Submitted:
28 May 2024
Posted:
29 May 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Experimental Design
| Extraction method | Sample Type | |
|---|---|---|
| Dry Sample (D) | Fresh Sample (F) | |
| Cetyltrimethylammonium bromide (CTAB 2X) (Kit 1) | T1. CTAB-D | T2. CTAB-F |
| GeneJET Plant Genomic DNA Purification Kit (Kit 2) | T3. GenJET-D | T4. GenJET-F |
| EZ-10 Spin Column Genomic | DNA Minipreps Kit (Plant) (Kit 3) | T5. EZGen-D | T6. EZGen-F |
2.1. Materials
- Fresh leaves sample of Inga feuillei
- Cetyltrimethylammonium bromide - CTAB (Promega Corporation, Madison, USA)
- β-mercaptoetanol (Central Drug House, New Delhi, India)
- Tris-HCl 1M (Promega Corporation, Madison, USA)
- EDTA 0.5M (Promega Corporation, Madison, USA)
- NaCl (Sigma-Milipore, Saint Louis, MO, USA).
- PVP-40 (Caisson Labs, Smithfield, UT, USA)
- Chloroform (J.T. Baker, Phillipsburg, NJ, USA)
- Iso amyl alcohol (Central Drug House, New Delhi, India)
- Isopropanol (Central Drug House, New Delhi, India)
- Absolute ethanol (Merck, Darmstadt, Germany)
- Rnase A solution (Thermo Fisher Scientific, Waltham, MA, USA)
- Buffer TE solution (Promega Corporation, Madison, USA)
- Agarose (Promega Corporation, Madison, USA)
- Loading dye 6X (Thermo Fisher Scientific, Waltham, MA, USA)
- GeneRuler 1 Kb DNA Ladder (Thermo Fisher Scientific, Waltham, MA, USA)
2.2. Equipment
- Biological safety cabinet (Labconco, Kansas, MO, USA)
- TissueLyser II (Qiagen, Hilden, Germany)
- Microcentrifuge air-cooled (Labnet International, Edison, NJ, USA)
- Thermomixer C (Eppendorf, Hamburg, Germany)
- Biospectrometer (Eppendorf, Hamburg, Germany)
- Enduro Gel documentation Systems (Labnet International, Edison, NJ, USA)
3. Procedure
3.1. Sample Collection
3.2. Sample Homogenization
- The sample of plant material (leaves) is collected in a 2 mL tube with 750 µL extraction buffer (2X CTAB extraction buffer and 2µL of β-mercaptoethanol).
- The tubes are then opened, a steel bead is added to each, closed, and placed in the TissueLyser II disruptor kit.
- The samples are ground for eight minutes at a speed of 30 rps. The tubes are then removed to continue the extraction.

3.3. DNA Extraction
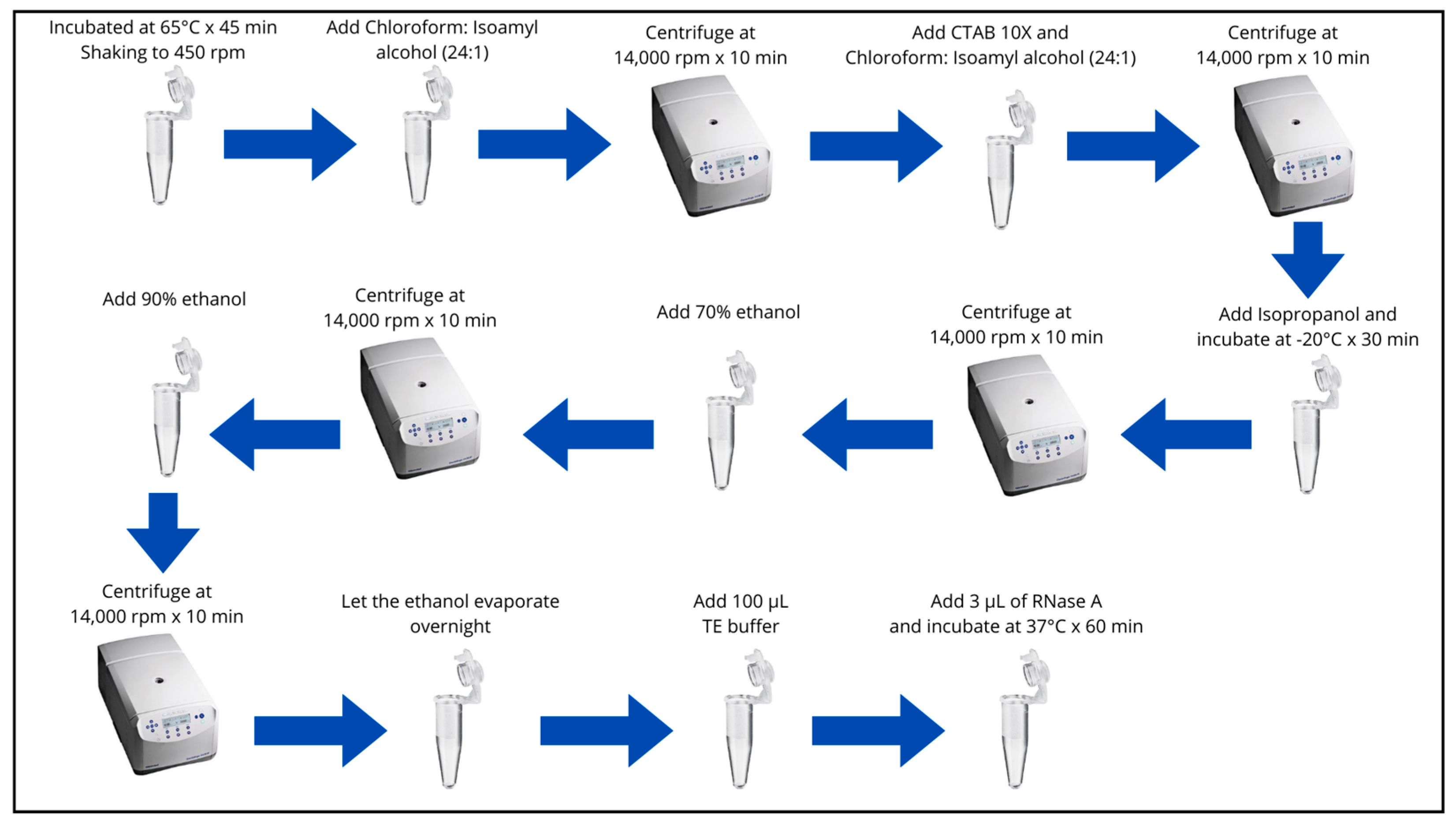
- Incubate samples at 65°C for 45 minutes to 1 hour. Shaking the tubes constantly (by inversion) every 20 minutes in a bain-marie or at 450 rpm in a Thermomixer C (Eppendorf, Hamburg, Germany).
- Remove the samples and let them cool at room temperature for 4 min.
- Add 750 µL of Chloroform: Isoamyl Alcohol (24:1), shake the tubes slightly (by inversion approximately 40 times) until the content is homogenized (milky green).
- Centrifuge at 14,000 rpm for 10 minutes.
- Transfer 600 µL of the supernatant to new 2 mL microtubes, previously labeled.
- Add 150 uL of CTAB 10X (preheated to 65°C), homogenize by inversion (10 times) and add 750 µL of Chloroform: Isoamyl Alcohol (24:1) and homogenize (by inversion approximately 40 times).
- Centrifuge at 14,000 rpm for 10 minutes.
- Transfer 500 µL of the supernatant to 1.5 mL microtubes, previously labeled.
- Add approximately 2/3 volume of Isopropanol (350 µL), shake gently by inversion (10 times) and incubate at -20°C for 30 minutes.
- Centrifuge at 14,000 rpm for 10 minutes.
- Decant the Isopropanol being careful not to lose the pellet.
- Add 500 µL of 70% ethanol. Remove the pellet from the bottom of the tube and gently shake the tube.
- Centrifuge at 14,000 rpm for 10 minutes.
- Decant the 70% ethanol, taking care not to lose the pellet.
- Add 500 µL of 90% Ethanol. Remove the pellet from the bottom of the tube and gently shake the tube.
- Centrifuge at 14,000 rpm for 10 minutes.
- Decant the 90% ethanol, taking care not to lose the pellet.
- Let the ethanol evaporate until the next day or leave it upside down for 2 to 3 hours to ensure that there is no ethanol left in the tube.
- Add 100 µL T10E1. Gently homogenize the DNA sample.
- Add 3 µL of RNase A and incubate in the thermomixer at 37°C. for 60 minutes at 300 rpm.
3.4. Quantification of DNA
- The ratio of absorbances A260/A280 helps determine if the obtained DNA is contaminated by the presence of aromatic compounds, as these absorb at a wavelength of 280 nm.
- DNA is considered of optimal quality when the 260/280 ratio is greater than 1.8. A ratio of A260/280 > 2.1 could indicate the presence of RNA in the sample. Conversely, if this ratio is low (A260/280 < 1.6), it could be indicative of contamination by proteins or phenols.
- A second assessment of nucleic acid purity is the 260/230 ratio, as at 230 nm, the maximum absorbance of salts present in the solution, carbohydrates, or other possible contaminants is detected.
- Accepted values fall within the range of 1.5 to 2.2; if the ratio is less than 1.5, it indicates the presence of contaminants in the sample.
3.5. Electrophoresis of DNA
- Dispense 1 µl of 6X loading buffer (Loading dye 6X, Thermo scientific) per sample to migrate on a parafilm sheet.
- Add 2 µL of the DNA sample. Mingle.
- Add 1X TAE buffer to the electrophoretic chamber and place the mold with the already polymerized agarose gel.
- From the second well, load the samples previously prepared with the 6X loading buffer.
- Place 2-3 µL of a 1 Kb DNA molecular weight marker (1 kb MP) in the first well of each row of the 1% agarose gel.
- Connect the electrodes correctly and migrate to 100 V for 30 minutes
- See the results with the Enduro Gel documentation Systems (Labnet International, Edison, NJ, USA).
3.6. DNA Amplification by Polymerase Chain Reation (PCR)
4. Expected Results
Reagents Setup
- Tris-HCl 1M, pH 8.0 100 mM
- EDTA 0.5M, pH 8.0 20 mM
- NaCl 0.7 M
- CTAB 10 %
- Tris-HCl 1M, pH 8.0 100 mM
- EDTA 0.5M, pH 8.0 20 mM
- NaCl 1.4 M
- PVP-40 1 % (w/v)
- CTAB 2 %
- Tris- pH=8.0 1M 10 mM
- EDTA-8.0 0,5 M 1 mM
- 1M TRIS -pH 8.0 (adjust pH 8.0 with HCl)
- Tris base pH=8.0 1 M
- 0.5M EDTA -pH 8.0 (adjust pH with NaOH)
5. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Abdel-Latif, A., & Osman, G. (2017). Comparison of three genomic DNA extraction methods to obtain high DNA quality from maize. Plant Methods, 13(1), 1. https://doi.org/10.1186/s13007-016-0152-4. [CrossRef]
- Aboul-Maaty, N. A.-F., & Oraby, H. A.-S. (2019). Extraction of high-quality genomic DNA from different plant orders applying a modified CTAB-based method. Bulletin of the National Research Centre, 43(1), 25. [CrossRef]
- Arbi, G., Naceur, B., Chokri, M., & Mohamed, B. (s. f.). A simple, rapid and efficient method for the extraction of genomic DNA from Allium roseum L. (Alliaceae).
- Doyle JJ, Doyle JL. (1990). Isolation of plant DNA from fresh tissue. Focus. 3: 12(13).
- Ferreira, C. F., Gutierrez, D. L., Kreuze, J. F., Iskra-Caruana, M. L., Chabannes, M., Barbosa, A. C. O., Santos, T. A., Silva, A. G. S., Santos, R. M. F., Amorim, E. P., De Oliveira, S. A. S., & Jesus, O. N. (2019). Brief Note Rapid plant DNA and RNA extraction protocol using a bench drill. Genetics and Molecular Research, 18(3). [CrossRef]
- Gupta, N. (2019). DNA extraction and polymerase chain reaction. Journal of Cytology, 36(2), 116. [CrossRef]
- Helmersen, K., & Aamot, H. V. (2020). DNA extraction of microbial DNA directly from infected tissue: An optimized protocol for use in nanopore sequencing. Scientific Reports, 10(1), 2985. [CrossRef]
- Inglis, P. W., Pappas, M. D. C. R., Resende, L. V., & Grattapaglia, D. (2018). Fast and inexpensive protocols for consistent extraction of high quality DNA and RNA from challenging plant and fungal samples for high-throughput SNP genotyping and sequencing applications. PLOS ONE, 13(10), e0206085. [CrossRef]
- Porebski, S., Bailey, L. G., & Baum, B. R. (1997). Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Molecular Biology Reporter, 15(1), 8-15. [CrossRef]
- Sahu, S. K., Thangaraj, M., & Kathiresan, K. (2012). DNA Extraction Protocol for Plants with High Levels of Secondary Metabolites and Polysaccharides without Using Liquid Nitrogen and Phenol. ISRN Molecular Biology, 2012, 1-6. [CrossRef]
- Montaño, Roberto. Tratamientos pregerminativos y sustratos en la germinacion de copoasu (Theobroma grandiflorum (Willd. ex Spreng.) Schum.) y pacay (Inga edulis Martius) en la comunidad de Santa Rosa del Abuna, Pando. Bachelor's tesis. Universidad Mayor de San Andres. Bolivia. 2006. https://repositorio.umsa.bo/handle/123456789/10773.
- Villacorta Santos and Vasquez Angel. Efecto antiinflamatorio y analgésico del extracto etanólico de la cascara Inga feuilleei DC. Pacay en ratones. Universidad Norbert Wiener. Peru. 2022. https://repositorio.uwiener.edu.pe/handle/20.500.13053/6258.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





