Submitted:
07 June 2024
Posted:
07 June 2024
You are already at the latest version
Abstract
Keywords:
1. Instruction
2. Materials and Methods
2.1. Patients
2.2. Follow Up
2.3. Detection of Preoperative Serum CEA
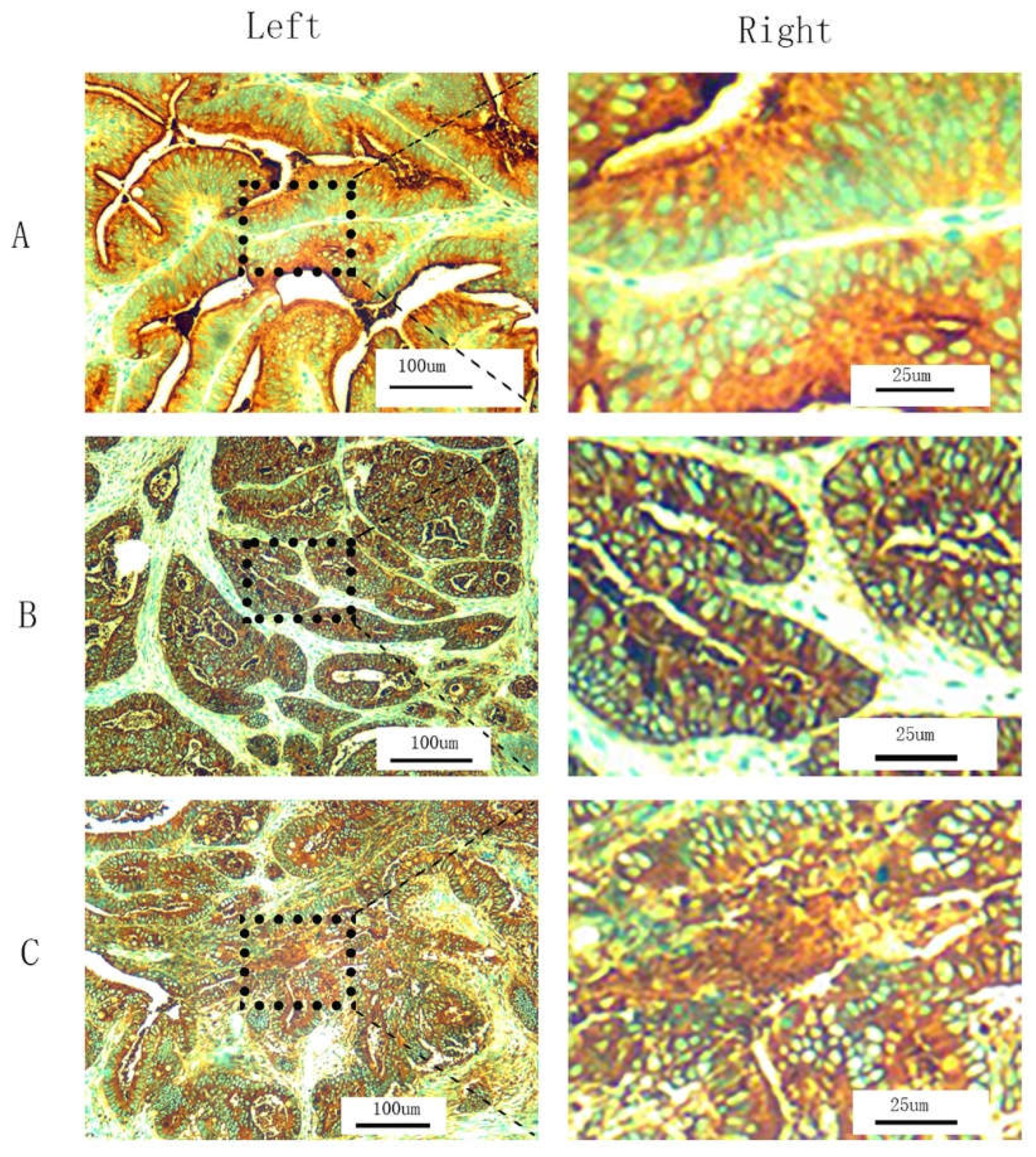
2.4. T-CEA Immunohistochemistry
2.5. Combined CEA Classification
2.6. Receiver Operating Characteristic Curve (ROC) Analysis
2.7. Statistical Analysis
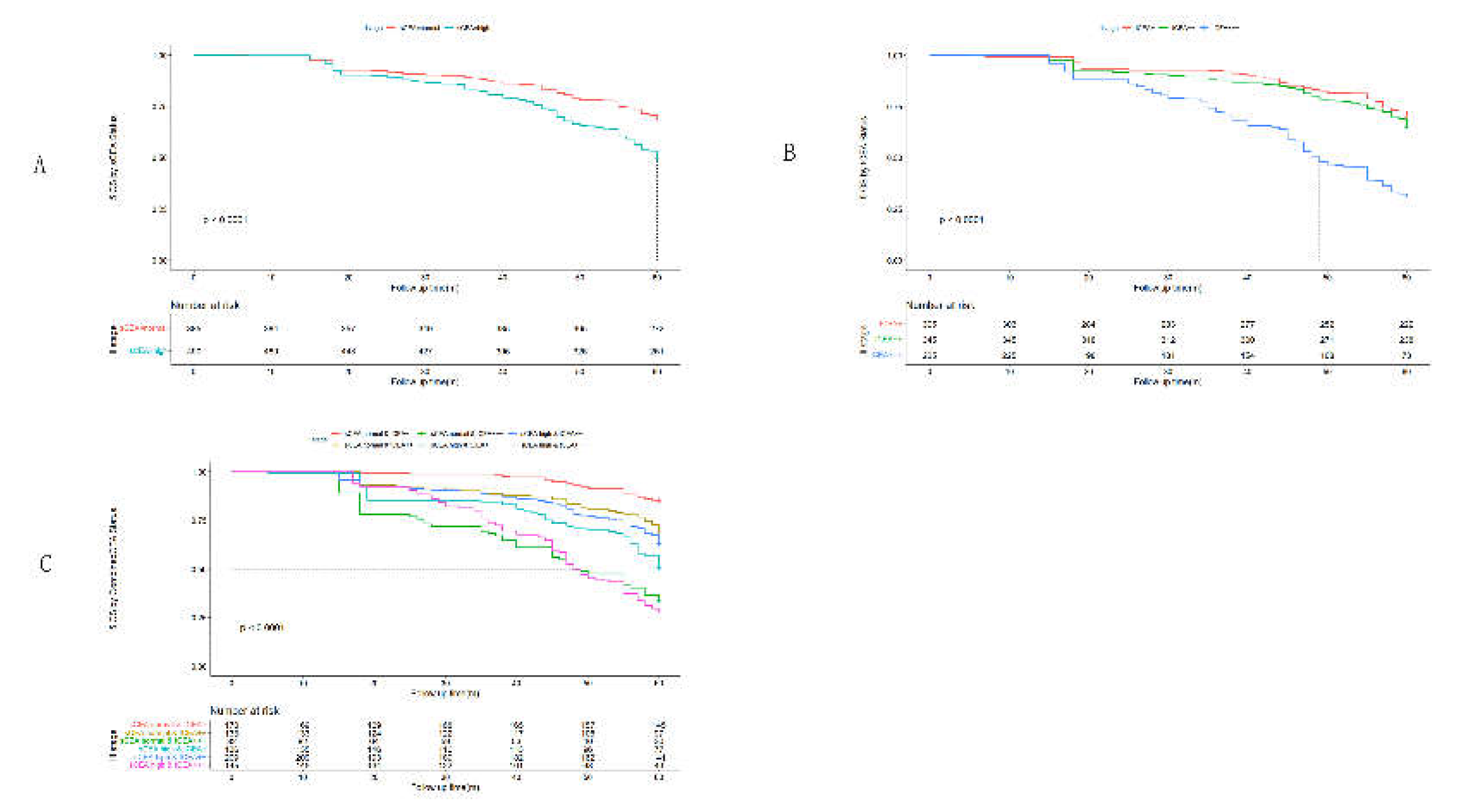
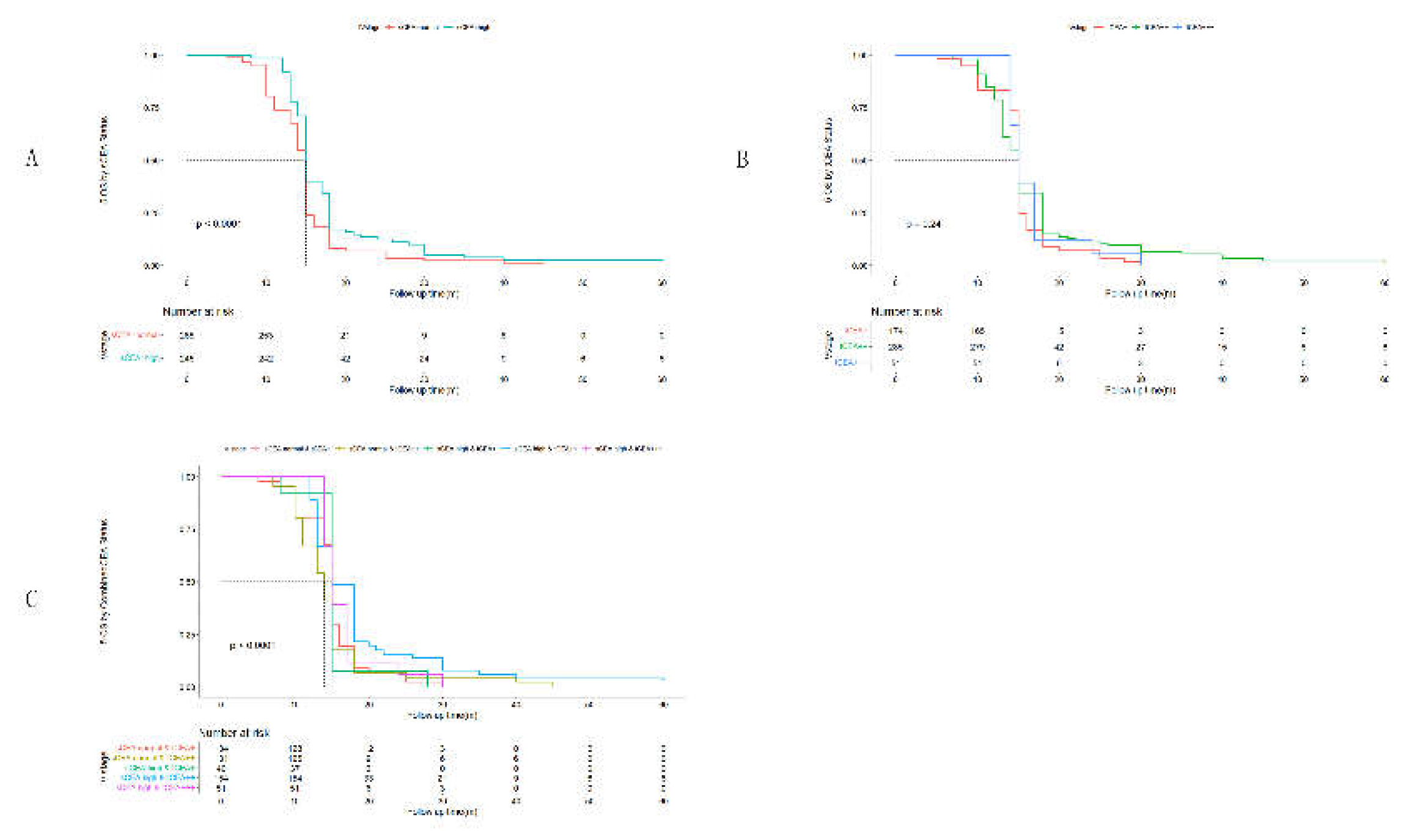
3. Results
3.1. Clinicopathological Features by Combined CEA
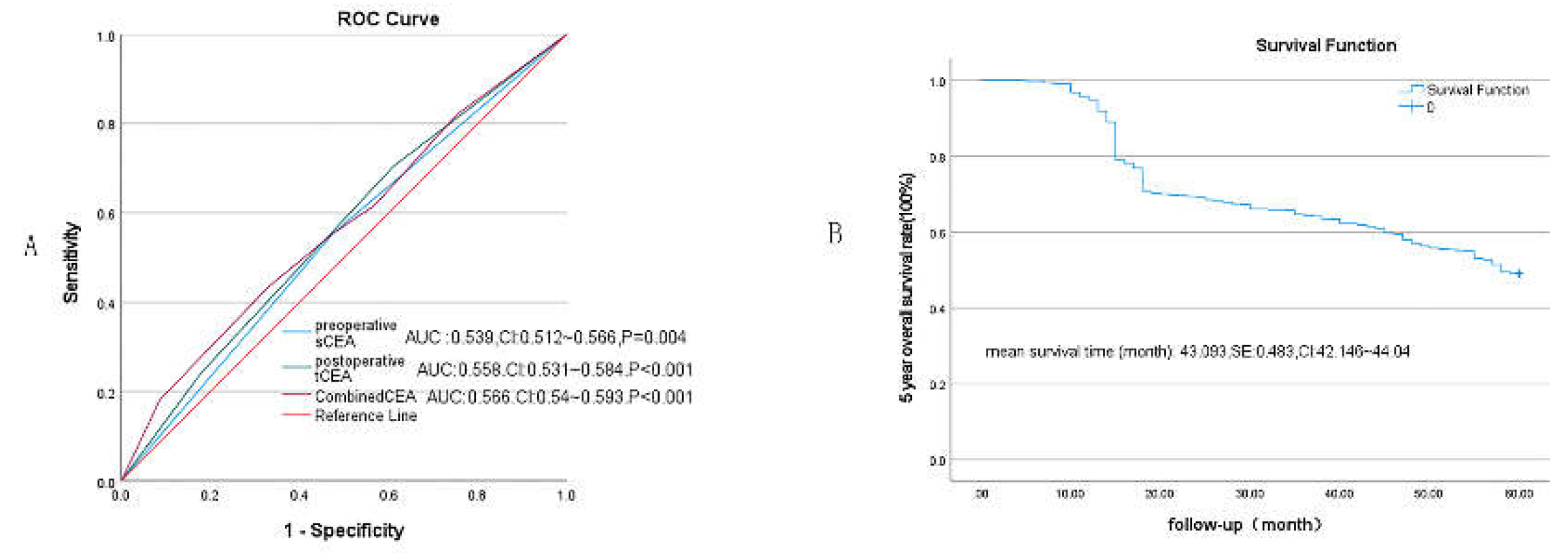
3.2. Receiver Operating Characteristic Curve (ROC) Analysis
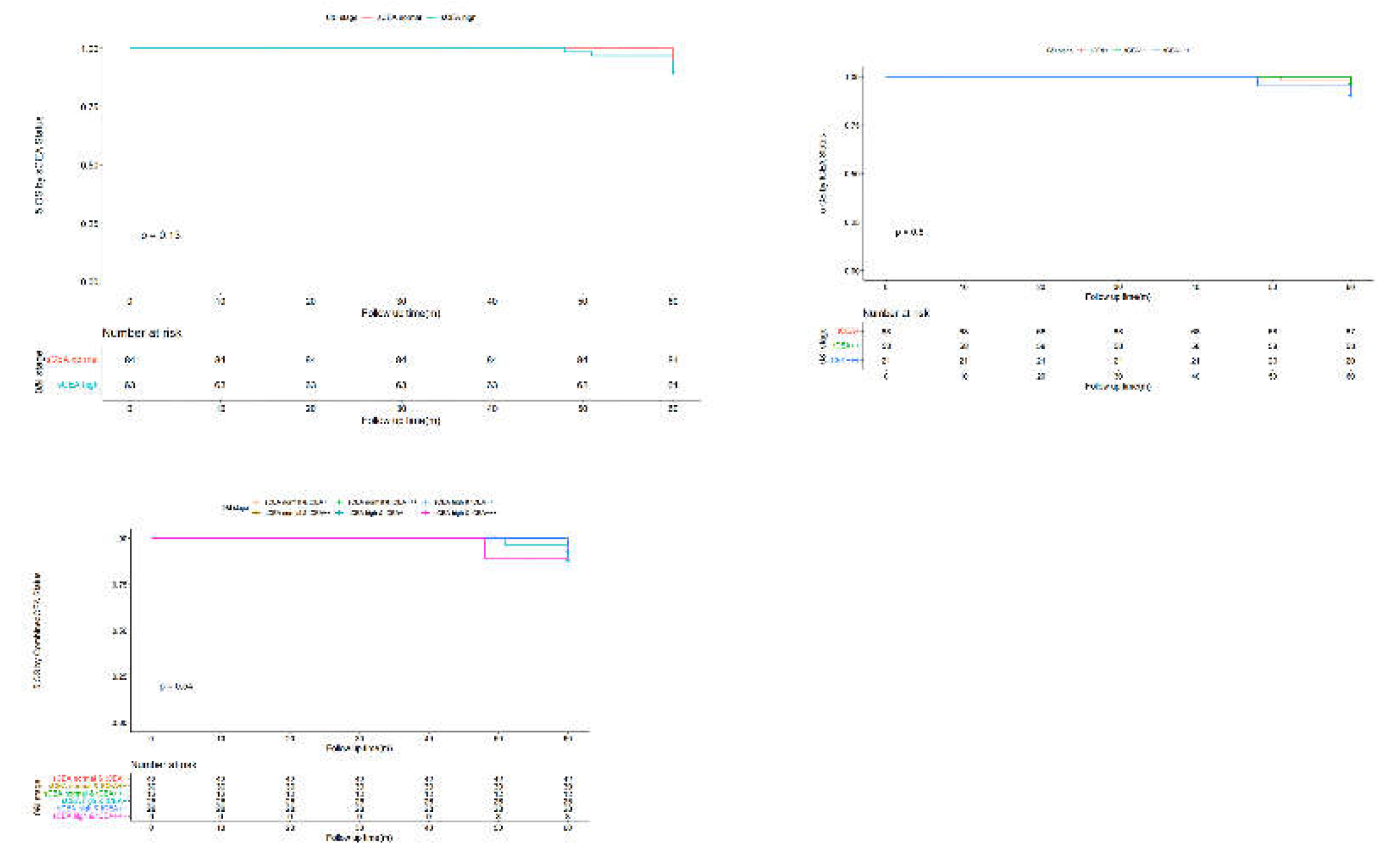
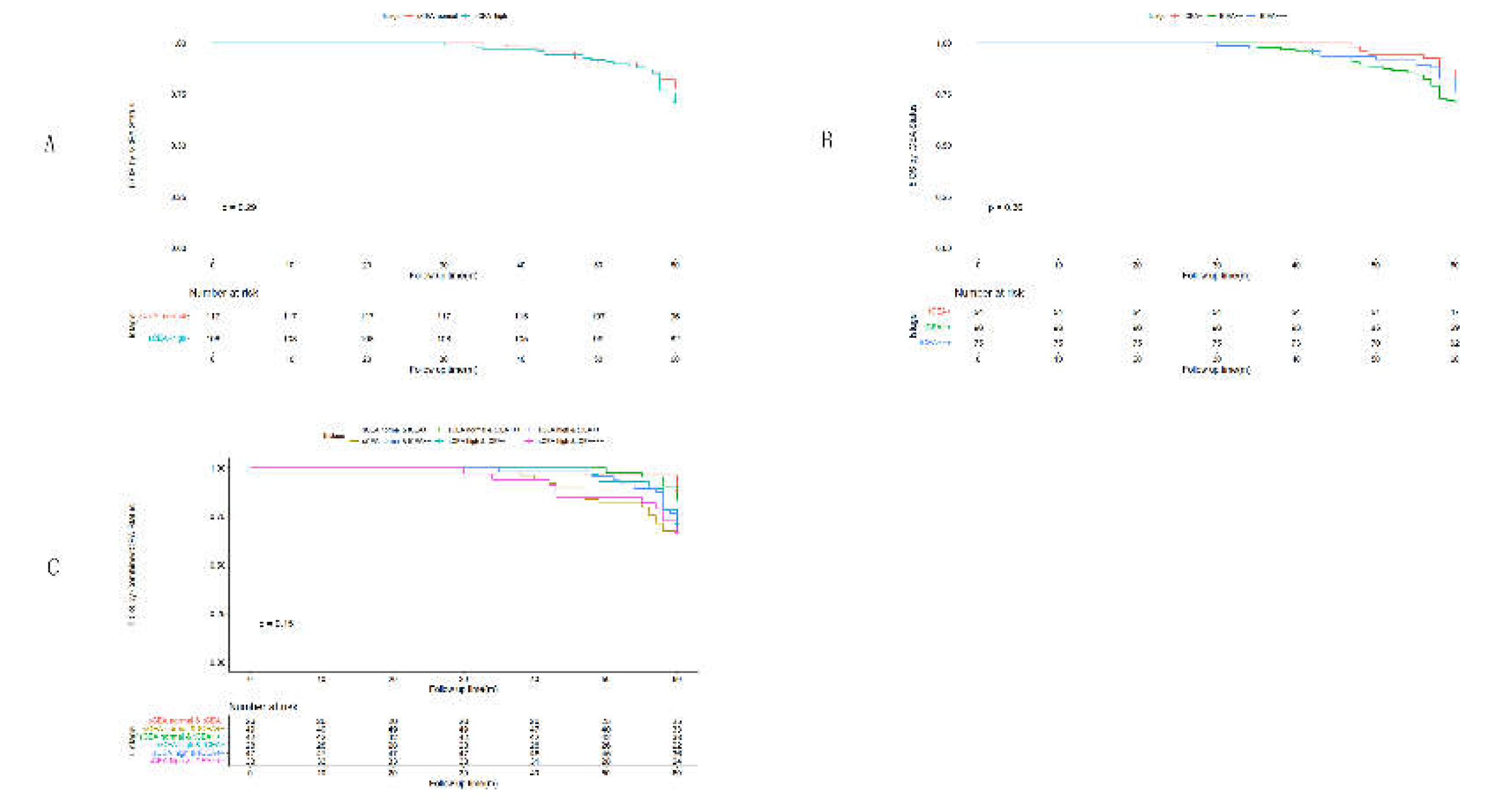
3.4. Univariate Analysis by Cox Regression for Clinicopathological Features
3.5. Multivariate Analysis by Cox Regression for Clinicopathological Features
4. Discussion
5. Conclusion
Funding
Acknowledgments
Authors’ contributions
Ethics statement
Availability of data and materials
Consent for publiciation
Conflicts of Interests
References
- Wang L, Lin S, Yang C, Cai S, Li W: Effect of KRAS mutations and p53 expression on the postoperative prognosis of patients with colorectal cancer. Molecular genetics & genomic medicine 2022, 10(7):e1905.
- Bennedsen ALB, Furbo S, Bjarnsholt T, Raskov H, Gögenur I, Kvich L: The gut microbiota can orchestrate the signaling pathways in colorectal cancer. APMIS : acta pathologica, microbiologica, et immunologica Scandinavica 2022, 130(3):121-139.
- Ren G, Li R, Zheng G, Du K, Dan H, Wu H, Dou X, Duan L, Xie Z, Niu L et al: Prognostic value of normal levels of preoperative tumor markers in colorectal cancer. Scientific reports 2023, 13(1):22830.
- Goldstein MJ, Mitchell EP: Carcinoembryonic antigen in the staging and follow-up of patients with colorectal cancer. Cancer investigation 2005, 23(4):338-351.
- Li C, Zhang D, Pang X, Pu H, Lei M, Fan B, Lv J, You D, Li Z, Zhang T: Trajectories of Perioperative Serum Tumor Markers and Colorectal Cancer Outcomes: A Retrospective, Multicenter Longitudinal Cohort Study. EBioMedicine 2021, 74:103706.
- Li X, Stassen L, Schrotz-King P, Zhao Z, Cardoso R, Raut JR, Bhardwaj M, Brenner H: Potential of Fecal Carcinoembryonic Antigen for Noninvasive Detection of Colorectal Cancer: A Systematic Review. Cancers (Basel) 2023, 15(23).
- Cao H, Zhu L, Li L, Wang W, Niu X: Serum CA724 has no diagnostic value for gastrointestinal tumors. Clinical and experimental medicine 2023, 23(6):2433-2442.
- Tong G, Xu W, Zhang G, Liu J, Zheng Z, Chen Y, Niu P, Xu X: The role of tissue and serum carcinoembryonic antigen in stages I to III of colorectal cancer-A retrospective cohort study. Cancer medicine 2018, 7(11):5327-5338.
- Ma R, Gong M, Sun T, Su L, Li K: The prognostic role of γδ T cells in colorectal cancer based on nomogram. European journal of medical research 2023, 28(1):467.
- Aldilaijan AF, Kim YI, Kim CW, Yoon YS, Park IJ, Lim SB, Kim J, Ro JS, Kim JC: Clinical implication of tissue carcinoembryonic antigen expression in association with serum carcinoembryonic antigen in colorectal cancer. Scientific reports 2023, 13(1):7616.
- Zhang Z, Liu X, Yang X, Jiang Y, Li A, Cong J, Li Y, Xie Q, Xu C, Liu D: Identification of faecal extracellular vesicles as novel biomarkers for the non-invasive diagnosis and prognosis of colorectal cancer. Journal of extracellular vesicles 2023, 12(1):e12300.
- Ma L, Li W, Liu N, Ding Z, Cai J, Zhang Y: Prothrombin time (PT) and CEA as prognostic predictive biomarkers for postoperative recurrence after curative resection in patients with stage I-III colorectal cancer: a retrospective cohort study. Updates in surgery 2022, 74(3):999-1009.
- Tiernan JP, Perry SL, Verghese ET, West NP, Yeluri S, Jayne DG, Hughes TA: Carcinoembryonic antigen is the preferred biomarker for in vivo colorectal cancer targeting. Br J Cancer 2013, 108(3):662-667.
- Tormo BR, Gavilondo JV, Dominguez C, Freyre M, Rodriguez T, Biberfeld P: CEA in colonic adenocarcinomas and precancerous lesions. An immunohistochemical study with a novel monoclonal antibody. APMIS : acta pathologica, microbiologica, et immunologica Scandinavica 1989, 97(12):1073-1080.
- Polivka J, Windrichova J, Pesta M, Houfkova K, Rezackova H, Macanova T, Vycital O, Kucera R, Slouka D, Topolcan O: The Level of Preoperative Plasma KRAS Mutations and CEA Predict Survival of Patients Undergoing Surgery for Colorectal Cancer Liver Metastases. Cancers (Basel) 2020, 12(9).
- Rao H, Wu H, Huang Q, Yu Z, Zhong Z: Clinical Value of Serum CEA, CA24-2 and CA19-9 in Patients with Colorectal Cancer. Clinical laboratory 2021, 67(4).
- Kuang J, Gong Y, Xie H, Yan L, Huang S, Gao F, Tang S, Gan J: The prognostic value of preoperative serum CA724 for CEA-normal colorectal cancer patients. PeerJ 2020, 8:e8936.
- Kemper M, Hentschel W, Graß JK, Stüben BO, Konczalla L, Rawnaq T, Ghadban T, Izbicki JR, Reeh M: Serum Midkine is a clinical significant biomarker for colorectal cancer and associated with poor survival. Cancer Med 2020, 9(6):2010-2018.
- Li Z, Zhu H, Pang X, Mao Y, Yi X, Li C, Lei M, Cheng X, Liang L, Wu J et al: Preoperative serum CA19-9 should be routinely measured in the colorectal patients with preoperative normal serum CEA: a multicenter retrospective cohort study. BMC Cancer 2022, 22(1):962.
- Tong G, Zhang G, Hu Y, Xu X, Wang Y: Correlation between mismatch repair statuses and the prognosis of stage I–IV colorectal cancer. Frontiers in oncology 2024, 13.
- Zhang G, He F, Zhao G, Huang Z, Li X, Xia X, Guo Y, Xu W, Xiong S, Ma Y et al: Combining Serum DNA Methylation Biomarkers and Protein Tumor Markers Improved Clinical Sensitivity for Early Detection of Colorectal Cancer. International journal of genomics 2021, 2021:6613987.
- Huang SC, Chang SC, Liao TT, Yang MH: Detection and clinical significance of CEACAM5 methylation in colorectal cancer patients. Cancer Sci 2024, 115(1):270-282.
- Gold P, Freedman SO: DEMONSTRATION OF TUMOR-SPECIFIC ANTIGENS IN HUMAN COLONIC CARCINOMATA BY IMMUNOLOGICAL TOLERANCE AND ABSORPTION TECHNIQUES. The Journal of experimental medicine 1965, 121(3):439-462.
- Duffy MJ: Carcinoembryonic antigen as a marker for colorectal cancer: is it clinically useful? Clinical chemistry 2001, 47(4):624-630.
- Hammarström S: The carcinoembryonic antigen (CEA) family: structures, suggested functions and expression in normal and malignant tissues. Seminars in cancer biology 1999, 9(2):67-81.
- Kim JC, Roh SA, Lee KH, Namgung H, Kim JR, Kim JS: Genetic and pathologic changes associated with lymphovascular invasion of colorectal adenocarcinoma. Clinical & experimental metastasis 2005, 22(5):421-428.
- Hardiman KM, Felder SI, Friedman G, Migaly J, Paquette IM, Feingold DL: The American Society of Colon and Rectal Surgeons Clinical Practice Guidelines for the Surveillance and Survivorship Care of Patients After Curative Treatment of Colon and Rectal Cancer. Diseases of the colon and rectum 2021, 64(5):517-533.
- Nazato DM, Matos LL, Waisberg DR, Souza JR, Martins LC, Waisberg J: Prognostic value of carcinoembryonic antigen distribution in tumor tissue of colorectal carcinoma. Arquivos de gastroenterologia 2009, 46(1):26-31.
- Saito G, Sadahiro S, Okada K, Tanaka A, Suzuki T, Kamijo A: Relation between Carcinoembryonic Antigen Levels in Colon Cancer Tissue and Serum Carcinoembryonic Antigen Levels at Initial Surgery and Recurrence. Oncology 2016, 91(2):85-89.
- Park JW, Chang HJ, Kim BC, Yeo HY, Kim DY: Clinical validity of tissue carcinoembryonic antigen expression as ancillary to serum carcinoembryonic antigen concentration in patients curatively resected for colorectal cancer. Colorectal disease : the official journal of the Association of Coloproctology of Great Britain and Ireland 2013, 15(9):e503-511.
- Becerra AZ, Probst CP, Tejani MA, Aquina CT, González MG, Hensley BJ, Noyes K, Monson JR, Fleming FJ: Evaluating the Prognostic Role of Elevated Preoperative Carcinoembryonic Antigen Levels in Colon Cancer Patients: Results from the National Cancer Database. Ann Surg Oncol 2016, 23(5):1554-1561.
- Kim CW, Yoon YS, Park IJ, Lim SB, Yu CS, Kim JC: Elevation of preoperative s-CEA concentration in stage IIA colorectal cancer can also be a high risk factor for stage II patients. Ann Surg Oncol 2013, 20(9):2914-2920.
- Kankanala VL, Mukkamalla SKR: Carcinoembryonic Antigen. In: StatPearls. edn. Treasure Island (FL) ineligible companies. Disclosure: Shiva Kumar Mukkamalla declares no relevant financial relationships with ineligible companies.: StatPearls Publishing Copyright © 2024, StatPearls Publishing LLC.; 2024.
- Hou S, Jing J, Wang Y, Du L, Tian B, Xu X, Sun T, Shi Y: Evaluation of Clinical Diagnostic and Prognostic Value of Preoperative Serum Carcinoembryonic Antigen, CA19-9, and CA24-2 for Colorectal Cancer. Alternative therapies in health and medicine 2023, 29(6):192-197.
- Björkman K, Jalkanen S, Salmi M, Mustonen H, Kaprio T, Kekki H, Pettersson K, Böckelman C, Haglund C: A prognostic model for colorectal cancer based on CEA and a 48-multiplex serum biomarker panel. Scientific reports 2021, 11(1):4287.
- Luo H, Shen K, Sun H, Li R, Wang Z, Xie Z: Correlation study between serum neuro-specific enolase and gastric and colorectal cancers. Medicine (Baltimore) 2020, 99(16):e19796.
- You W, Sheng N, Yan L, Chen H, Gong J, He Z, Zheng K, Chen Z, Wang Y, Tan G et al: The difference in prognosis of stage II and III colorectal cancer based on preoperative serum tumor markers. J Cancer 2019, 10(16):3757-3766.
- Peng HX, Yang L, He BS, Pan YQ, Ying HQ, Sun HL, Lin K, Hu XX, Xu T, Wang SK: Combination of preoperative NLR, PLR and CEA could increase the diagnostic efficacy for I-III stage CRC. Journal of clinical laboratory analysis 2017, 31(5).
- Wang J, Wang X, Yu F, Chen J, Zhao S, Zhang D, Yu Y, Liu X, Tang H, Peng Z: Combined detection of preoperative serum CEA, CA19-9 and CA242 improve prognostic prediction of surgically treated colorectal cancer patients. International journal of clinical and experimental pathology 2015, 8(11):14853-14863.
- Chen L, Jiang B, Wang Z, Liu M, Yang H, Xing J, Zhang C, Yao Z, Zhang N, Cui M et al: Combined preoperative CEA and CD44v6 improves prognostic value in patients with stage I and stage II colorectal cancer. Clinical & translational oncology : official publication of the Federation of Spanish Oncology Societies and of the National Cancer Institute of Mexico 2014, 16(3):285-292.
| Variables | sCEA:normal & tCEA+ | sCEA:normal & tCEA++ | sCEA:normal & tCEA+++ | sCEA:high & tCEA+ | sCEA: high & tCEA++ | sCEA:high & tCEA+++ | F or x2 test | P |
| Gender | 12.22 | 0.032* | ||||||
| Male | 186(10.6) | 165(9.4) | 74(4.2) | 111(6.3) | 209(11.9) | 143(8.1) | ||
| Femail | 186(10.6) | 178(10.1) | 62(3.5) | 119(6.8) | 229(13.0) | 95(5.4) | ||
| Age(year) | 67(16) | 65(15) | 67(21.75) | 65(19.5) | 67(21) | 67(15) | 5.37 | <0.001*** |
| Location | 202.11 | <0.001*** | ||||||
| Ileocecum | 41(2.3) | 33(1.9) | 18(1.0) | 11(0.6) | 7(0.4) | 37(2.1) | ||
| Right colon | 30(1.7) | 76(4.3) | 8(0.5) | 12(0.7) | 38(2.2) | 8(0.5) | ||
| Transverse colon | 66(3.8) | 39(2.2) | 21(1.2) | 42(2.4) | 86(4.9) | 23(1.3) | ||
| Left colon | 84(4.8) | 32(1.8) | 36(2.0) | 38(2.2) | 87(5.0) | 47(2.7) | ||
| Sigmoid colon | 35(2.0) | 45(2.6) | 12(0.7) | 31(1.8) | 18(1.0) | 26(1.5) | ||
| Rectum | 116(6.6) | 118(6.7) | 41(2.3) | 96(5.5) | 202(11.5) | 97(5.5) | ||
| Tumor size(cm) | 3.7(1.1) | 4.1(1) | 3.6(1) | 3.5(1.2) | 3.5(0.9) | 4.1(1.33) | 18.60 | <0.001*** |
| Blood loss(ml) | 180(110) | 160(150) | 180(115) | 180(160) | 180(110) | 160(52.5) | 3.51 | 0.004** |
| T stage | 452.82 | <0.001a*** | ||||||
| Tis | 9(0.5) | 3(0.2) | 3(0.2) | 1(0.1) | 0(0) | 0(0) | ||
| T1 | 23(1.3) | 37(2.1) | 5(0.3) | 21(1.2) | 20(1.1) | 7(0.4) | ||
| T2 | 106(6.0) | 26(1.5) | 30(1.7) | 24(1.4) | 91(5.2) | 37(2.1) | ||
| T3 | 53(3.0) | 157(8.9) | 73(4.2) | 117(6.7) | 177(10.1) | 53(3.0) | ||
| T4a | 23(1.3) | 29(1.7) | 24(1.4) | 66(3.8) | 92(5.2) | 45(2.6) | ||
| T4b | 158(9.0) | 91(5.2) | 1(0.1) | 1(0.1) | 58(3.3) | 96(5.5) | ||
| Differentiation | 22.25 | <0.001*** | ||||||
| well | 26(1.5) | 66(3.8) | 19(1.1) | 31(1.8) | 59(3.4) | 24(1.4) | ||
| moderate | 234(13.3) | 251(14.3) | 75(4.3) | 136(7.7) | 350(19.9) | 90(5.1) | ||
| poor or undifferentiation | 112(6.4) | 26(1.5) | 42(2.4) | 63(3.6) | 29(1.7) | 124(7.1) | ||
| Harvested Lymph node(no.) | 14(3) | 13(3) | 14(3) | 14(2) | 14(2) | 14(3) | 6.83 | <0.001*** |
| Positive Lymph node(no.) | 2(2) | 2(6) | 0(2) | 2(6) | 3(5) | 2(5) | 22.17 | <0.001*** |
| Chemotherapy | 22.25 | <0.001*** | ||||||
| Yes | 319(18.2) | 295(16.8) | 117(6.7) | 200(11.4) | 402(22.9) | 227(12.9) | ||
| No | 53(3.0) | 48(2.7) | 19(1.1) | 30(1.7) | 36(2.0) | 11(0.6) | ||
| TNM stage | 160.92 | <0.001a*** | ||||||
| 0&Ⅰ | 42(2.4) | 30(1.7) | 12(0.7) | 26(1.5) | 28(1.6) | 9(0.5) | ||
| Ⅱ | 26(1.5) | 49(2.8) | 42(2.4) | 28(1.6) | 47(2.7) | 33(1.9) | ||
| Ⅲ | 170(9.7) | 133(7.6) | 82(4.7) | 136(7.7) | 209(11.9) | 145(8.3) | ||
| Ⅳ | 134(7.6) | 131(7.5) | 0(0) | 40(2.3) | 154(8.8) | 51(2.9) | ||
| Complication | 58.60 | <0.001*** | ||||||
| No | 357(20.3) | 284(16.2) | 121(6.9) | 214(12.2) | 421(12.2) | 215(12.2) | ||
| Yes | 15(0.9) | 59(3.4) | 15(0.9) | 16(0.9) | 17(1.0) | 23(1.3) |
| Factor | N | Hazard Ratio(HR) | Mean and 95%CI for survival time(60months) | 5-year OS(%) | P value |
| Gender | 0.296 | ||||
| M | 888 | Ref. | 42.78(41.45~44.09) | 47.5 | |
| F | 869 | 1.072 | 43.42(42.06~44.78) | 50.5 | |
| Location | <0.001*** | ||||
| Ileocecum | 147 | Ref. | 36.93(33.42~40.49) | 38.1 | |
| Right colon | 172 | 1.488 | 35.63(32.42~38.85) | 37.8 | |
| Transverse colon | 277 | 1.521 | 47.40(45.24~49.56) | 57.0 | |
| Left colon | 324 | 0.777 | 43.28(41.13~45.44) | 48.5 | |
| Sigmoid colon | 167 | 1.019 | 45.99(42.95~49.03) | 57.5 | |
| Rectum | 670 | 0.815 | 43.77(42.27~45.27) | 49.1 | |
| T stage | <0.001*** | ||||
| Tis | 16 | Ref. | 60(60~60) | 93.8 | |
| T1 | 113 | 0.064 | 57.97(56.93~59.02) | 81.4 | |
| T2 | 314 | 0.197 | 42.29(40.04~44.53) | 46.8 | |
| T3 | 630 | 0.759 | 40.37(38.75~41.99) | 39.8 | |
| T4a | 279 | 0.911 | 52.96(51.37~54.54) | 69.9 | |
| T4b | 405 | 0.345 | 36.34(34.26~38.42) | 39.8 | |
| Differentiation | <0.001*** | ||||
| well | 225 | Ref. | 52.88(50.77~64.98) | 76.9 | |
| moderate | 1136 | 0.159 | 45.27(44.11~46.43) | 56.2 | |
| poor or undifferentiation | 396 | 0.355 | 31.30(29.53~33.08) | 12.6 | |
| Chemotherapy | <0.001*** | ||||
| Yes | 1560 | Ref. | 41.09(40.07~42.11) | 44.0 | |
| No | 197 | 6.78 | 58.98(58.46~59.50) | 88.3 | |
| TNM stage | <0.001*** | ||||
| 0&Ⅰ | 147 | Ref. | 59.86(59.61~60.00) | ||
| Ⅱ | 225 | 0.003 | 58.08(57.40~58.76) | ||
| Ⅲ | 875 | 0.027 | 51.98(51.08~52.88) | ||
| Ⅳ | 510 | 0.060 | 16.40(15.76~17.04) | ||
| Complication | 0.85 | ||||
| No | 1612 | Ref. | 42.95(41.96~43.94) | 49 | |
| Yes | 145 | 1.023 | 44.70(41.60~47.79) | 49 | |
| sCEA | 0.55 | ||||
| normal | 851 | Ref. | 42.91(41.49~44.33) | 53.0 | |
| high | 906 | 0.879 | 43.27(42.00~44.53) | 45.3 | |
| tCEA | 0.002** | ||||
| + | 601 | Ref. | 44.30(42.65~45.94) | ||
| ++ | 784 | 0.762 | 41.27(39.8~42.75) | ||
| +++ | 372 | 0.985 | 44.99(43.22~46.76) | ||
| CombinedCEA | <0.001*** | ||||
| sCEA:normal & tCEA+ | 372 | Ref. | 42.92(40.73~45.10) | 57.0 | |
| sCEA:normal & tCEA++ | 343 | 0.413 | 40.06(37.74~42.37) | 46.6 | |
| sCEA:normal & tCEA+++ | 136 | 0.833 | 50.07(47.43~52.72) | 58.1 | |
| sCEA:high & tCEA+ | 230 | 0.509 | 46.57(44.14~48.99) | 53.9 | |
| sCEA: high & tCEA++ | 438 | 0.599 | 42.30(40.40~44.21) | 48.2 | |
| sCEA:high & tCEA+++ | 238 | 0.742 | 41.85(39.60~44.11) | 31.5 |
| Factor | HR | 95%CI for HR | Ward | P |
| Location | 29.05 | <0.001*** | ||
| Ileocecum | Ref. | |||
| Right colon | 0.850 | 0.623~1.159 | ||
| Transverse colon | 0.577 | 0.427~0.779 | ||
| Left colon | 0.722 | 0.547~0.954 | ||
| Sigmoid colon | 0.480 | 0.341~0.675 | ||
| Rectum | 0.821 | 0.636~1.061 | ||
| T stage | 95.93 | <0.001*** | ||
| Tis | Ref. | |||
| T1 | 0.174 | 0.015~1.986 | ||
| T2 | 0.280 | 0.025~3.118 | ||
| T3 | 0.565 | 0.050~6.399 | ||
| T4a | 0.306 | 0.027~4.472 | ||
| T4b | 0.220 | 0.020~2.470 | ||
| Differentiation | 190.18 | <0.001*** | ||
| well | Ref. | |||
| moderate | 2.142 | 1.572~2.917 | ||
| poor or undifferentiation | 6.794 | 4.806~9.605 | ||
| Chemotherapy | 0.61 | 0.433 | ||
| Yes | Ref. | |||
| No | 0.779 | 0.417~1.454 | ||
| TNM stage | 954.18 | <0.001*** | ||
| 0&Ⅰ | Ref. | |||
| Ⅱ | 4.789 | 1.062~21.604 | ||
| Ⅲ | 9.632 | 2.194~42.292 | ||
| Ⅳ | 267.44 | 60.944~1173.58 | ||
| tCEA | 4.68 | 0.096 | ||
| + | Ref. | |||
| ++ | 4.831 | 0.664~35.174 | ||
| +++ | 1.635 | 0.142~18.789 | ||
| CombinedCEA | 32.67 | <0.001*** | ||
| sCEA:normal & tCEA+ | Ref. | |||
| sCEA:normal & tCEA++ | 0.475 | 0.065~3.488 | ||
| sCEA:normal & tCEA+++ | 1.176 | 0.100~13.824 | ||
| sCEA:high & tCEA+ | 1.242 | 0.921~1.674 | ||
| sCEA: high & tCEA++ | 0.266 | 0.036~1.958 | ||
| sCEA:high & tCEA+++ | 1.275 | 0.111~14.606 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




