Submitted:
09 June 2024
Posted:
11 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Expression Levels of miRNA in GC Patients Based on The Cancer Genome Atlas
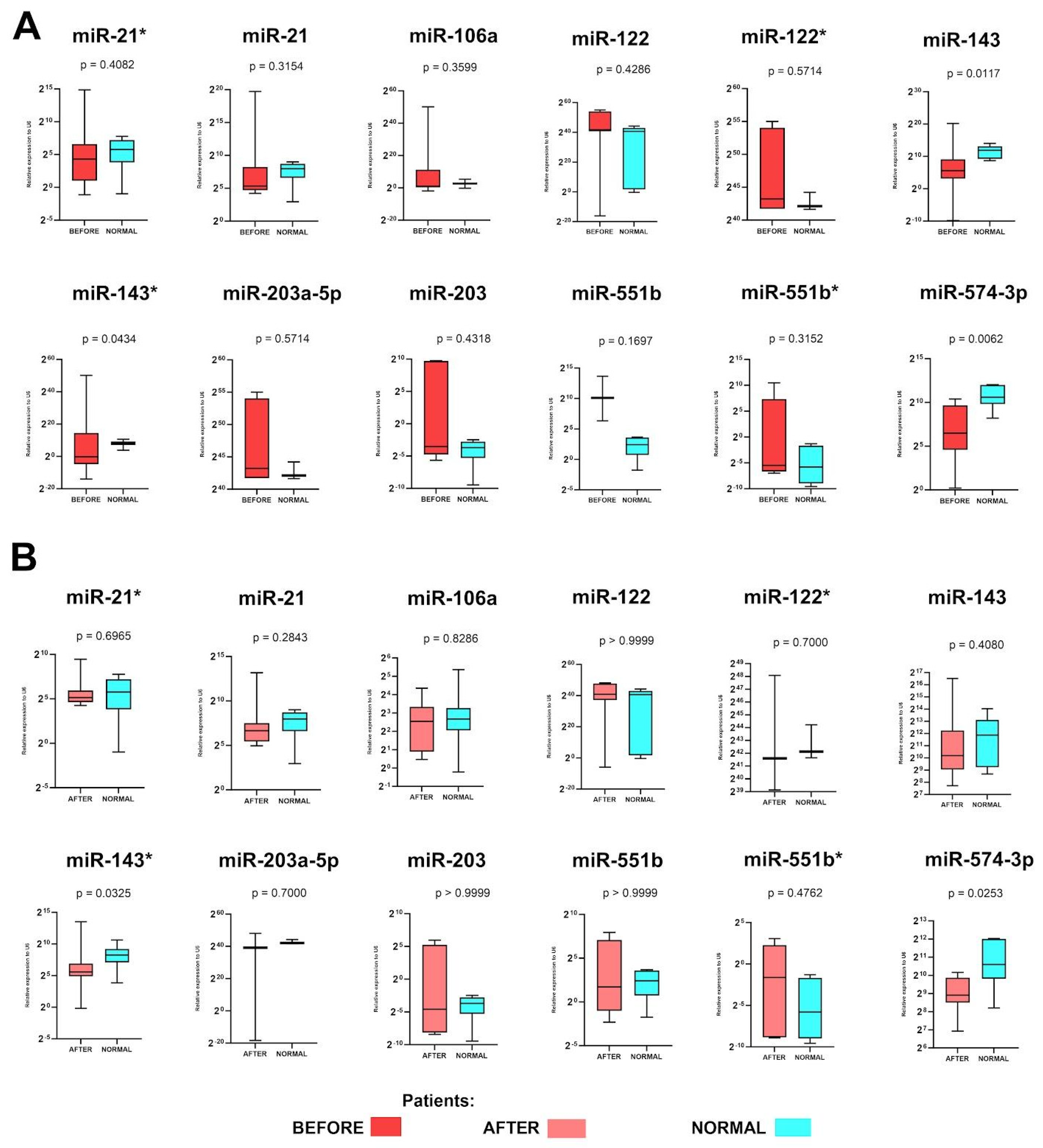
2.2. Expression Levels of miR-143, miR-143* and miR-574-3p before and miR-143* and miR-574-3p after Surgery Are Up-Regulated in GC Patients
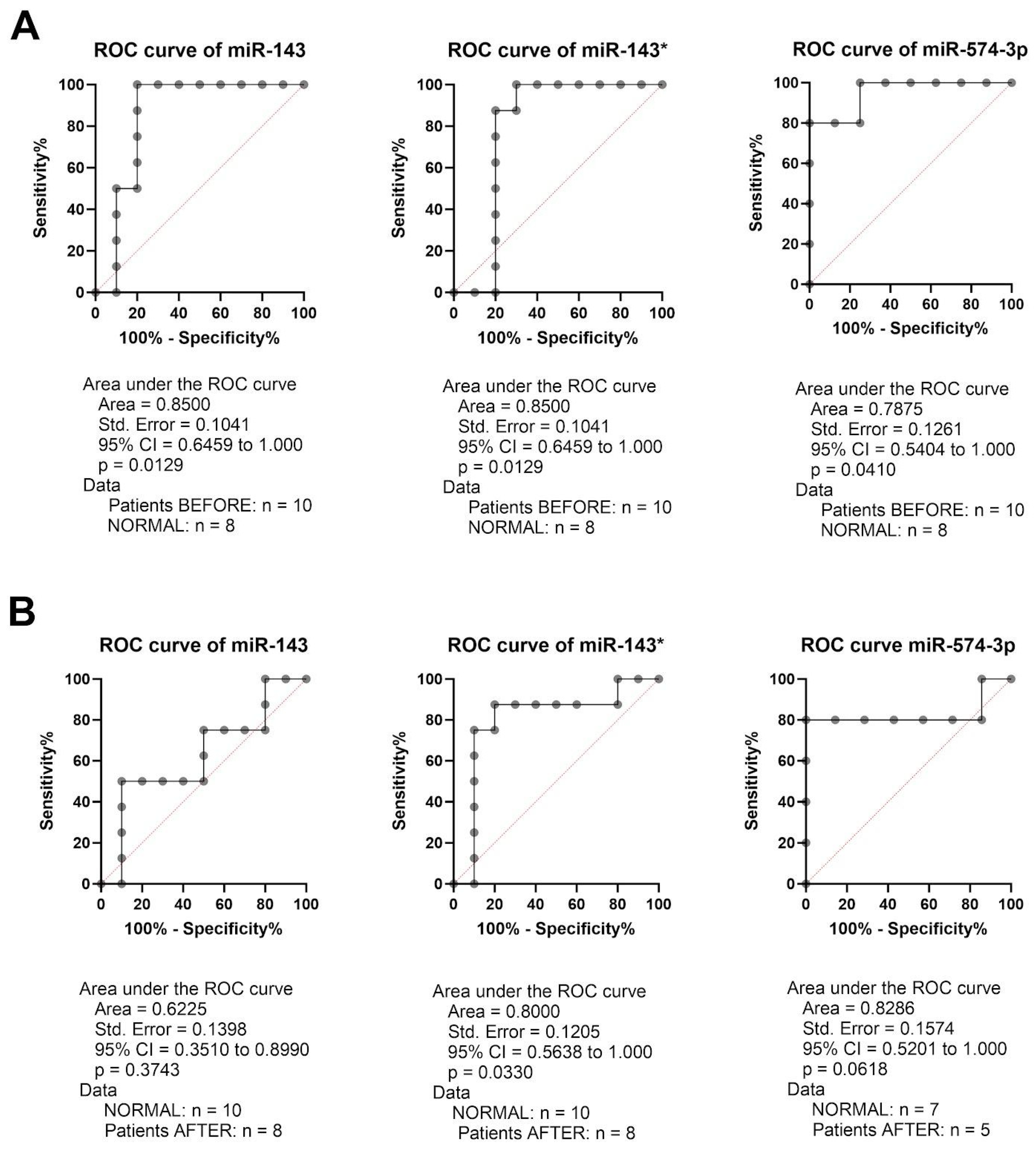
2.3. miR-143, miR-143* and miR-574-3p Have Potential as a Diagnostic Marker in GC Patients
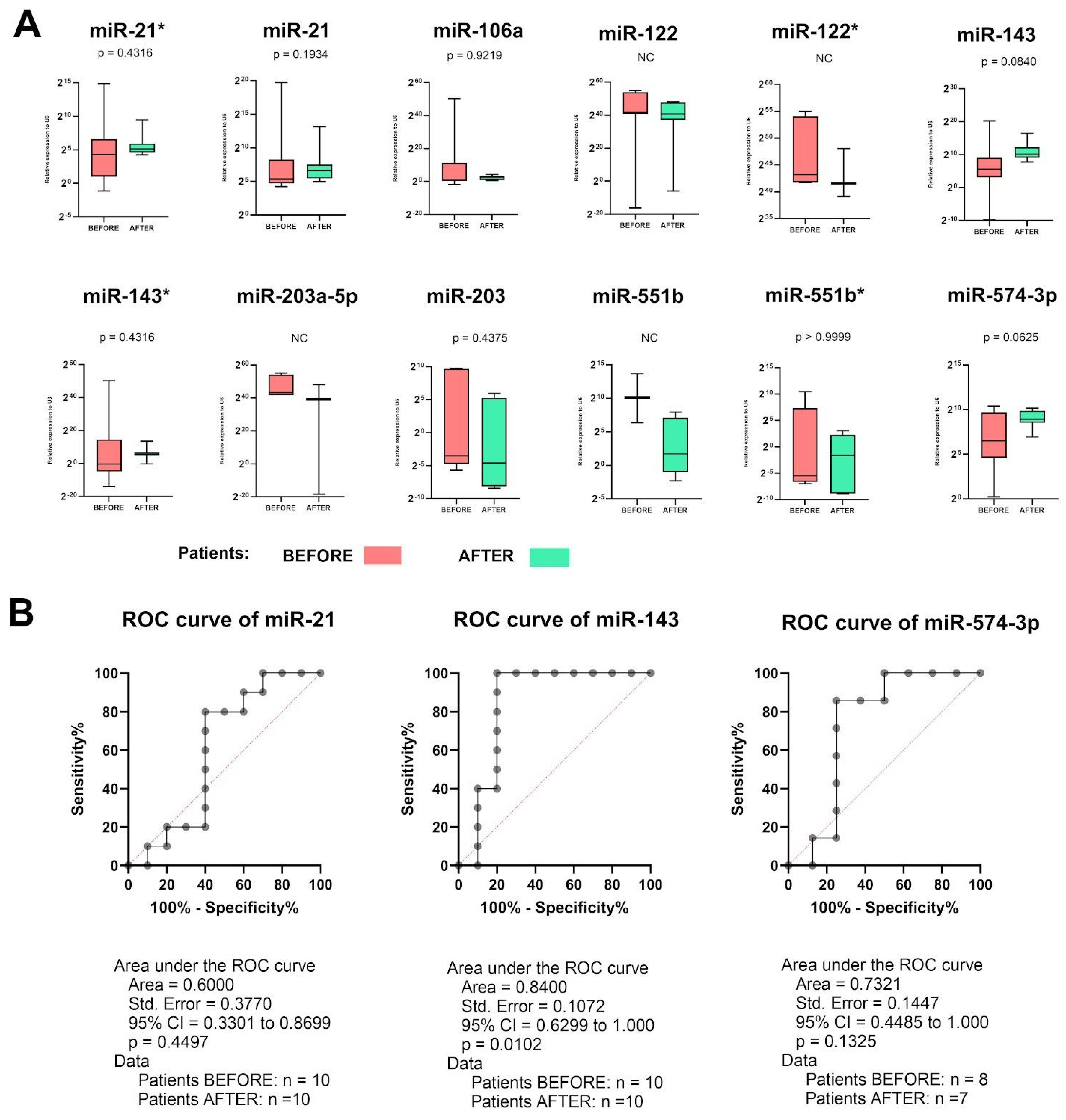
2.4. miR-143 Could Be Used for Assessment of Therapy Response in GC Patients
2.5. miR-551b* Could Be Used for Assessment of Therapy Response in GC Patients—Better Than CT Scan?
3. Discussion
3.1. miRNA-21
3.2. miRNA-106a-5p
3.3. miRNA-122
3.4. miRNA-143
3.5. miRNA-203a
3.6. miRNA-551b
3.7. miRNA-574
4. Materials and Methods
4.1. Patients’ Criteria Included in the Study and Samples Preparation
| Variable | All patients (n = 10) |
|
|---|---|---|
| Age [mean] | 61 years | |
| Sex | Female | 4 (40%) |
| Male | 6 (60%) | |
| Localization | Corpus | 5 (50%) |
| Cardia | 5 (50%) | |
| Grade before chemotherapy | G1 | 1 (10%) |
| G2 | 4 (40%) | |
| G3 | 5 (50%) | |
| Grade after chemotherapy | G1 | 3 (30%) |
| G2 | 1 (10%) | |
| G3 | 3 (30%) | |
| Gx | 5 (50%) | |
| Chemotherapy | FLOT | 6 (60%) |
| FLO | 4 (40%) | |
| Variable | cTNM | ycTNM | ypTNM | |
|---|---|---|---|---|
| Stage | T0 T1 |
0 0 |
2 (20%) 0 |
5 (50%) 2 (20%) |
| T2 | 1 (10%) | 0 | 0 | |
| T3 | 4 (40%) | 4 (40%) | 3 (30%) | |
| T4 | 5 (50%) | 4 (40%) | 0 | |
| Lymph nodes | N0 | 3 (30%) | 6 (60%) | 7 (70%) |
| N1 | 3 (30%) | 2 (20%) | 2 (20%) | |
| N2 | 3 (30%) | 1 (10%) | 0 | |
| N3 | 1 (10%) | 1 (10%) | 1 (10%) | |
| Metastasis | M0 | 10 (100%) | 10 (100%) | 10 (100%) |
| M1 | 0 | 0 | 0 | |
4.2. Ethical Issues
4.3. Sample Preparation
4.4. Total RNA Isolation
4.5. Assessment of miRNA Expression Levels
4.6. miRNA Calculation
4.7. Databases
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F.; Bsc, M.F.B.; Me, J.F.; Soerjomataram, M.I.; et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Allemani, C.; Matsuda, T.; Di Carlo, V.; Harewood, R.; Matz, M.; Nikšić, M.; Bonaventure, A.; Valkov, M.; Johnson, C.J.; Estève, J.; et al. Global surveillance of trends in cancer survival 2000–14 (CONCORD-3): Analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet 2018, 391, 1023–1075. [Google Scholar] [CrossRef]
- Arnold, M.; Park, J.Y.; Camargo, M.C.; Lunet, N.; Forman, D.; Soerjomataram, I. Is gastric cancer becoming a rare disease? A global assessment of predicted incidence trends to 2035. Gut 2020, 69, 823–829. [Google Scholar] [CrossRef]
- Arnold, M.; Rutherford, M.J.; Bardot, A.; Ferlay, J.; Andersson, T.M.L.; Myklebust, T.Å.; et al. Progress in cancer survival, mortality, and incidence in seven high-income countries 1995–2014 (ICBP SURVMARK-2): A population-based study. Lancet Oncol. 2019, 20, 1493–1505. [Google Scholar] [CrossRef]
- Johnston, F.M.; Beckman, M. Updates on Management of Gastric Cancer. Curr. Oncol. Rep. 2019, 21, 67. [Google Scholar] [CrossRef]
- Kabel, A.M.; Marghalani, A.M.; Bin Salman, T.; Faqeeh, F.J.; Asiri, M.K. Gastric carcinoma: Insights into risk factors, methods of diagnosis, possible lines of management, and the role of primary care. J. Fam. Med. Prim. Care 2020, 9, 2659–2663. [Google Scholar] [CrossRef]
- Roviello, G.; Catalano, M.; D’angelo, A.; Palmieri, V.E. Second line of treatment for HER2-positive gastric cancer: An evolving issue. Rep. Pr. Oncol. Radiother. 2021, 26, 316–317. [Google Scholar] [CrossRef]
- Cunningham, D.; Allum, W.H.; Stenning, S.P.; Thompson, J.N.; Van de Velde, C.J.; Nicolson, M.; Scarffe, J.H.; Lofts, F.J.; Falk, S.J.; Iveson, T.J.; et al. Perioperative Chemotherapy versus Surgery Alone for Resectable Gastroesophageal Cancer. New Engl. J. Med. 2006, 355, 11–20. [Google Scholar] [CrossRef]
- Al-Batran, S.-E.; Homann, N.; Pauligk, C.; Goetze, T.O.; Meiler, J.; Kasper, S.; Kopp, H.-G.; Mayer, F.; Haag, G.M.; Luley, K.; et al. Perioperative chemotherapy with fluorouracil plus leucovorin, oxaliplatin, and docetaxel versus fluorouracil or capecitabine plus cisplatin and epirubicin for locally advanced, resectable gastric or gastro-oesophageal junction adenocarcinoma (FLOT4): A randomised, phase 2/3 trial. Lancet 2019, 393, 1948–1957. [Google Scholar] [CrossRef]
- Al-Batran, S.-E.; Homann, N.; Schmalenberg, H.; Kopp, H.-G.; Haag, G.M.; Luley, K.B.; Schmiegel, W.H.; Folprecht, G.; Probst, S.; Prasnikar, N.; et al. Perioperative chemotherapy with docetaxel, oxaliplatin, and fluorouracil/leucovorin (FLOT) versus epirubicin, cisplatin, and fluorouracil or capecitabine (ECF/ECX) for resectable gastric or gastroesophageal junction (GEJ) adenocarcinoma (FLOT4-AIO): A multicenter, randomized phase 3 trial. J. Clin. Oncol. 2017, 35, 4004. [Google Scholar] [CrossRef]
- Mansouri, H.; Zemni, I.; Achouri, L.; Mahjoub, N.; Ayedi, M.A.; Safta, I.B.; Dhiab, T.B.; Chargui, R. Chemoradiotherapy or chemotherapy as adjuvant treatment for resected gastric cancer: Should we use selection criteria? Reports of Practical Oncology and Radiotherapy 2021, 26, 266–280. [Google Scholar] [CrossRef]
- Duque-Santana, V.; Campos, F.L.; Martín, M.; Pelari, L.; Hernandez, A.; Valero, M.; Galindo, J.; Priego, P.; Cuadrado, M.; Longo, F.; et al. Dose-escalated neoadjuvant chemoradiotherapy for locally advanced oesophageal or oesophagogastric junctional adenocarcinoma. Rep. Pr. Oncol. Radiother. 2022, 27, 500–508. [Google Scholar] [CrossRef]
- van Putten, M.; Lemmens, V.E.P.P.; van Laarhoven, H.W.M.; Pruijt, H.F.M.; Nieuwenhuijzen, G.A.P.; Verhoeven, R.H.A. Poor compliance with perioperative chemotherapy for resectable gastric cancer and its impact on survival. European Journal of Surgical Oncology 2019, 45, 1926–1933. [Google Scholar] [CrossRef]
- Drake, J.A.; Stiles, Z.E.; Tsao, M.W.; Deneve, J.L.; Glazer, E.S.; Yakoub, D.; Grothey, A.; Somer, B.G.; Dickson, P.V. Analysis of the Survival Impact of Postoperative Chemotherapy After Preoperative Chemotherapy and Resection for Gastric Cancer. Ann. Surg. Oncol. 2021, 28, 1417–1427. [Google Scholar] [CrossRef]
- Bukowski, K.; Kciuk, M.; Kontek, R. Mechanisms of Multidrug Resistance in Cancer Chemotherapy. Int. J. Mol. Sci. 2020, 21, 3233. [Google Scholar] [CrossRef]
- Plotnikova, O.; Baranova, A.; Skoblov, M. Comprehensive Analysis of Human microRNA–mRNA Interactome. Front. Genet. 2019, 10, 933. [Google Scholar] [CrossRef]
- Kolenda, T.; Paszkowska, A.; Braska, A.; Kozłowska-Masłoń, J.; Guglas, K.; Poter, P.; Wojtczak, P.; Bliźniak, R.; Lamperska, K.; Teresiak, A. Host gene and its guest: Short story about relation of long-noncoding MIR31HG transcript and microRNA miR-31. Rep. Pr. Oncol. Radiother. 2023, 28, 114–134. [Google Scholar] [CrossRef]
- Guglas, K.; Kozłowska-Masłoń, J.; Kolenda, T.; Paszkowska, A.; Teresiak, A.; Bliźniak, R.; et al. Midsize noncoding RNAs in cancers: A new division that clarifies the world of noncoding RNA or an unnecessary chaos? Reports of Practical Oncology and Radiotherapy, 2022; 27, 1077–1093. [Google Scholar] [CrossRef]
- Kolenda, T.; Guglas, K.; Kopczyńska, M.; Sobocińska, J.; Teresiak, A.; Bliźniak, R.; Lamperska, K. Good or not good: Role of miR-18a in cancer biology. Rep. Pr. Oncol. Radiother. 2020, 25, 808–819. [Google Scholar] [CrossRef]
- Kipkeeva, F.; Muzaffarova, T.; Korotaeva, A.; Nikulin, M.; Grishina, K.; Mansorunov, D.; Apanovich, P.; Karpukhin, A. MicroRNA in Gastric Cancer Development: Mechanisms and Biomarkers. Diagnostics 2020, 10, 891. [Google Scholar] [CrossRef]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Vol. 5, Nature Reviews Genetics. 2004.
- Kozłowska-Masłoń, J.; Guglas, K.; Kolenda, T.; Lamperska, K.; Makałowska, I. miRNA in head and neck squamous cell carcinomas: Promising but still distant future of personalized oncology. Rep. Pr. Oncol. Radiother. 2023, 28, 681–697. [Google Scholar] [CrossRef]
- Kolenda, T.; Guglas, K.; Baranowski, D.; Sobocińska, J.; Kopczyńska, M.; Teresiak, A.; et al. cfRNAs as biomarkers in oncology – still experimental or applied tool for personalized medicine already? Reports of Practical Oncology and Radiotherapy 2020, 25, 783–792. [Google Scholar] [CrossRef]
- Kolenda, T.; Przybyła, W.; Kapałczyńska, M.; Teresiak, A.; Zajączkowska, M.; Bliźniak, R.; Lamperska, K.M. Tumor microenvironment – Unknown niche with powerful therapeutic potential. Rep. Pr. Oncol. Radiother. 2018, 23, 143–153. [Google Scholar] [CrossRef]
- Lamperska, K.M.; Kolenda, T.; Teresiak, A.; Kowalik, A.; Kruszyna-Mochalska, M.; Jackowiak, W.; Bliźniak, R.; Przybyła, W.; Kapałczyńska, M.; Kozlowski, P. Different levels of let-7d expression modulate response of FaDu cells to irradiation and chemotherapeutics. PLoS ONE 2017, 12, e0180265. [Google Scholar] [CrossRef]
- Guglas, K.; Kolenda, T.; Kozłowska-Masłoń, J.; Severino, P.; Teresiak, A.; Bliźniak, R.; Lamperska, K. The Impact of YRNAs on HNSCC and HPV Infection. Biomedicines 2023, 11, 681. [Google Scholar] [CrossRef]
- https://www.thermofisher.com/es/es/home/life-science/pcr/real-time-pcr/real-time-pcr-assays/mirna-ncrna-taqman-assays.html?gclid=Cj0KCQjwsPCyBhD4ARIsAPaaRf2gjNp3MXKK0M3WXnwJtno3nH5JKHS5Dxt7gPsOiNHdVlV9WSGJVtoaAjlnEALw_wcB&ef_id=Cj0KCQjwsPCyBhD4ARIsAPaaRf2gjNp3MXKK0M3WXnwJtno3nH5JKHS5Dxt7gPsOiNHdVlV9WSGJVtoaAjlnEALw_wcB:G:s&s_kwcid=AL!3652!3!606132911486!p!!g!!microrna%20assay!17574808760!139287690658&cid=gsd_pcr_sbu_r03_co_cp1491_pjt9622_gsd00000_0se_gaw_rs_pur_&gad_source=1.
- Kolenda, T.; Śmiełowska, M.I.; Lipowicz, J.; Ostapowicz, J.; Pacześna, P.; Rosochowicz, M.; et al. The RNA world: From experimental laboratory to “in silico” approach. Part 1: User friendly RNA expression databases portals. Reports of Practical Oncology and Radiotherapy 2024, 29, 245–257. [Google Scholar] [CrossRef]
- Kolenda, T.; Poter, P.; Guglas, K.; Kozłowska-Masłoń, J.; Braska, A.; Kazimierczak, U.; Teresiak, A. Biological role and diagnostic utility of ribosomal protein L23a pseudogene 53 in cutaneous melanoma. Rep. Pr. Oncol. Radiother. 2023, 28, 255–270. [Google Scholar] [CrossRef]
- Li, W.; Ng, J.M.K.; Wong, C.C.; Ng, E.K.W.; Yu, J. Molecular alterations of cancer cell and tumour microenvironment in metastatic gastric cancer. Oncogene 2018, 37, 4903–4920. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Q.; Tang, X. Long non-coding RNA XIST contributes into drug resistance of gastric cancer cell. Minerva Medica 2019, 110, 270–272. [Google Scholar] [CrossRef]
- Zeng, Z.; Wang, J.; Zhao, L.; Hu, P.; Zhang, H.; Tang, X.; He, D.; Tang, S.; Zeng, Z. Potential Role of microRNA-21 in the Diagnosis of Gastric Cancer: A Meta-Analysis. PLoS ONE 2013, 8, e73278. [Google Scholar] [CrossRef]
- Chan, S.-H.; Wu, C.-W.; Li, A.F.Y.; Chi, C.-W.; Lin, W.-C. miR-21 microRNA expression in human gastric carcinomas and its clinical association. Anticancer Res. 2008, 28, 907–911. [Google Scholar]
- Meng, L.; Chen, Z.; Jiang, Z.; Huang, T.; Hu, J.; Luo, P.; Zhang, H.; Huang, M.; Huang, L.; Chen, Y.; et al. MiR-122-5p suppresses the proliferation, migration, and invasion of gastric cancer cells by targeting LYN. Acta Biochim. et Biophys. Sin. 2019, 52, 49–57. [Google Scholar] [CrossRef]
- Song, A.; Zhao, L.; Wang, Y.; He, D.; Li, Y. Retracted: Chemoresistance in gastric cancer is attributed to the overexpression of excision repair cross-complementing 1 (ERCC1) caused by microRNA-122 dysregulation. J. Cell. Physiol. 2019, 234, 22485–22492. [Google Scholar] [CrossRef]
- Hosseinahli, N.; Zeinali, T.; Hosseinahli, N.; Karimi, L.; Shanehbandi, D.; Mansoori, B.; Mohammadi, A.; Kazemi, T.; Hajiasgharzadeh, K.; Baradaran, B. Restoration of miRNA-143 Expression Inhibits Growth and Migration of MKN-45 Gastric Cancer Cell Line. Adv. Pharm. Bull. 2022, 12, 183–190. [Google Scholar] [CrossRef]
- Chiang, Y.; Song, Y.; Wang, Z.; Chen, Y.; Yue, Z.; Xu, H.; Xing, C.; Liu, Z. Aberrant Expression of miR-203 and Its Clinical Significance in Gastric and Colorectal Cancers. J. Gastrointest. Surg. 2011, 15, 63–70. [Google Scholar] [CrossRef]
- Xiao, B.; Guo, J.; Miao, Y.; Jiang, Z.; Huan, R.; Zhang, Y.; Li, D.; Zhong, J. Detection of miR-106a in gastric carcinoma and its clinical significance. Clin. Chim. Acta 2008, 400, 97–102. [Google Scholar] [CrossRef]
- Ju, Y.; Choi, G.-E.; Lee, M.W.; Jeong, M.; Kwon, H.; Kim, D.H.; Kim, J.; Jin, H.; Lee, K.E.; Hyun, K.-Y.; et al. Identification of miR-143-3p as a diagnostic biomarker in gastric cancer. BMC Med Genom. 2023, 16, 1–16. [Google Scholar] [CrossRef]
- Zhuang, M.; Shi, Q.; Zhang, X.; Ding, Y.; Shan, L.; Shan, X.; Qian, J.; Zhou, X.; Huang, Z.; Zhu, W.; et al. Involvement of miR-143 in cisplatin resistance of gastric cancer cells via targeting IGF1R and BCL2. Tumor Biol. 2014, 36, 2737–2745. [Google Scholar] [CrossRef]
- Du, F.; Feng, Y.; Fang, J.; Yang, M. MicroRNA-143 enhances chemosensitivity of Quercetin through autophagy inhibition via target GABARAPL1 in gastric cancer cells. Biomed. Pharmacother. 2015, 74, 169–177. [Google Scholar] [CrossRef]
- Wang, F.; Liu, J.; Zou, Y.; Jiao, Y.; Huang, Y.; Fan, L.; Li, X.; Yu, H.; He, C.; Wei, W.; et al. MicroRNA-143-3p, up-regulated in H. pylori-positive gastric cancer, suppresses tumor growth, migration and invasion by directly targeting AKT2. Oncotarget 2017, 8, 28711–28724. [Google Scholar] [CrossRef]
- Wang, Z.; Zhao, Z.; Yang, Y.; Luo, M.; Zhang, M.; Wang, X.; Liu, L.; Hou, N.; Guo, Q.; Song, T.; et al. MiR-99b-5p and miR-203a-3p Function as Tumor Suppressors by Targeting IGF-1R in Gastric Cancer. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef]
- Jiang, Y.; Jin, S.; Tan, S.; Shen, Q.; Xue, Y. MiR-203 acts as a radiosensitizer of gastric cancer cells by directly targeting ZEB1. OncoTargets Ther. 2019, 12, 6093–6104. [Google Scholar] [CrossRef]
- Guo, X.; Wang, X.; Li, S.; Lu, Y.; Xie, T.; Qiu, Z.; et al. LncRNA-GC1 contributes to gastric cancer chemo-resistance through inhibition of miR-551b-3p and the overexpression of dysbindin. Annals of Oncology 2019, 30, v8. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, R.; Xu, R.; Shang, J.; He, H.; Yang, Q. MicroRNA-574-5p in gastric cancer cells promotes angiogenesis by targeting protein tyrosine phosphatase non-receptor type 3 (PTPN3). Gene 2020, 733, 144383. [Google Scholar] [CrossRef]
- Bruick, R.K.; McKnight, S.L. A Conserved Family of Prolyl-4-Hydroxylases That Modify HIF. Science 2001, 294, 1337–1340. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, R.; Zhang, S.; Xu, R.; Yang, Q. MicroRNA-574-3p regulates epithelial mesenchymal transition and cisplatin resistance via targeting ZEB1 in human gastric carcinoma cells. Gene 2019, 700, 110–119. [Google Scholar] [CrossRef]
- Xu, X.; Yang, X.; Xing, C.; Zhang, S.; Cao, J. miRNA: The nemesis of gastric cancer (Review). Oncol. Lett. 2013, 6, 631–641. [Google Scholar] [CrossRef]
- Ji, Z.; Wang, X.; Liu, Y.; Zhong, M.; Sun, J.; Shang, J. MicroRNA-574-3p Regulates HIF-α Isoforms Promoting Gastric Cancer Epithelial-Mesenchymal Transition via Targeting CUL2. Dig. Dis. Sci. 2021, 67, 3714–3724. [Google Scholar] [CrossRef]
- Matuszcak, C.; Haier, J.; Hummel, R.; Lindner, K. MicroRNAs: Promising chemoresistance biomarkers in gastric cancer with diagnostic and therapeutic potential. World Journal of Gastroenterology. 2014, 20, 13658–13666. [Google Scholar] [CrossRef]
| miRNA | Cancer samples [n] | Normal samples [n] | Median expression level in cancer samples | Median expression level in normal samples | Fold change | P-value | FDR |
|---|---|---|---|---|---|---|---|
| miRNA-21-3p | 372 | 32 | 3285.16 | 1165.75 | 2.82 | 6.8e-24 | 2.5e-21 |
| miRNA-21-5p | 372 | 32 | 282109.88 | 64062.21 | 4.4 | 1.83e-68 | 4.7e-65 |
| miRNA-106a-5p | 372 | 32 | 18.72 | 7.85 | 2.38 | 0.00099 | 0.0068 |
| miRNA-122-3p | 372 | 32 | 0.21 | 0.01 | 20.55 | 0.25 | 0.6 |
| miRNA-122-5p | 372 | 32 | 17.3 | 0.57 | 29.92 | 0.54 | 0.83 |
| miRNA-143-3p | 372 | 32 | 193518.49 | 443164.16 | 0.44 | 3.6e-10 | 9.3e-9 |
| miRNA-143-5p | 372 | 32 | 121.59 | 229.49 | 0.53 | 0.011 | 0.054 |
| miRNA-203a-3p | 372 | 32 | - | - | - | - | - |
| miRNA-203a-5p | 372 | 32 | - | - | - | - | - |
| miRNA-551b-3p | 372 | 32 | 1.53 | 07.02 | 0.22 | 0.037 | 0.15 |
| miRNA-551b-5p | 372 | 32 | 0.02 | 0.03 | 0.56 | 0.13 | 0.39 |
| miRNA-574-3p | 372 | 32 | 81.06 | 85.97 | 0.94 | 0.2 | 0.54 |
| Parameter | Groups | miRNA-21 | miRNA-106a | miRNA-122 | miRNA-143 | miRNA-203a | miRNA-551b | miRNA-574 |
|---|---|---|---|---|---|---|---|---|
| Sample type | Normal vs Primary | <1E-12 | 0,00008 | 0,14658 | 0,00000 | 0,16578 | 0,00242 | 0,69254 |
| Race | Caucasian vs African American | 0,59606 | 0,65550 | 0,19030 | 0,05878 | 0,66776 | 0,05402 | 0,05402 |
| Caucasian vs Asian | 0,30630 | 0,01604 | 0,17484 | 0,02172 | 0,31662 | 0,63522 | 0,63522 | |
| African American vs Asian | 0,29902 | 0,56788 | 0,62328 | 0,29880 | 0,99920 | 0,13424 | 0,13424 | |
| Gender | Male vs Female | 0,74084 | 0,53014 | 0,24308 | 0,32242 | 0,28556 | 0,56872 | 0,56872 |
| Age | 21-40 vs 41-60 | 0,14104 | 0,04318 | 0,18056 | 0,43436 | 0,22830 | 0,41534 | 0,41534 |
| 21-40 vs 61-80 | 0,25226 | 0,00808 | 0,12376 | 0,76562 | 0,77240 | 0,05157 | 0,05157 | |
| 21-40 vs 81-100 | 0,05878 | 0,34576 | 0,90334 | 0,55958 | 0,13628 | 0,08584 | 0,08584 | |
| 41-60 vs 61-80 | 0,08270 | 0,23986 | 0,20184 | 0,00918 | 0,00672 | 0,93720 | 0,93720 | |
| 41-60 vs 81-100 | 0,49906 | 0,15064 | 0,17968 | 0,56066 | 0,61330 | 0,55180 | 0,55180 | |
| 61-80 vs 81-100 | 0,11083 | 0,01720 | 0,10224 | 0,43432 | 0,06366 | 0,63018 | 0,63018 | |
| Cancer stage | I vs II | 0,15316 | 0,16144 | 0,29706 | 0,47704 | 0,43726 | 0,54456 | 0,54456 |
| I vs III | 0,01124 | 0,60368 | 0,32454 | 0,54468 | 0,05878 | 0,56204 | 0,56204 | |
| I vs IV | 0,23102 | 0,53414 | 0,36276 | 0,40824 | 0,07544 | 0,75392 | 0,75392 | |
| II vs III | 0,15882 | 0,23550 | 0,47678 | 0,87786 | 0,12802 | 0,90778 | 0,90778 | |
| II vs IV | 0,88272 | 0,49128 | 0,25600 | 0,68498 | 0,15774 | 0,28022 | 0,28022 | |
| III vs IV | 0,48102 | 0,81706 | 0,58708 | 0,62610 | 0,89714 | 0,35976 | 0,35976 | |
| Tumor grade | G1 vs G2 | 0,67776 | 0,74114 | 0,30394 | 0,79310 | 0,60826 | 0,22676 | 0,22676 |
| G1 vs G3 | 0,65476 | 0,90388 | 0,09204 | 0,57692 | 0,17936 | 0,13166 | 0,13166 | |
| G2 vs G3 | 0,86524 | 0,34396 | 0,46276 | 0,00182 | 0,02424 | 0,83656 | 0,83656 | |
| Nodal metastasis status | N0 vs N1 | 0,30396 | 0,88494 | 0,27724 | 0,78988 | 0,75946 | 0,83018 | 0,83018 |
| N0 vs N2 | 0,29180 | 0,71396 | 0,29278 | 0,71558 | 0,80560 | 0,72304 | 0,72304 | |
| N0 vs N3 | 0,71348 | 0,49888 | 0,42276 | 0,28858 | 0,00474 | 0,17108 | 0,17108 | |
| N1 vs N2 | 0,96318 | 0,88320 | 0,55314 | 0,56090 | 0,98958 | 0,89646 | 0,89646 | |
| N1 vs N3 | 0,78694 | 0,48110 | 0,35356 | 0,24474 | 0,02912 | 0,15342 | 0,15342 | |
| N2 vs N3 | 0,75010 | 0,39538 | 0,42578 | 0,41616 | 0,09393 | 0,14404 | 0,14404 | |
| Tumor histology | AC NOS vs AC Diffuse | 0,52732 | 0,75522 | 0,22130 | 0,21224 | 0,20154 | 0,13558 | 0,13558 |
| AC NOS vs AC Signet Ring | 0,12324 | 0,13950 | 0,65222 | 0,62216 | 0,00184 | 0,10358 | 0,10358 | |
| AC NOS vs IAC NOS | 0,45426 | 0,55860 | 0,36642 | 0,11122 | 0,66980 | 0,92958 | 0,92958 | |
| AC NOS vs IAC Mucinous | 0,39568 | 0,02212 | 0,28880 | 0,43058 | 0,33528 | 0,12252 | 0,12252 | |
| AC NOS vs IAC Papillary | 0,55152 | 0,84612 | 0,94758 | 0,09892 | 0,27432 | 0,47208 | 0,47208 | |
| AC NOS vs IAC Tubular | 0,09716 | 0,55192 | 0,32894 | 0,00000 | 0,04314 | 0,09158 | 0,09158 | |
| AC Diffuse vs AC Signet Ring | 0,07644 | 0,29140 | 0,33310 | 0,36696 | 0,06108 | 0,53796 | 0,53796 | |
| AC Diffuse vs IAC NOS | 0,21652 | 0,46906 | 0,27970 | 0,02094 | 0,37026 | 0,14580 | 0,14580 | |
| AC Diffuse vs IAC Mucinous | 0,66654 | 0,06158 | 0,30358 | 0,89524 | 0,93058 | 0,78986 | 0,78986 | |
| AC Diffuse vs IAC Papillary | 0,40010 | 0,96520 | 0,16990 | 0,00822 | 0,19376 | 0,90458 | 0,90458 | |
| AC Diffuse vs IAC Tubular | 0,04388 | 0,86194 | 0,31354 | 0,00004 | 0,00562 | 0,78714 | 0,78714 | |
| AC Signet Ring vs IAC NOS | 0,22388 | 0,22494 | 0,52088 | 0,81776 | 0,00520 | 0,11222 | 0,11222 | |
| AC Signet Ring vs IAC Mucinous | 0,10170 | 0,11510 | 0,35920 | 0,42808 | 0,09626 | 0,67820 | 0,67820 | |
| AC Signet Ring vs IAC Papillary | 0,61504 | 0,45914 | 0,56958 | 0,37434 | 0,11228 | 0,62612 | 0,62612 | |
| AC Signet Ring vs IAC Tubular | 0,42724 | 0,38018 | 0,37260 | 0,33454 | 0,00010 | 0,58822 | 0,58822 | |
| IAC NOS vs IAC Mucinous | 0,19918 | 0,17518 | 0,28926 | 0,09030 | 0,56498 | 0,13216 | 0,13216 | |
| IAC NOS vs IAC Papillary | 0,75650 | 0,64798 | 0,35850 | 0,29404 | 0,24576 | 0,63370 | 0,63370 | |
| IAC NOS vs IAC Tubular | 0,41558 | 0,26374 | 0,29396 | 0,02458 | 0,02216 | 0,09702 | 0,09702 | |
| IAC Mucinous vs IAC Papillary | 0,35352 | 0,22480 | 0,23418 | 0,07300 | 0,20372 | 0,78462 | 0,78462 | |
| IAC Mucinous vs IAC Tubular | 0,07514 | 0,00634 | 0,78488 | 0,00846 | 0,01260 | 0,92004 | 0,92004 | |
| IAC Papillary vs IAC Tubular | 0,95976 | 0,97014 | 0,20794 | 0,84260 | 0,55078 | 0,79666 | 0,79666 | |
| TP53 mutation status | Mutant vs Non-mutant | 0,64328 | 0,28594 | 0,21586 | 0,00600 | 0,26196 | 0,35738 | 0,35738 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





