadiya, S.; Lahkar, M. Protective effect of methanolic extract of Garcinia indica fruits in 6-OHDA rat model of Parkinson’s disease. Indian journal of pharmacology. 2012, 44, 683–687. [Google Scholar] [CrossRef]
- Vassou, S.L.; Kusuma, G.; Parani, M. DNA barcoding for spe¬cies identification from dried and powdered plant parts: a case study with authentication of the raw drug market samples of Sida cordifolia. Gene. 2015, 559, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Shanmughanandhan, D.; Ragupathy, S.; Newmaster, S.G.; Mohana¬sundaram, S.; Sathishkumar, R. Estimating Herbal Product Authentication and Adulteration in India using a Vouchered, DNA-Based Biological Reference Material Library. Drug Saf. 2016, 39, 1211–1227. [Google Scholar] [CrossRef]
- Nithaniyal, S.; Vassou, S.L.; Poovitha, S.; Balaji, R.; Parani, M. Identification of species adulteration in traded medicinal plant raw drugs using DNA barcoding. Genome. 2017, 60, 139–146. [Google Scholar] [CrossRef]
- Urumarudappa, S.K.J.; Tungphatthong, C.; Sukrong, S. Miti¬gating the impact of admixtures in Thai Herbal products. Front. Pharmacol. 2019, 10, 01205. [Google Scholar] [CrossRef]
- Amritha, N.; Bhooma, V.; Parani, M. Authentication of the market samples of Ashwagandha by DNA barcoding reveals that powders are significantly more adulterated than roots. J. Ethnopharmacol. 2020, 256, 112725. [Google Scholar] [CrossRef] [PubMed]
- Balaji, R.; Parani, M. DNA barcoding of the market samples of single-drug Herbal Powders reveals adulteration with Taxo¬nomically unrelated plant species. Diversity. 2022, 14, 495. [Google Scholar] [CrossRef]
- Seethapathy, G.S.; Tadesse, M.; Urumarudappa, S.K.J.; Gunaga, V.S.; Vasudeva, R.; Malterud, K.E.; Shaanker, R.U.; de Boer, H.J.; Ravikanth, G.; Wangensteen, H. Authentication of Garcinia fruits and food supplements using DNA barcoding and NMR spectroscopy. Scientific reports. 2018, 8, 10561. [Google Scholar] [CrossRef] [PubMed]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017, 45, e18. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.J.; Yu, W.B.; Yang, J.B.; Song, Y.; dePamphilis, C.W.; Yi, T.S.; Li, D.Z. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef] [PubMed]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq—versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Poczai, P.; Hyvönen, J.; Tang, J.; Amiryousefi, A. Chloroplot: An Online Program for the Versatile Plotting of Organelle Genomes. Front. Genet. 2020, 11, 576124. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Ni, Y.; Li, J.; Zhang, X.; Yang, H.; Chen, H.; Liu, C. CPGView: A package for visualizing detailed chloroplast genome structures. Molecular Ecology Resources. 2023, 00, 1–11. [Google Scholar] [CrossRef]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: a web server for microsatellite prediction. Bioinformatics, 2017, 33, 25832585. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S. E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Molecular biology and evolution. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Amiryousefi, A.; Hyvönen, J.; Poczai, P. IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics. 2018. 34, 3030–3031. [CrossRef]
- Biying, Y.; Jipu, S. The complete chloroplast genome sequence of Garcinia anomala (Clusiaceae) from Yunnan Province, China. Mitochondrial DNA Part B, 2021, 6, 7–1899. [Google Scholar] [CrossRef]
- Xiang, M.; Weifang, C.; Liang, T. The complete chloroplast genome sequence of Garcinia oblongifolia (Clusiaceae). Mitochondrial DNA Part B. 2020, 5, 3206–3207. [Google Scholar] [CrossRef]
- Dejun, Y.; Qiong, Q.; Linhong, X.; Yumei, X.; Yi, W. The complete chloroplast genome sequence of Garcinia pedunculata. Mitochondrial DNA Part B. 2020, 5, 220–221. [Google Scholar] [CrossRef]
- Pei-Dong, C.; Da-Juan, C.; Xiu-Rong, K.; Zhi-Xin, Z.; Hua-Feng, W. The complete plastome of Garcinia subelliptica, Merr. 1909 (Clusiaceae). Mitochondrial DNA Part B. 2022, 7, 331–332. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple sequence Alignment Software Version 7: improvements in performance and usability. Mol Biol Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11, Molecular Biology and Evolution. 2021, 38, 3022–3027. [CrossRef]
- Downie, S.R.; Palmer, J.D. 1992. Use of Chloroplast DNA Rearrangements in Reconstructing Plant Phylogeny. In: Soltis, P.S., Soltis, D.E., Doyle, J.J. (eds) Molecular Systematics of Plants. Springer, Boston, MA. [CrossRef]
- Downie, S.R.; Jansen, R.K. A comparative analysis of whole plastid genomes from the Apiales: Expansion and contraction of the inverted repeat, mitochondrial to plastid transfer of DNA, and identification of highly divergent noncoding regions. Syst. Bot. 2015, 40, 336–351. [Google Scholar] [CrossRef]
- Cheon, S-H.; Woo, M-A.; Jo, S.; Kim, Y-K.; Kim, K-J. The Chloroplast Phylogenomics and Systematics of Zoysia (Poaceae). Plants. 2021, 10, 1517. [CrossRef]
- Wang, R.J.; Cheng, C.L.; Chang, C.C.; Wu, C.L.; Su, T.M. , Chaw, S.M. Dynamics and evolution of the inverted repeat-large single copy junctions in the chloroplast genomes of monocots. BMC Evol Biol. 2008, 8, 36. [Google Scholar] [CrossRef]
- Hoch, B.; Maier, R.; Appel, K.; Igloi, G.L.; Kossel, H. Editing of a chloroplast mRNA by creation of an initiation codon. Nature 1991, 353, 178–180. [Google Scholar] [CrossRef]
- Kuroda, H.; Suzuki, H.; Kusumegi, T.; Hirose, T.; Yukawa, Y.; Sugiura, M. Translation of psbC mRNAs starts from the downstream GUG, not the upstream AUG, and requires the extended Shine-Dalgarno sequence in tobacco chloroplasts. Plant & cell physiology. 2007, 48, 1374–1378. [Google Scholar] [CrossRef]
- Po, L.Q.; Zhong, X.Q. Codon usage in the chloroplast genome of rice (Oryza sativa L. ssp. japonica). Acta Agron. Sin. 2004, 30, 1220–1224. [Google Scholar]
- Xue, Y.; Liu, R.; Xue, J.; Wang, S.; Zhang, X. Genetic diversity and relatedness analysis of nine wild species of tree peony based on simple sequence repeats markers. Hortic. Plant J. 2021, 7, 579–588. [Google Scholar] [CrossRef]
- Somaratne, Y.; Guan, D.L.; Wang, W.Q.; Zhao, L.; Xu, S.Q. Complete chloroplast genome sequence of Xanthium sibiricum provides useful DNA barcodes for future species identification and phylogeny. Plant Syst Evol. 2019, 305, 949–960. [Google Scholar] [CrossRef]
- Akhunov, E.D.; Akhunova, A.R.; Anderson, O.D.; et al. Nucleotide diversity maps reveal variation in diversity among wheat genomes and chromosomes. BMC Genomics. 2010, 11, 702. [Google Scholar] [CrossRef] [PubMed]
- Anerao, J.; Jha, V.; Shaikh, N.; et al. DNA barcoding of important fruit tree species of agronomic interest in the genus Garcinia L. from the Western Ghats. Genet. Resour. Crop Evol. 2021, 68, 3161–3177. [Google Scholar] [CrossRef]
- Balaji, R.; Parani, M. Development of an allele-specific PCR (AS-PCR) method for identifying high-methyl eugenol-containing purple Tulsi (Ocimum tenuiflorum L.) in market samples. Mol Biol Rep. 2024, 51, 439. [Google Scholar] [CrossRef]
- Shaibi, M.; Balaji, R.; Parani, M. Molecular differentiation of the green and purple Tulsi (Ocimum tenuiflorum L.) and its application in authentication of market samples. J. Plant Biochem. Biotechnol. 2024, 33, 265–269. [Google Scholar] [CrossRef]
| Amino acid | Codon | Count | RSCU | Amino acid residue (%) | Amino acid | Codon | Count | RSCU | Amino acid residue (%) |
|---|---|---|---|---|---|---|---|---|---|
| Ala | GCA(A) | 4.5 | 1.1 | 4.49 | Ile | AUA(I) | 10 | 0.95 | 8.63 |
| Ala | GCC(A) | 2.7 | 0.65 | Ile | AUC(I) | 5.9 | 0.56 | ||
| Ala | GCG(A) | 1.8 | 0.45 | Ile | AUU(I) | 15.6 | 1.49 | ||
| Ala | GCU(A) | 7.4 | 1.81 | Lys | AAA(K) | 16.5 | 1.48 | 6.11 | |
| Arg | AGA(R) | 7.5 | 2.03 | 6.14 | Lys | AAG(K) | 5.8 | 0.52 | |
| Arg | AGG(R) | 2.9 | 0.78 | Met | AUG(M) | 7.8 | 1 | 2.14 | |
| Arg | CGA(R) | 4.8 | 1.29 | Phe | UUC(F) | 7.6 | 0.67 | 6.19 | |
| Arg | CGC(R) | 1.3 | 0.34 | Phe | UUU(F) | 15 | 1.33 | ||
| Arg | CGG(R) | 1.9 | 0.51 | Pro | CCA(P) | 4 | 1.14 | 3.86 | |
| Arg | CGU(R) | 4 | 1.06 | Pro | CCC(P) | 2.5 | 0.7 | ||
| Asn | AAC(N) | 4.2 | 0.45 | 5.12 | Pro | CCG(P) | 2 | 0.58 | |
| Asn | AAU(N) | 14.5 | 1.55 | Pro | CCU(P) | 5.6 | 1.58 | ||
| Asp | GAC(D) | 3.1 | 0.44 | 3.81 | Ser | AGC(S) | 2.4 | 0.48 | 8.14 |
| Asp | GAU(D) | 10.8 | 1.56 | Ser | AGU(S) | 5.5 | 1.12 | ||
| Cys | UGC(C) | 1.4 | 0.55 | 1.37 | Ser | UCA(S) | 6.1 | 1.23 | |
| Cys | UGU(C) | 3.6 | 1.45 | Ser | UCC(S) | 4.6 | 0.93 | ||
| Gln | CAA(Q) | 9.4 | 1.56 | 3.29 | Ser | UCG(S) | 3.1 | 0.64 | |
| Gln | CAG(Q) | 2.6 | 0.44 | Ser | UCU(S) | 8 | 1.61 | ||
| Glu | GAA(E) | 14.1 | 1.5 | 5.15 | Stop | UAA(*) | 2 | 1.13 | 1.48 |
| Glu | GAG(E) | 4.7 | 0.5 | Stop | UAG(*) | 1.5 | 0.82 | ||
| Gly | GGA(G) | 9.1 | 1.63 | 6.14 | Stop | UGA(*) | 1.9 | 1.05 | |
| Gly | GGC(G) | 2.7 | 0.48 | Thr | ACA(T) | 5.8 | 1.32 | 4.82 | |
| Gly | GGG(G) | 3.8 | 0.68 | Thr | ACC(T) | 2.8 | 0.64 | ||
| Gly | GGU(G) | 6.8 | 1.21 | Thr | ACG(T) | 2 | 0.45 | ||
| His | CAC(H) | 2 | 0.45 | 2.41 | Thr | ACU(T) | 7 | 1.59 | |
| His | CAU(H) | 6.8 | 1.55 | Trp | UGG(W) | 6 | 1 | 1.64 | |
| Leu | CUA(L) | 5.1 | 0.81 | 10.33 | Tyr | UAC(Y) | 2.8 | 0.41 | 3.75 |
| Leu | CUC(L) | 2.5 | 0.4 | Tyr | UAU(Y) | 10.9 | 1.59 | ||
| Leu | CUG(L) | 2.4 | 0.38 | Val | GUA(V) | 6.5 | 1.43 | 4.96 | |
| Leu | CUU(L) | 8.2 | 1.3 | Val | GUC(V) | 2.4 | 0.53 | ||
| Leu | UUA(L) | 11.9 | 1.89 | Val | GUG(V) | 2.6 | 0.58 | ||
| Leu | UUG(L) | 7.6 | 1.21 | Val | GUU(V) | 6.6 | 1.46 |
| SSR motifs | Number of Repeats |
|---|---|
| A | 48 |
| C | 1 |
| T | 48 |
| AT | 3 |
| TA | 5 |
| TTG | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





