Submitted:
05 June 2024
Posted:
06 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. RNA-seq Reads Statistics
2.2. Principal Component Analysis (PCA) Highlights the Development Program of the Single Berry
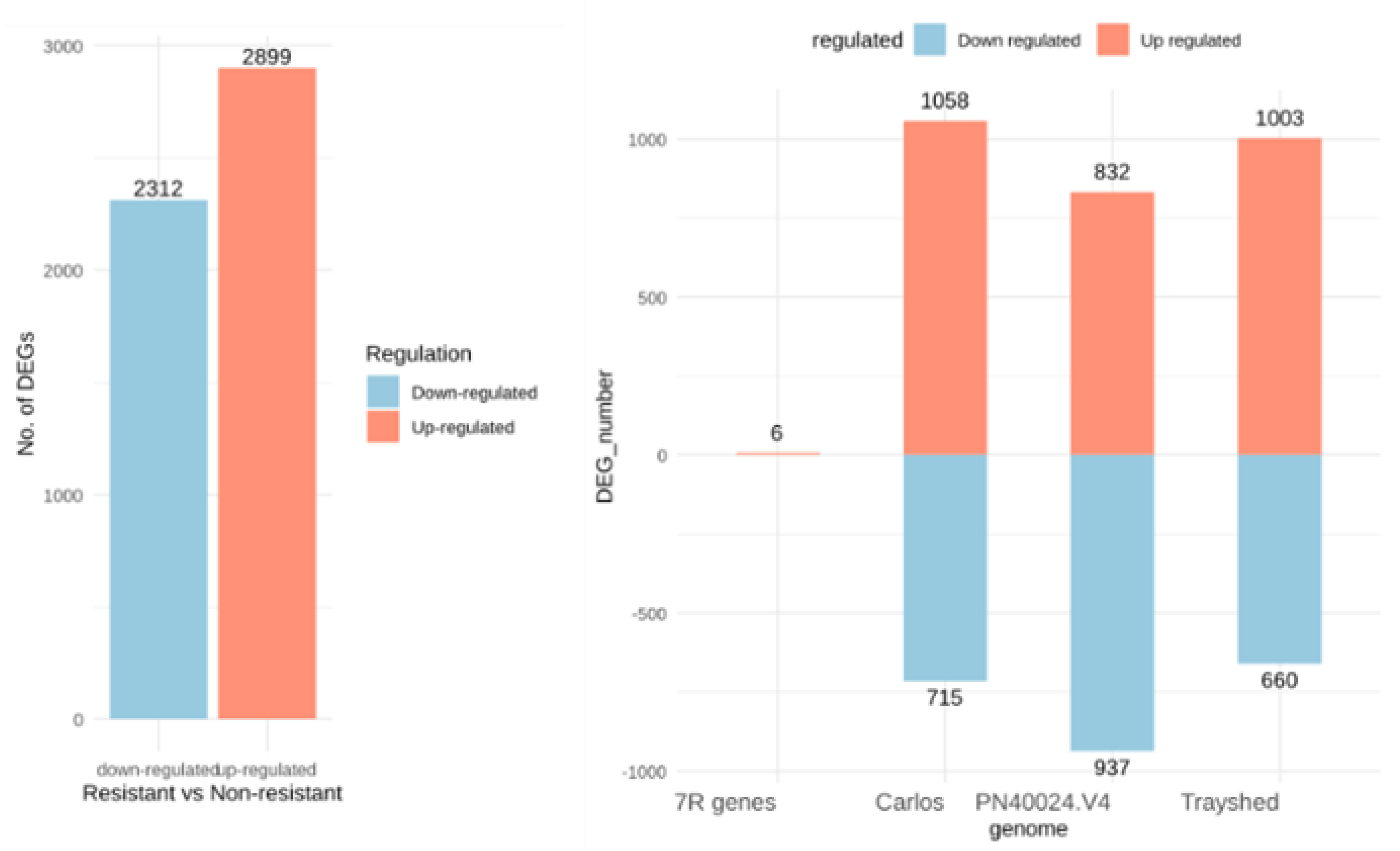
2.3. Identification of Differentially Expressed Genes
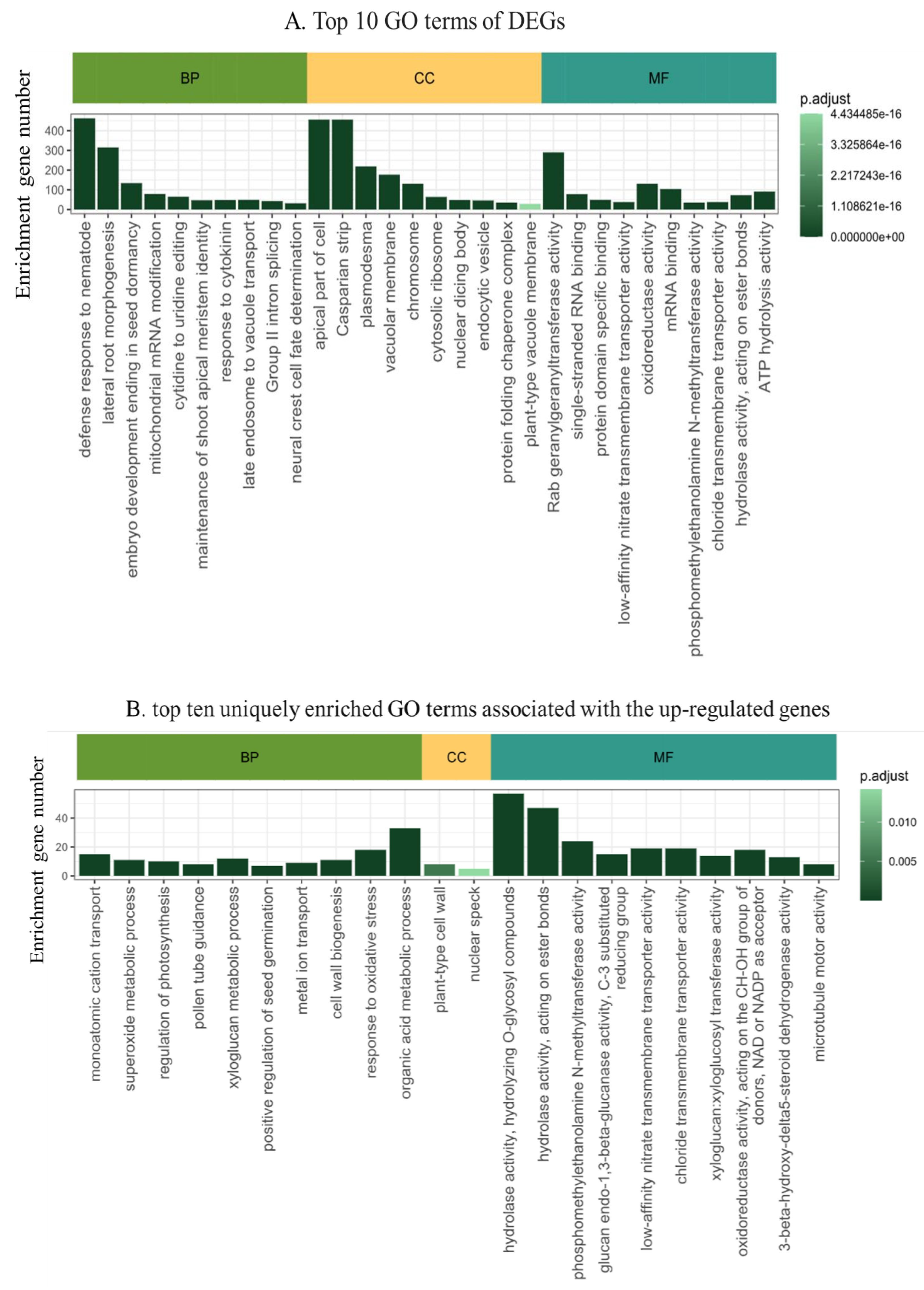
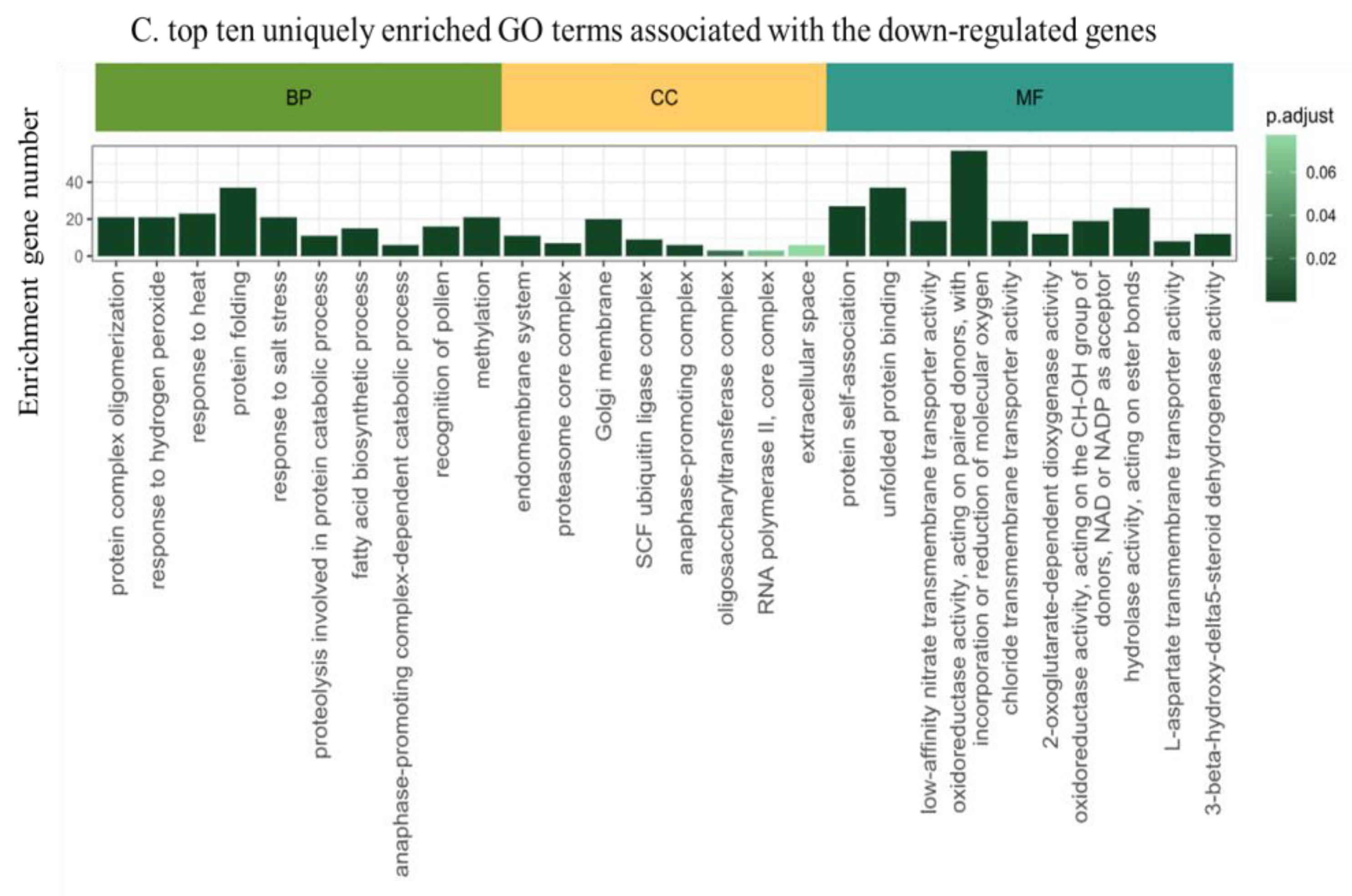
2.4. Functional Annotation of the Differentially Expressed Genes
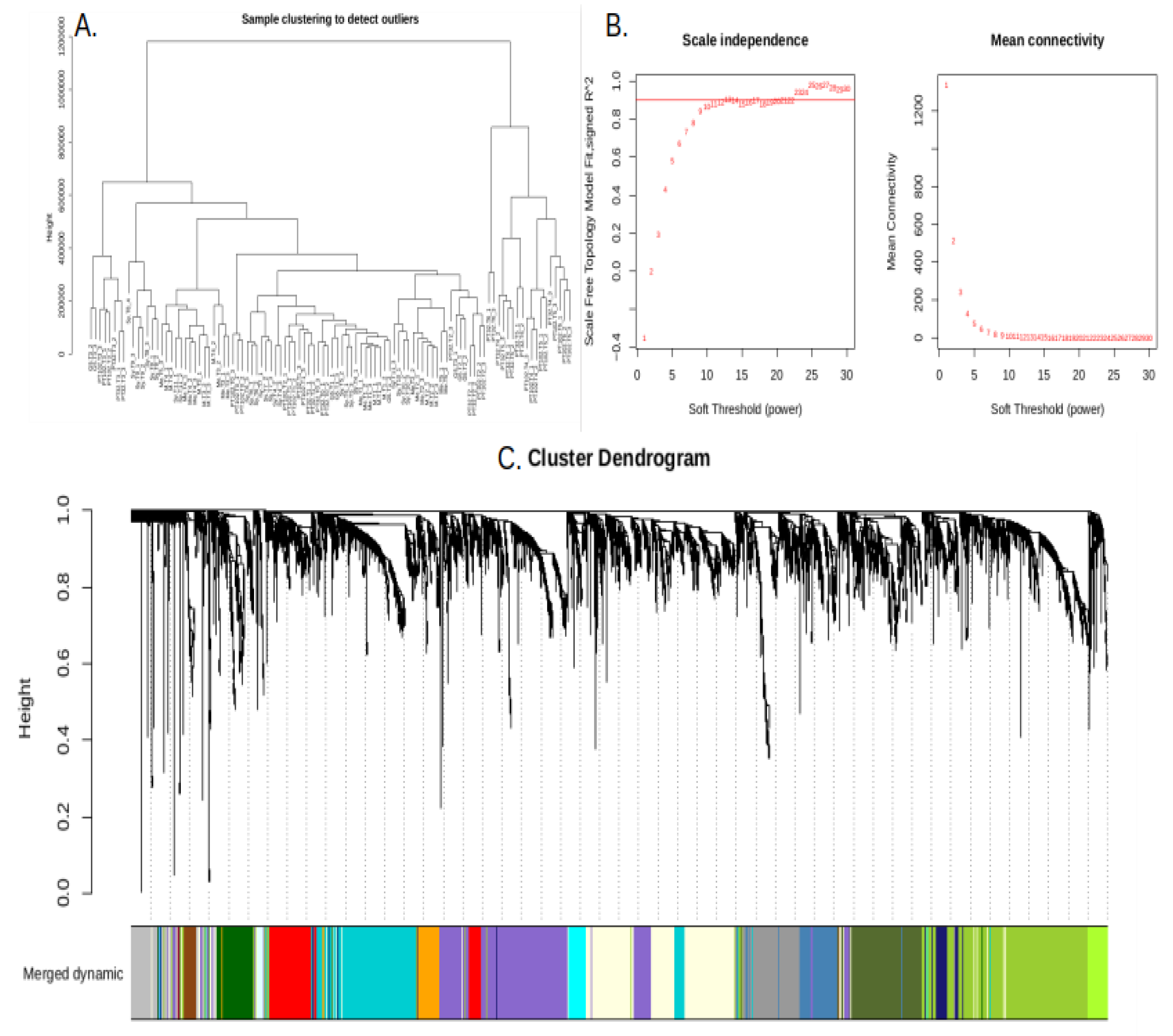
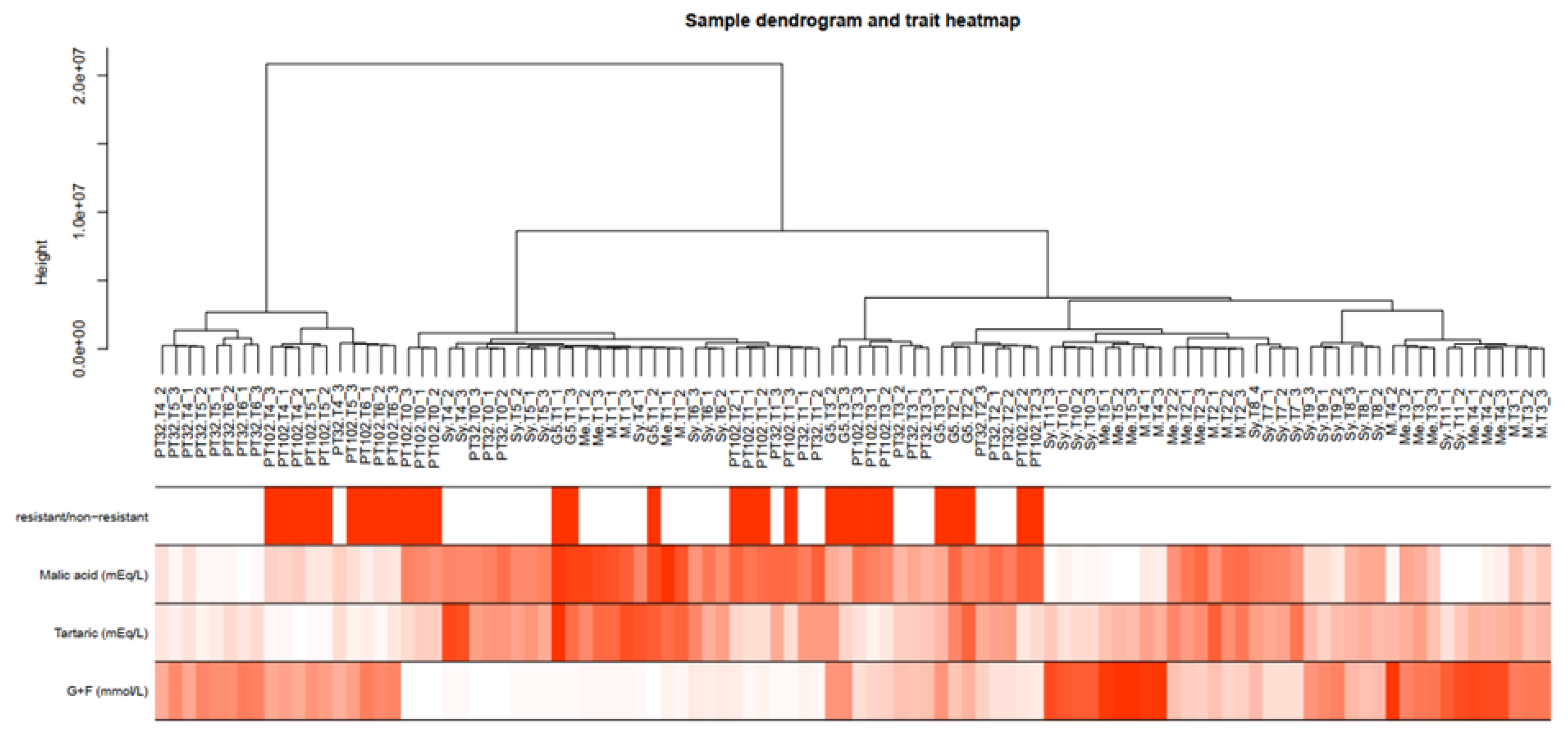
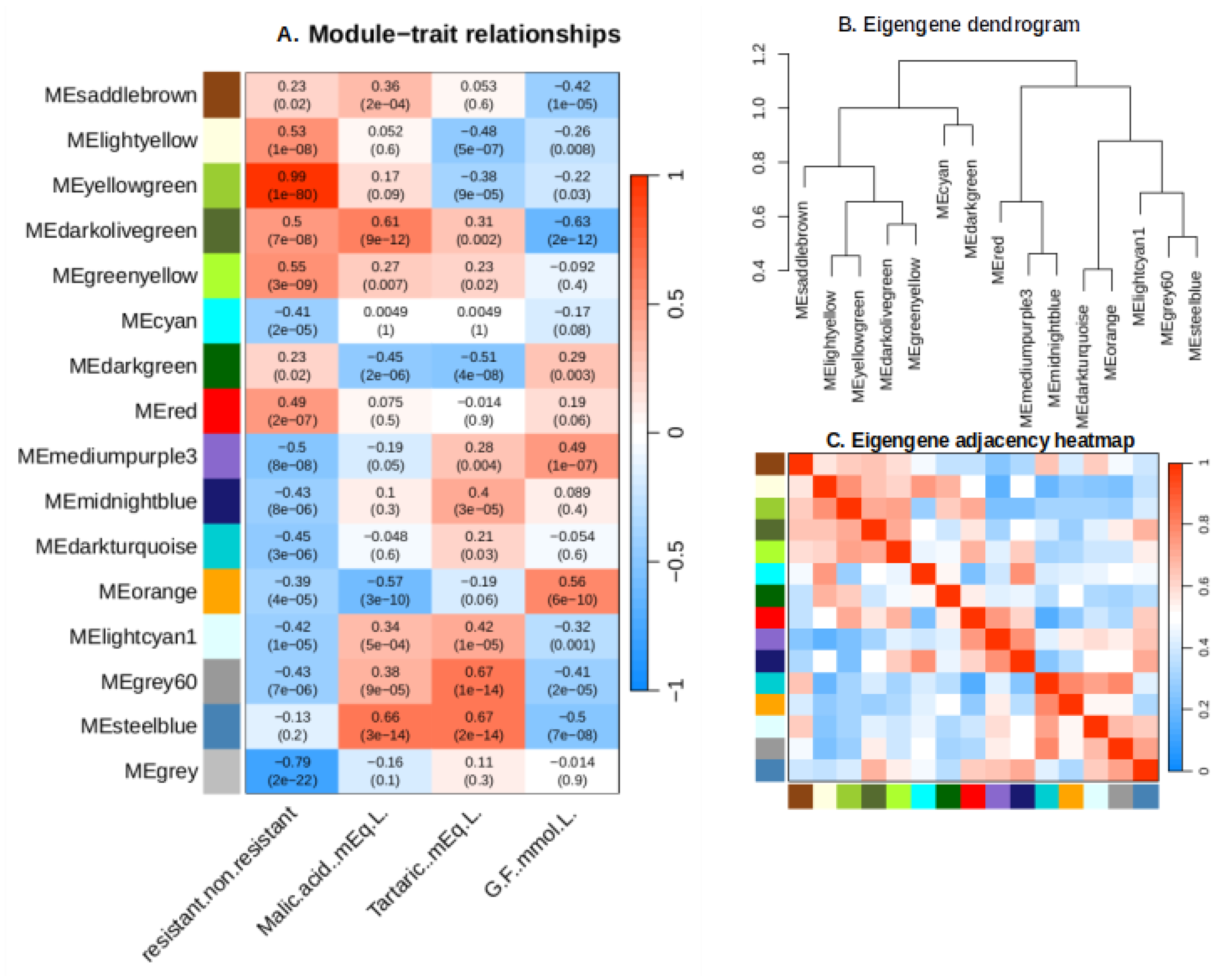
2.5. Weighted Gene Co-Expression Network Analysis (WGCNA)
2.6. Identification of Key Modules Associated with Traits
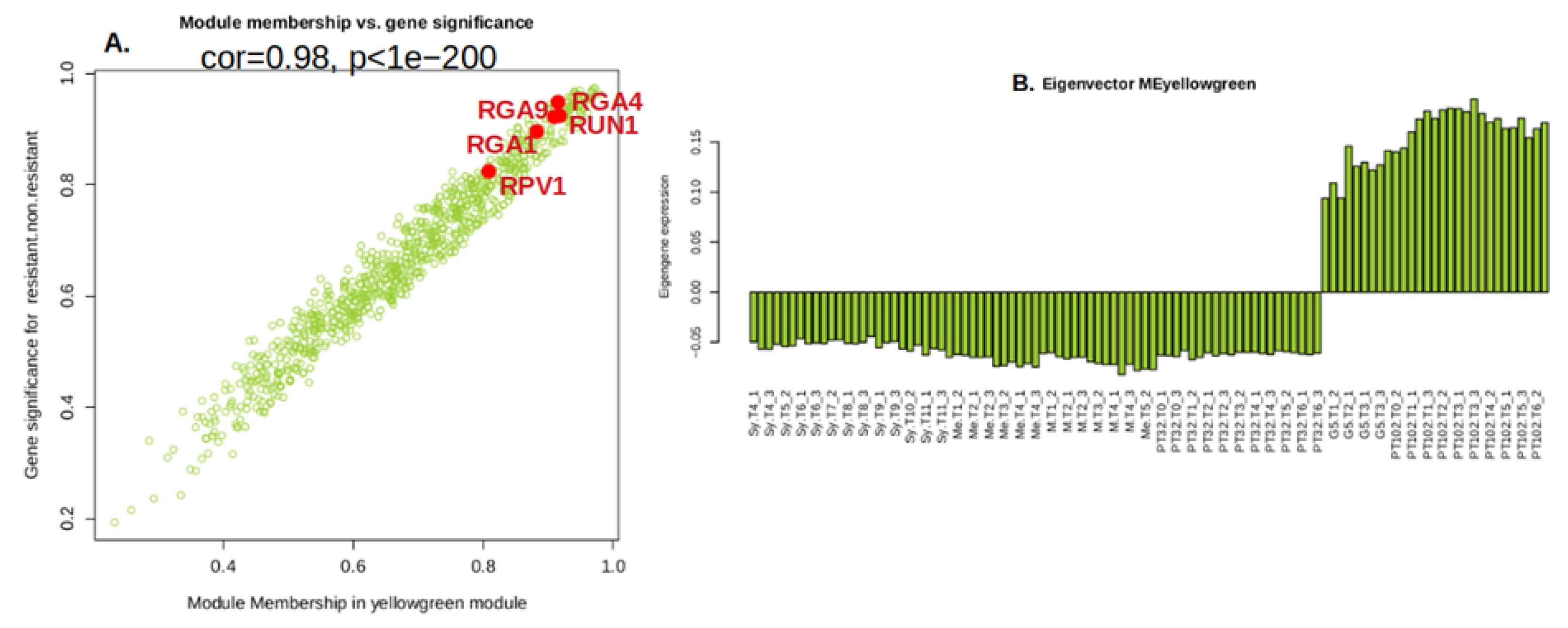
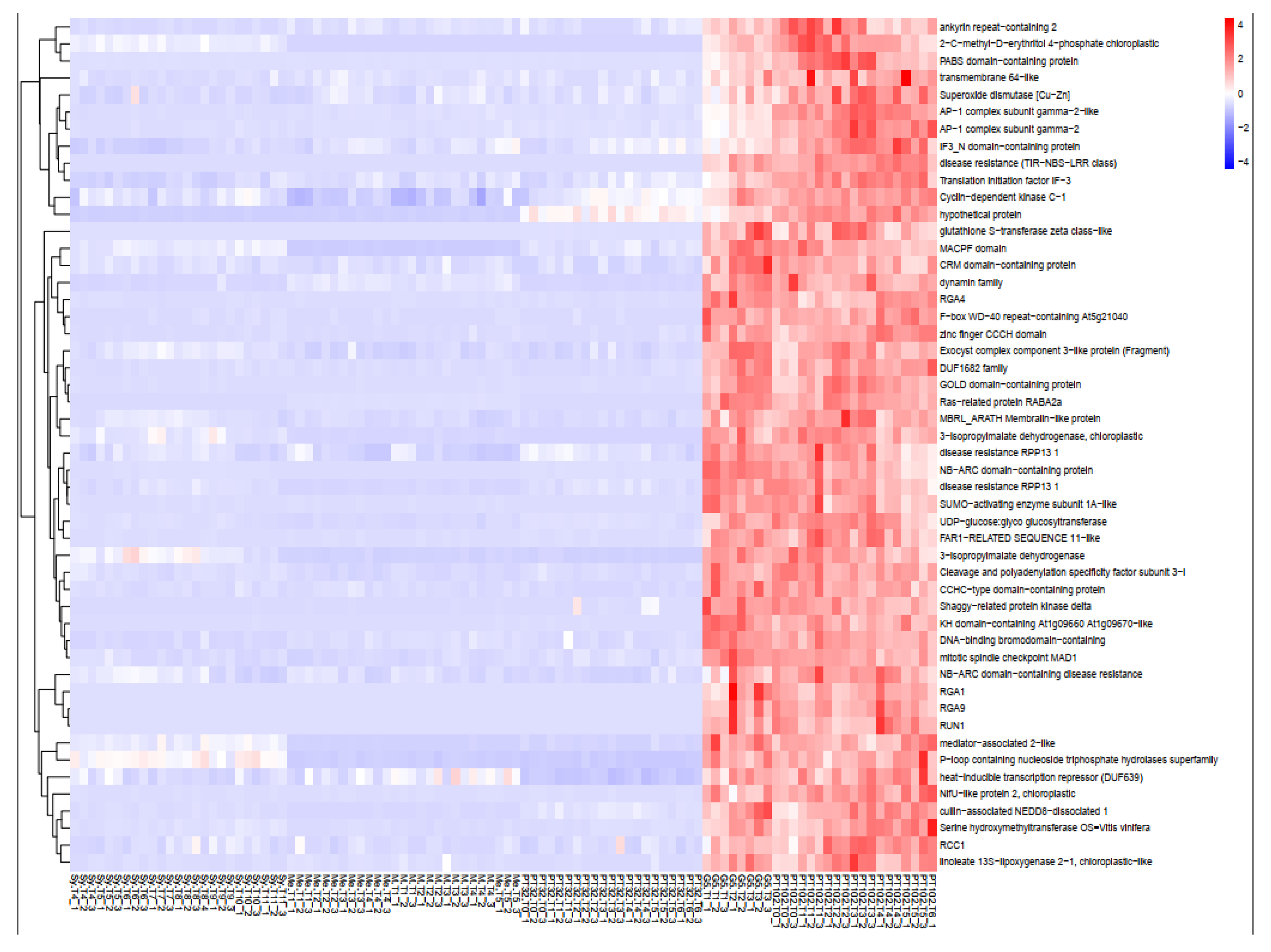
2.6.1. Yellow Green Module-Positively Associated with the Presence of MrRUN1/MrRPV1 Microsatellites Markers.
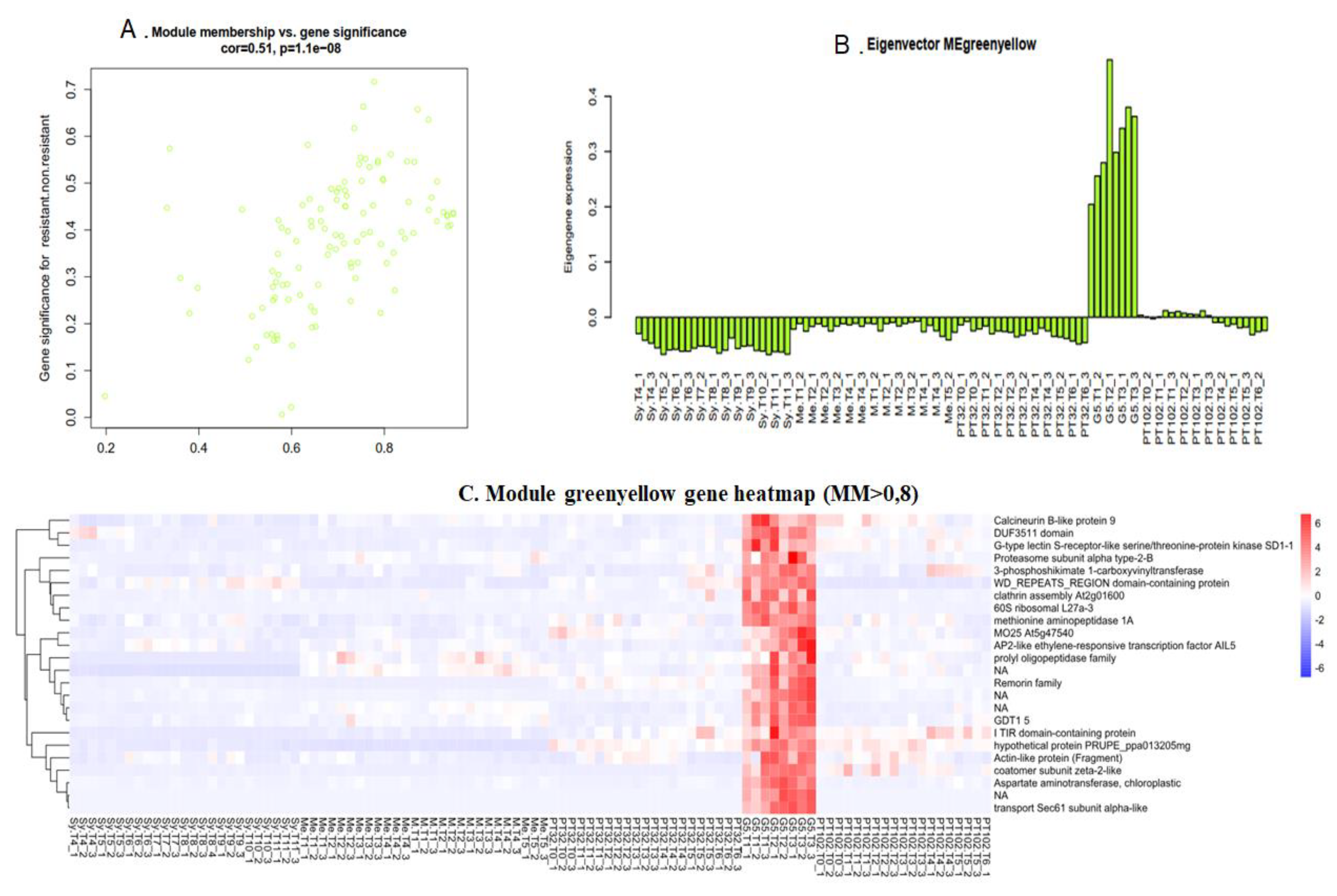
2.6.2. Green Yellow Module: G5 Specific Transcripts, almost Lost in the Next Backcross
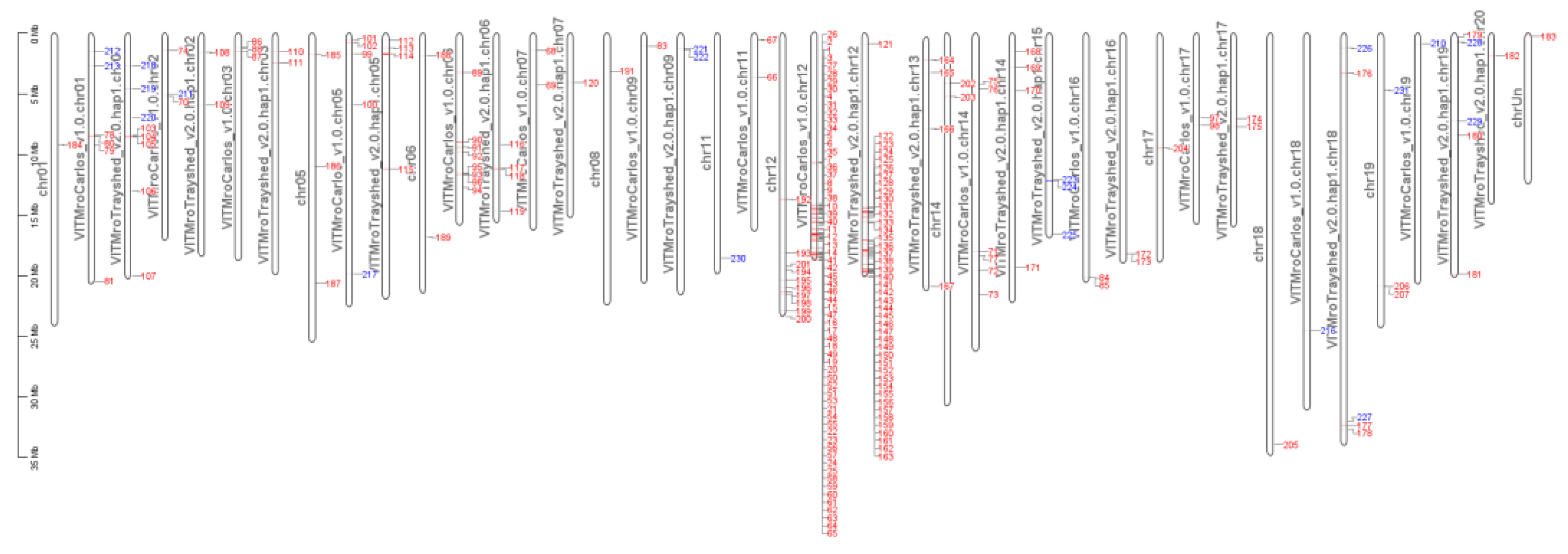
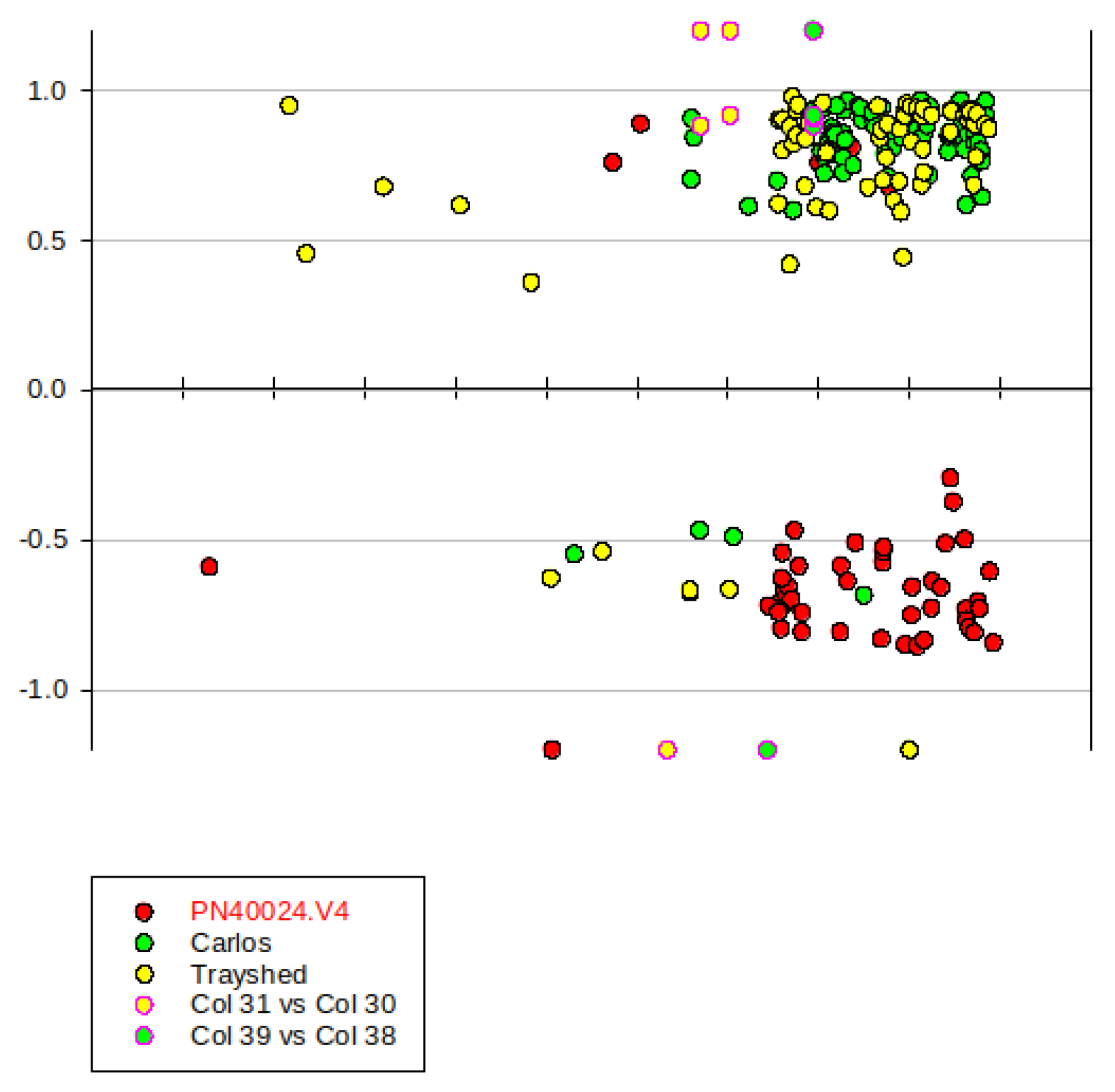
2.7. Gene Introgressed with MrRUN1/MrRPV1 on Chromosome 12.
3. Discussion
3.1. PCA and Gene Expression Profiles
3.2. Merged V. vinifera and V. rotundifolia Genome
3.3. Regulation of Genes Related to Pathogen Response
4. Materials and Methods
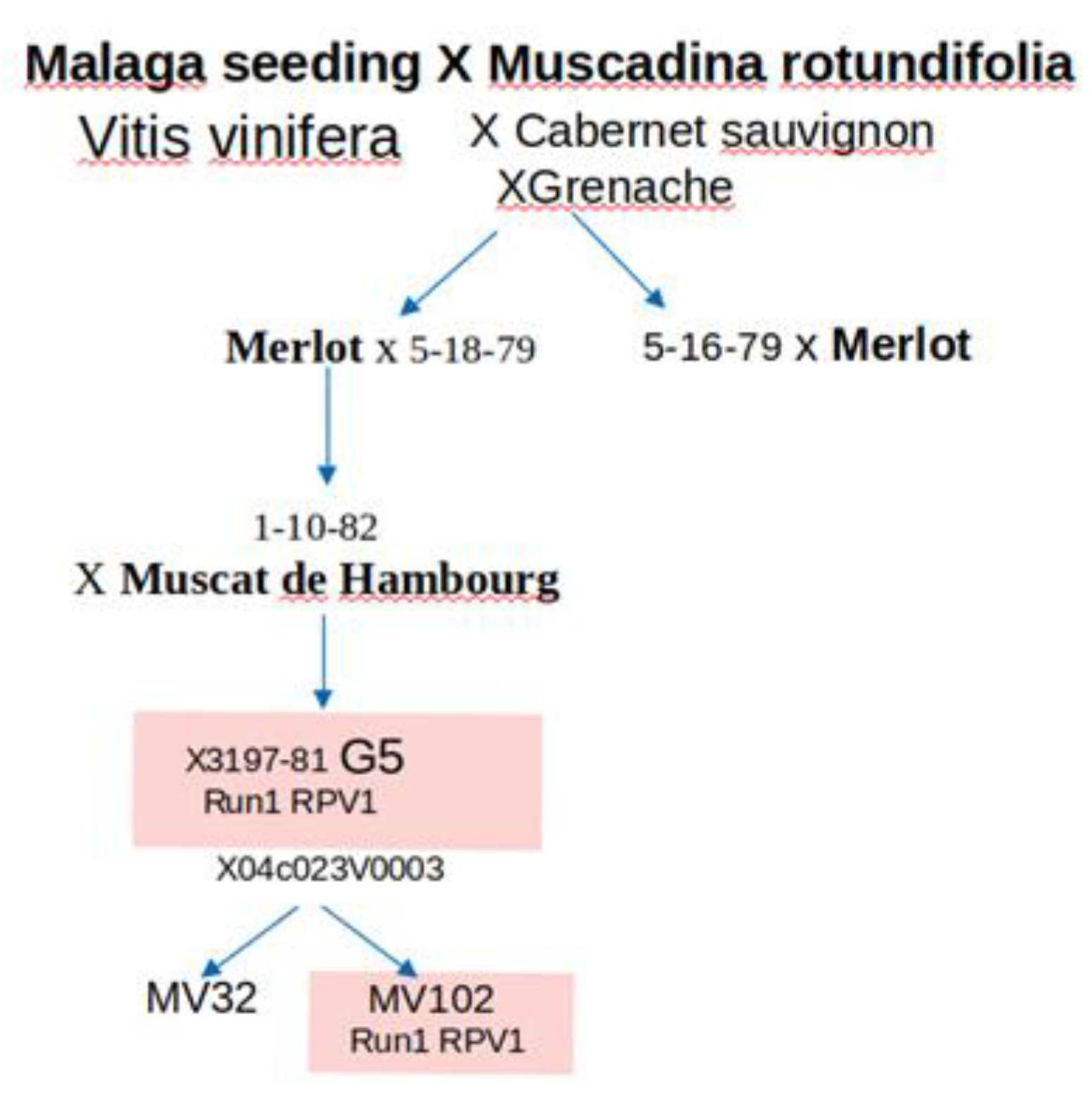
4.1. Grapevine Genotypes
4.2. Single Berry Sampling
4.3. HPLC Analysis of Primary Metabolites in Single Berries
4.4. RNA Extraction and Sequencing
4.5. Methodology for Transcriptome Analysis
4.6. GO Annotation Analysis
4.7. WGCNA Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- P. This, T. Lacombe, and M. R. Thomas, “Historical origins and genetic diversity of wine grapes,” Trends Genet. TIG, vol. 22, no. 9, pp. 511–519, Sep. 2006. [CrossRef]
- R. B. Ferreira, S. S. Monteiro, M. A. Piçarra-Pereira, and A. R. Teixeira, “Engineering grapevine for increased resistance to fungal pathogens without compromising wine stability,” Trends Biotechnol., vol. 22, no. 4, pp. 168–173, Apr. 2004. [CrossRef]
- M. C. Fontaine et al., “Europe as a bridgehead in the worldwide invasion history of grapevine downy mildew, Plasmopara viticola,” Curr. Biol., vol. 31, no. 10, pp. 2155-2166.e4, May 2021. [CrossRef]
- L. Ons, D. Bylemans, K. Thevissen, and B. P. A. Cammue, “Combining Biocontrol Agents with Chemical Fungicides for Integrated Plant Fungal Disease Control,” Microorganisms, vol. 8, no. 12, p. 1930, Dec. 2020. [CrossRef]
- J. Lu, L. Schell, and D. W. Ramming, “Interspecific hybridization between vitis rotundifolia and vitis vinifera and evaluation of the hybrids,” Acta Hortic., no. 528, pp. 481–486, May 2000. [CrossRef]
- V. A. Volynkin et al., “Introgressions of Vitis rotundifolia Michx. to obtain grapevine genotypes with complex resistance to biotic and abiotic stresses,” Vavilov J. Genet. Breed., vol. 25, no. 7, pp. 693–700, Nov. 2021. [CrossRef]
- J.-M. Salmon, H. Ojeda, and J.-L. Escudier, “Disease resistant grapevine varieties and quality: the case of Bouquet varieties,” OENO One, vol. 52, no. 3, Art. no. 3, Aug. 2018. [CrossRef]
- C. Da Silva et al., “The high polyphenol content of grapevine cultivar tannat berries is conferred primarily by genes that are not shared with the reference genome,” Plant Cell, vol. 25, no. 12, pp. 4777–4788, Dec. 2013. [CrossRef]
- L. Venturini et al., “De novo transcriptome characterization of Vitis vinifera cv. Corvina unveils varietal diversity,” BMC Genomics, vol. 14, p. 41, Jan. 2013. [CrossRef]
- L. Ma et al., “Transcriptome analysis of table grapes (Vitis vinifera L.) identified a gene network module associated with berry firmness,” PLoS ONE, vol. 15, no. 8, p. e0237526, Aug. 2020. [CrossRef]
- S. Savoi et al., “Transcriptome and metabolite profiling reveals that prolonged drought modulates the phenylpropanoid and terpenoid pathway in white grapes (Vitis vinifera L.),” BMC Plant Biol., vol. 16, Mar. 2016. [CrossRef]
- M. Rienth, L. Torregrosa, G. Sarah, M. Ardisson, J.-M. Brillouet, and C. Romieu, “Temperature desynchronizes sugar and organic acid metabolism in ripening grapevine fruits and remodels their transcriptome,” BMC Plant Biol., vol. 16, no. 1, p. 164, Jul. 2016. [CrossRef]
- Y. Ju, Z. Min, Y. Zhang, K. Zhang, M. Liu, and Y. Fang, “Transcriptome profiling provide new insights into the molecular mechanism of grapevine response to heat, drought, and combined stress,” Sci. Hortic., vol. 286, p. 110076, Aug. 2021. [CrossRef]
- S. Pilati et al., “Abscisic Acid Is a Major Regulator of Grape Berry Ripening Onset: New Insights into ABA Signaling Network,” Front. Plant Sci., vol. 8, 2017, Accessed: Feb. 05, 2024. [Online]. Available: https://www.frontiersin.org/journals/plant-science/articles/10.3389/fpls.2017.01093.
- Yan Lei et al., “Transcriptome Analysis of Berries of Spine Grape (Vitis davidii Föex) Infected by Colletotrichum viniferum during Symptom Development,” Horticulturae, vol. 8, no. 9, pp. 843–843, Sep. 2022. [CrossRef]
- N. Vigneron, J. Grimplet, E. Remolif, and M. Rienth, “Claros,” Sci. Rep., vol. 13, no. 1, p. 14664, Sep. 2023. [CrossRef]
- J. Qu, I. Dry, L. Liu, Z. Guo, and L. Yin, “Transcriptional profiling reveals multiple defense responses in downy mildew-resistant transgenic grapevine expressing a TIR-NBS-LRR gene located at the MrRUN1/MrRPV1 locus,” Hortic. Res., vol. 8, no. 1, pp. 1–12, Jul. 2021. [CrossRef]
- T. V. M. Fajardo and V. Quecini, “Comparative transcriptome analyses between cultivated and wild grapes reveal conservation of expressed genes but extensive rewiring of co-expression networks,” Plant Mol. Biol., vol. 106, no. 1–2, pp. 1–20, May 2021. [CrossRef]
- Q. Long et al., “Population comparative genomics discovers gene gain and loss during grapevine domestication,” Plant Physiol., p. kiae039, Jan. 2024. [CrossRef]
- V. Ferrero et al., “Complex patterns in tolerance and resistance to pests and diseases underpin the domestication of tomato,” New Phytol., vol. 226, no. 1, pp. 254–266, Apr. 2020. [CrossRef]
- J. Liu, L. Wang, S. Jiang, Z. Wang, H. Li, and H. Wang, “Mining of Minor Disease Resistance Genes in V. vinifera Grapes Based on Transcriptome,” Int. J. Mol. Sci., vol. 24, no. 20, Art. no. 20, Jan. 2023. [CrossRef]
- T. Possamai, D. Migliaro, M. Gardiman, R. Velasco, and B. De Nardi, “Rpv Mediated Defense Responses in Grapevine Offspring Resistant to Plasmopara viticola,” Plants, vol. 9, no. 6, p. 781, Jun. 2020. [CrossRef]
- M. Park et al., “Chromosome-level genome sequence assembly and genome-wide association study of Muscadinia rotundifolia reveal the genetics of 12 berry-related traits,” Hortic. Res., vol. 9, p. uhab011, Jan. 2022. [CrossRef]
- M. Sargolzaei et al., “Rpv29, Rpv30 and Rpv31: Three Novel Genomic Loci Associated With Resistance to Plasmopara viticola in Vitis vinifera,” Front. Plant Sci., vol. 11, 2020, Accessed: Feb. 14, 2024. [Online]. Available: https://www.frontiersin.org/journals/plant-science/articles/10.3389/fpls.2020.562432.
- J. Pauquet, A. Bouquet, P. This, and A.-F. Adam-Blondon, “Establishment of a local map of AFLP markers around the powdery mildew resistance gene Run1 in grapevine and assessment of their usefulness for marker assisted selection:,” Theor. Appl. Genet., vol. 103, no. 8, pp. 1201–1210, Dec. 2001. [CrossRef]
- C. L. Barker et al., “Genetic and physical mapping of the grapevine powdery mildew resistance gene, Run1, using a bacterial artificial chromosome library,” Theor. Appl. Genet., vol. 111, no. 2, pp. 370–377, Jul. 2005. [CrossRef]
- L. Cadle-Davidson, “Variation Within and Between Vitis spp. for Foliar Resistance to the Downy Mildew Pathogen Plasmopara viticola,” Plant Dis., vol. 92, no. 11, pp. 1577–1584, Nov. 2008. [CrossRef]
- I.B. Dry et al., “Molecular strategies to enhance the genetic resistance of grapevines to powdery mildew,” Aust. J. Grape Wine Res., vol. 16, pp. 94–105, Nov. 2009. [CrossRef]
- A. BOUQUET, J. Pauquet, A.-F. Adam-Blondon, L. Torregrosa, D. Merdinoglu, and S. Wiedemann-Merdinoglu, “Vers l’obtention de variétés de vigne résistantes à l’oïdium et au mildiou par les méthodes conventionnelles et biotechnologiques,” Bull. OIV, vol. 73, pp. 445–452, Jan. 2000.
- A. Feechan et al., “Genetic dissection of a TIR-NB-LRR locus from the wild North American grapevine species Muscadinia rotundifolia identifies paralogous genes conferring resistance to major fungal and oomycete pathogens in cultivated grapevine,” Plant J., vol. 76, no. 4, pp. 661–674, 2013. [CrossRef]
- S. Riaz, A. C. Tenscher, D. W. Ramming, and M. A. Walker, “Using a limited mapping strategy to identify major QTLs for resistance to grapevine powdery mildew (Erysiphe necator) and their use in marker-assisted breeding,” Theor. Appl. Genet., vol. 122, no. 6, pp. 1059–1073, Apr. 2011. [CrossRef]
- S. Savoi, L. Torregrosa, and C. Romieu, “Transcripts repressed at the stop of phloem unloading highlight the energy efficiency of sugar import in the ripening V. vinifera fruit.,” Plant Biology, preprint, Jan. 2021. [CrossRef]
- S. Savoi, L. Torregrosa, and C. Romieu, “Single berry development – a new phenotyping and transcriptomics paradigm,” VITIS – J. Grapevine Res., vol. 62, pp. 49–55, Oct. 2023. [CrossRef]
- J. Chaïb, L. Torregrosa, D. Mackenzie, P. Corena, A. Bouquet, and M. R. Thomas, “The grape microvine - a model system for rapid forward and reverse genetics of grapevines,” Plant J. Cell Mol. Biol., vol. 62, no. 6, pp. 1083–1092, Jun. 2010. [CrossRef]
- J. T. Robinson, H. Thorvaldsdottir, D. Turner, and J. P. Mesirov, “igv.js: an embeddable JavaScript implementation of the Integrative Genomics Viewer (IGV),” Bioinformatics, vol. 39, no. 1, p. btac830, Jan. 2023. [CrossRef]
- A. Velt et al., “An improved reference of the grapevine genome reasserts the origin of the PN40024 highly homozygous genotype,” G3 GenesGenomesGenetics, vol. 13, no. 5, p. jkad067, Mar. 2023. [CrossRef]
- G. Di Gaspero et al., “Evaluation of sensitivity and specificity in RNA-Seq-based detection of grapevine viral pathogens,” J. Virol. Methods, vol. 300, p. 114383, Feb. 2022. [CrossRef]
- C. Jiao, M. Gao, X. Wang, and Z. Fei, “Transcriptome characterization of three wild Chinese Vitis uncovers a large number of distinct disease related genes,” BMC Genomics, vol. 16, no. 1, p. 223, Mar. 2015. [CrossRef]
- C. Rispe et al., “De novo transcriptome assembly of the grapevine phylloxera allows identification of genes differentially expressed between leaf- and root-feeding forms,” BMC Genomics, vol. 17, no. 1, Art. no. 1, Dec. 2016. [CrossRef]
- G. Robertson et al., “De novo assembly and analysis of RNA-seq data,” Nat. Methods, vol. 7, no. 11, pp. 909–912, Nov. 2010. [CrossRef]
- M. G. Claros, R. Bautista, D. Guerrero-Fernández, H. Benzerki, P. Seoane, and N. Fernández-Pozo, “Why Assembling Plant Genome Sequences Is So Challenging,” Biology, vol. 1, no. 2, pp. 439–459, Sep. 2012. [CrossRef]
- M. Massonnet et al., “Ripening Transcriptomic Program in Red and White Grapevine Varieties Correlates with Berry Skin Anthocyanin Accumulation,” Plant Physiol., vol. 174, no. 4, pp. 2376–2396, Aug. 2017. [CrossRef]
- L. McHale, X. Tan, P. Koehl, and R. W. Michelmore, “Plant NBS-LRR proteins: adaptable guards,” Genome Biol., vol. 7, no. 4, p. 212, 2006. [CrossRef]
- N. Cochetel, A. Minio, M. Massonnet, A. M. Vondras, R. Figueroa-Balderas, and D. Cantu, “Diploid chromosome-scale assembly of the Muscadinia rotundifolia genome supports chromosome fusion and disease resistance gene expansion during Vitis and Muscadinia divergence,” G3 GenesGenomesGenetics, vol. 11, no. 4, Apr. 2021. [CrossRef]
- M. Huff et al., “Long-read, chromosome-scale assembly of Vitis rotundifolia cv. Carlos and its unique resistance to Xylella fastidiosa subsp. fastidiosa,” BMC Genomics, vol. 24, no. 1, p. 409, Jul. 2023. [CrossRef]
- K. Buck and M. Worthington, “Genetic Diversity of Wild and Cultivated Muscadine Grapes (Vitis rotundifolia Michx.),” Front. Plant Sci., vol. 13, p. 852130, Mar. 2022. [CrossRef]
- A. Morales-Cruz et al., “Introgression among North American wild grapes (Vitis) fuels biotic and abiotic adaptation,” Genome Biol., vol. 22, no. 1, p. 254, Sep. 2021. [CrossRef]
- S. Hou, Y. Yang, D. Wu, and C. Zhang, “Plant immunity,” Plant Signal. Behav., vol. 6, no. 6, pp. 794–799, Jun. 2011. [CrossRef]
- X. Wang et al., “Toll/interleukin-1 receptor (TIR) domain-containing proteins have NAD-RNA decapping activity,” Nat. Commun., vol. 15, no. 1, Art. no. 1, Mar. 2024. [CrossRef]
- S. Horsefield et al., “NAD+ cleavage activity by animal and plant TIR domains in cell death pathways,” Science, Aug. 2019. [CrossRef]
- J. Chen, X. Zhang, M. Bernoux, J. P. Rathjen, and P. N. Dodds, “Plant Toll/interleukin-1 receptor/resistance protein domains physically associate with enhanced disease susceptibility1 family proteins in immune signaling,” iScience, vol. 27, no. 2, p. 108817, Feb. 2024. [CrossRef]
- J. Ordon et al., “Disentangling cause and consequence: genetic dissection of the DANGEROUS MIX2 risk locus, and activation of the DM2h NLR in autoimmunity,” Plant J., vol. 106, no. 4, pp. 1008–1023, 2021. [CrossRef]
- D. C. J. Wong, P. Ariani, S. Castellarin, A. Polverari, and E. Vandelle, “Co-expression network analysis and cis-regulatory element enrichment determine putative functions and regulatory mechanisms of grapevine ATL E3 ubiquitin ligases,” Sci. Rep., vol. 8, no. 1, p. 3151, Feb. 2018. [CrossRef]
- B. Mauch-Mani and F. Mauch, “The role of abscisic acid in plant-pathogen interactions,” Curr. Opin. Plant Biol., vol. 8, no. 4, pp. 409–414, Aug. 2005. [CrossRef]
- V. Sichel et al., “Intravarietal diversity: an opportunity for climate change adaptation,” 2023, Accessed: Jan. 24, 2024. [Online]. Available: https://agris.fao.org/search/en/providers/122439/records/652f9decc1cd75198696e944.
- A. Brutus, F. Sicilia, A. Macone, F. Cervone, and G. De Lorenzo, “A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides,” Proc. Natl. Acad. Sci. U. S. A., vol. 107, no. 20, pp. 9452–9457, May 2010. [CrossRef]
- B. P. H. J. Thomma, T. Nürnberger, and M. H. A. J. Joosten, “Of PAMPs and Effectors: The Blurred PTI-ETI Dichotomy[OA],” Plant Cell, vol. 23, no. 1, pp. 4–15, Jan. 2011. [CrossRef]
- X.-L. Sun et al., “GsSRK, a G-type lectin S-receptor-like serine/threonine protein kinase, is a positive regulator of plant tolerance to salt stress,” J. Plant Physiol., vol. 170, no. 5, pp. 505–515, Mar. 2013. [CrossRef]
- C. Wang, J. Wu, Y. Zhang, and J. Lu, “Muscadinia rotundifolia ‘Noble’ defense response to Plasmopara viticola inoculation by inducing phytohormone-mediated stilbene accumulation,” Protoplasma, vol. 255, no. 1, pp. 95–107, Jan. 2018. [CrossRef]
- M. Zhou et al., “Transcription factors VviWRKY10 and VviWRKY30 co-regulate powdery mildew resistance in grapevine,” Plant Physiol., vol. 195, no. 1, pp. 446–461, May 2024. [CrossRef]
- A. Bigard, C. Romieu, H. Ojeda, and L. J.-M. Torregrosa, “The sugarless grape trait characterised by single berry phenotyping,” OENO One, vol. 56, no. 3, Art. no. 3, Jul. 2022. [CrossRef]
- L. J.-M. Torregrosa, M. Rienth, C. Romieu, and A. Pellegrino, “The microvine, a model for studies in grapevine physiology and genetics,” OENO One, vol. 53, no. 3, Art. no. 3, Jul. 2019. [CrossRef]
- S. Savoi, L. Torregrosa, and C. Romieu, “Transcripts switched off at the stop of phloem unloading highlight the energy efficiency of sugar import in the ripening V. vinifera fruit,” Hortic. Res., vol. 8, no. 1, pp. 1–15, Sep. 2021. [CrossRef]
- V. Sichel et al., “Intravarietal diversity: an opportunity for climate change adaptation,” 2023, Accessed: Jan. 24, 2024. [Online]. Available: https://agris.fao.org/search/en/providers/122439/records/652f9decc1cd75198696e944.
- O. Cassan, S. Lèbre, and A. Martin, “Inferring and analyzing gene regulatory networks from multi-factorial expression data: a complete and interactive suite,” BMC Genomics, vol. 22, no. 1, p. 387, May 2021. [CrossRef]
- P. Langfelder and S. Horvath, “WGCNA: an R package for weighted correlation network analysis,” BMC Bioinformatics, vol. 9, no. 1, p. 559, Dec. 2008. [CrossRef]

| Genotype | Traits | Sample Year | Location | Dates of Sampling | Sample Number |
| G5 | resistant | 2021 | Pech Rouge | 3 dates | 8 |
| MV102 (PT102) | resistant | 2018 | Greenhouse | 7 dates | 21 |
| MV32 (PT32) |
non-resistant | 2018 | Greenhouse | 7 dates | 21 |
| Syrah | non-resistant | 2018/2019 | SupAgro campus | 11 dates | 25 |
| Merlot clone1 | non-resistant | 2022 | Bordeaux | 4 dates | 12 |
| Merlot clone2 | non-resistant | 2022 | Bordeaux | 5 dates | 15 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





