Submitted:
13 May 2024
Posted:
13 May 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
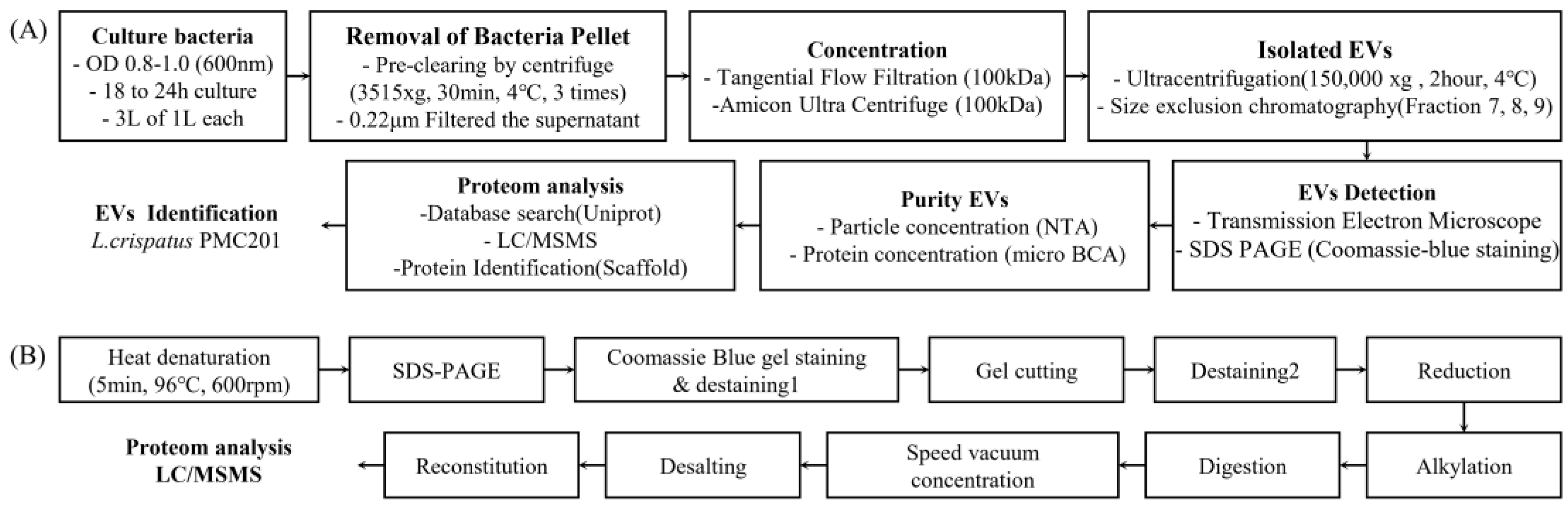
2. Materials and Methods
2.1. Optimized Culture Media for Isolating of EVs
2.2. Isolation of Bacterial EVs
2.3. Bio-TEM Imaging of EVs
2.4. NTA Measurements of EVs
2.5. Methanol/Chloroform Precipitation
2.6. SDS-PAGE and Coomassie Blue Staining
2.7. Proteomic Analysis of EVs Derived from L. crispatus PMC 201
2.8. Integrated Bioinformatics Analysis of protein Sequences from Proteomic Data
2.9. Statistical Analysis
3. Results
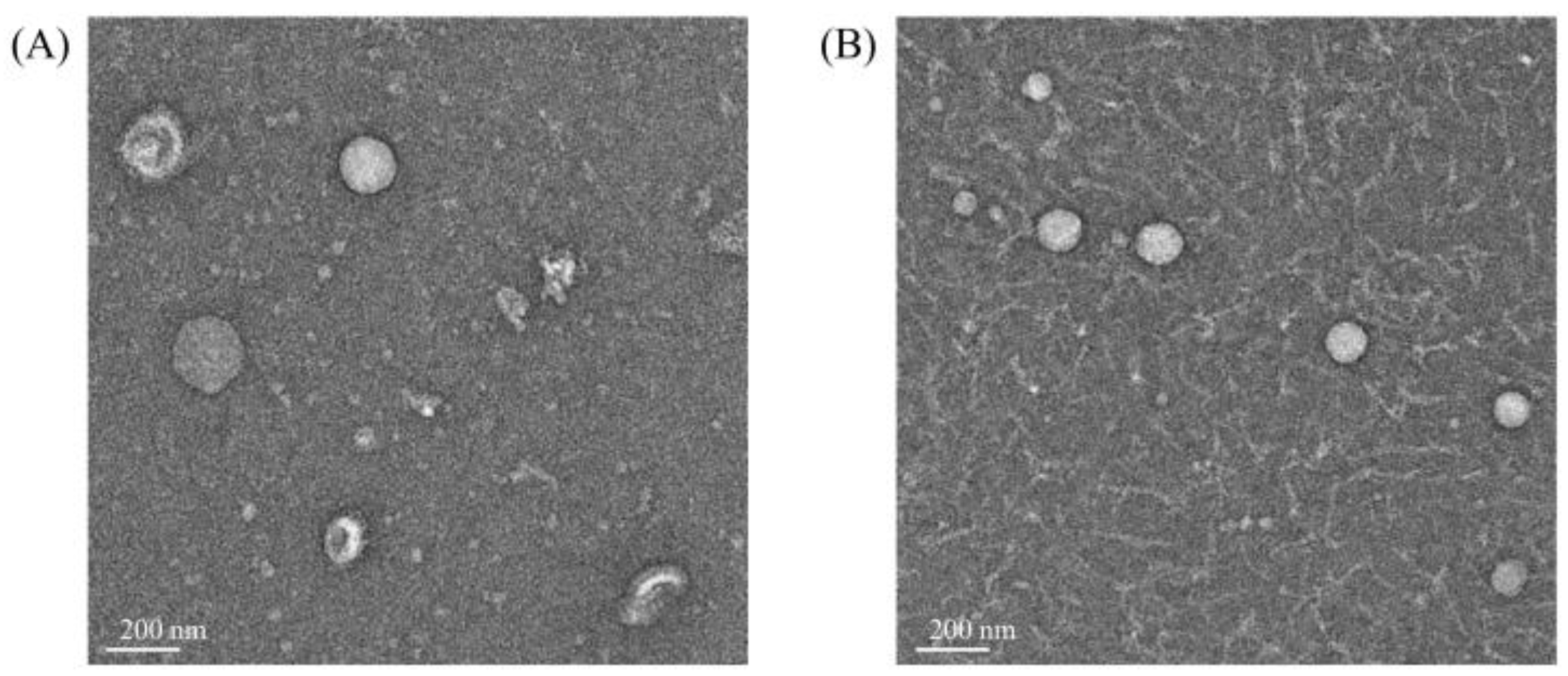
3.1. Isolation of EVs Derived from L. crispatus PMC201
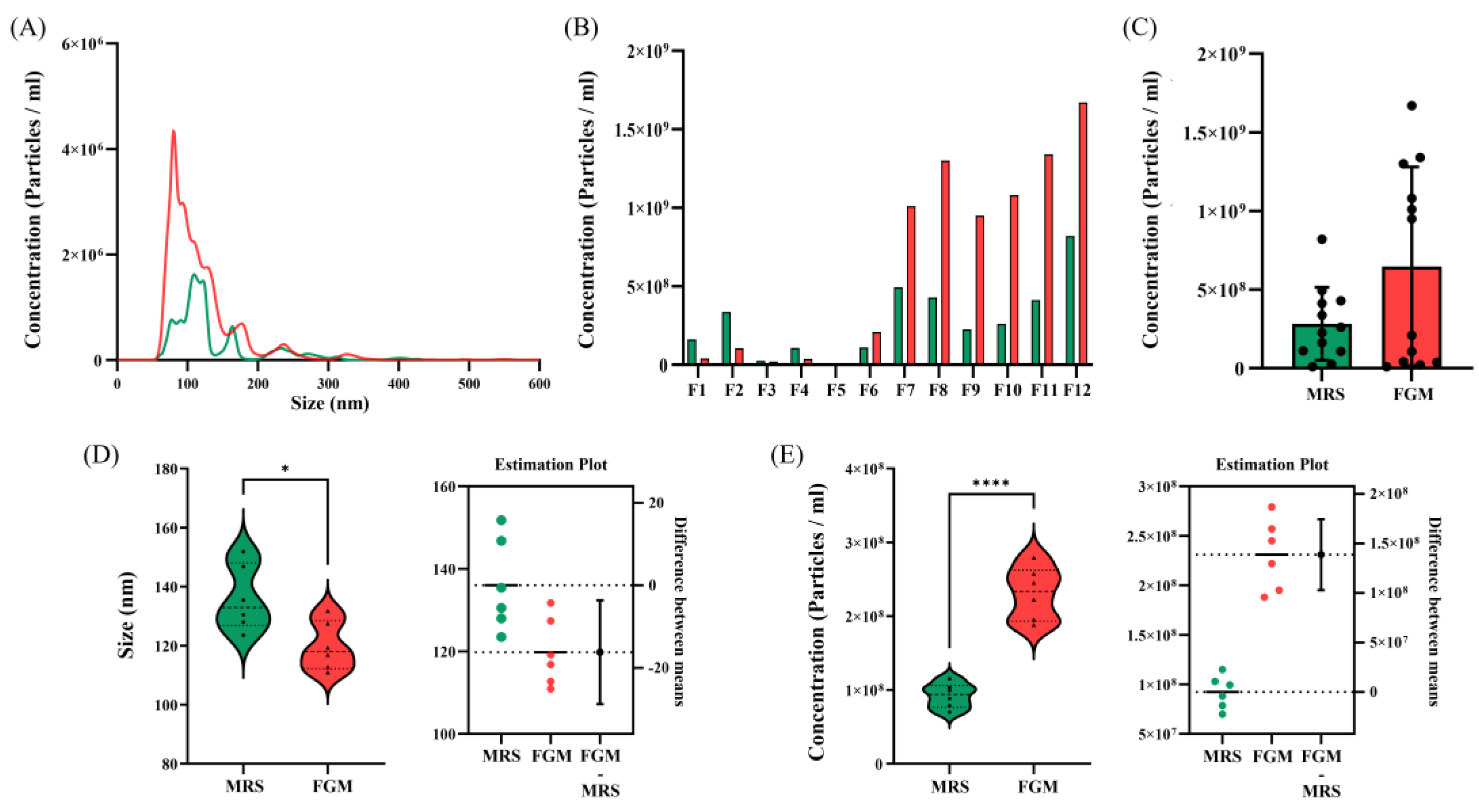
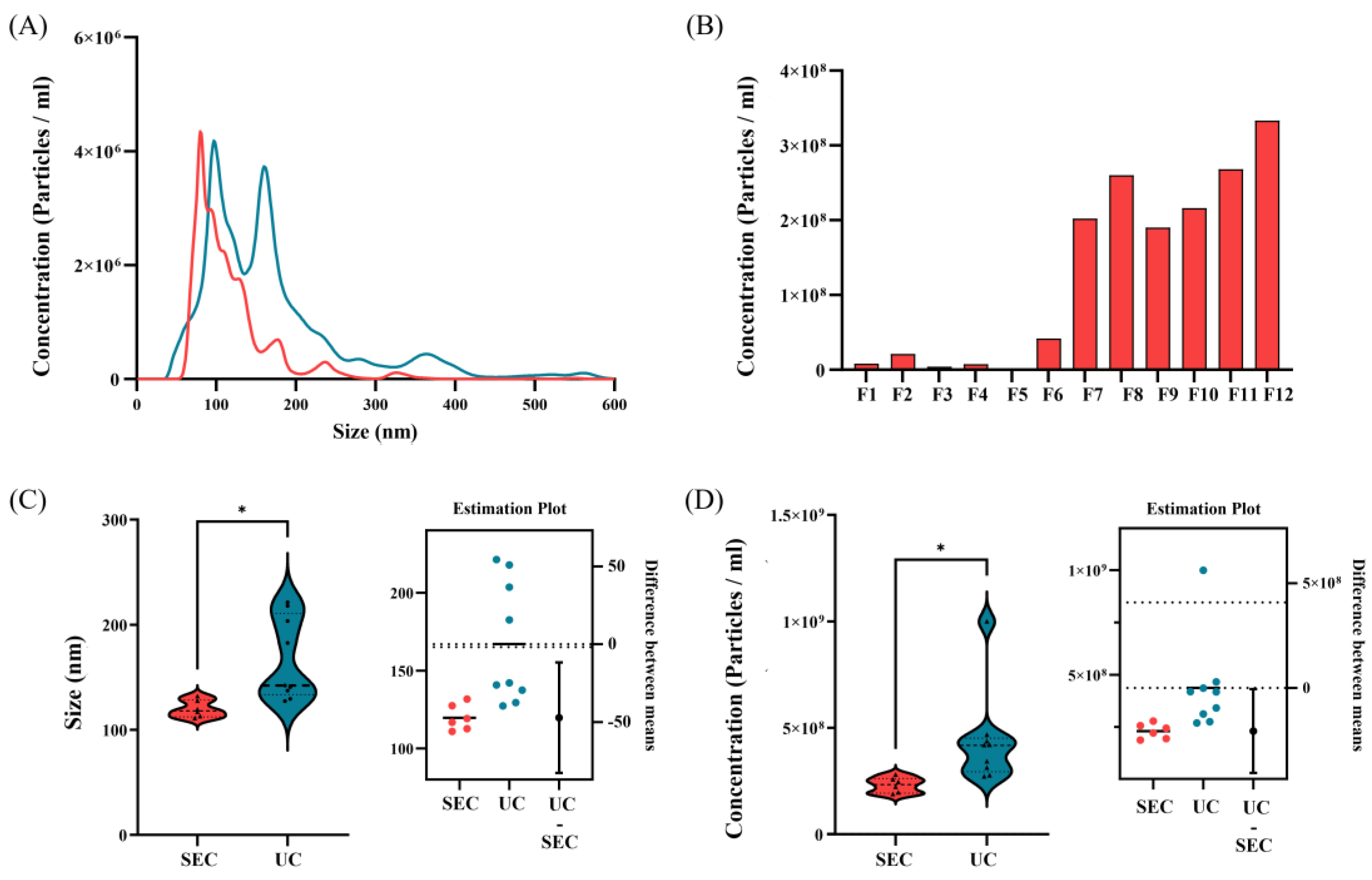
3.2. Comparison of EVs Size and Concentration
3.2.1. Comparison of EVs between Culture Media
3.2.2. Comparison of EVs According to Isolation Methods
3.3. EVs Derived from L. crispatus PMC201 Confirmed by TEM
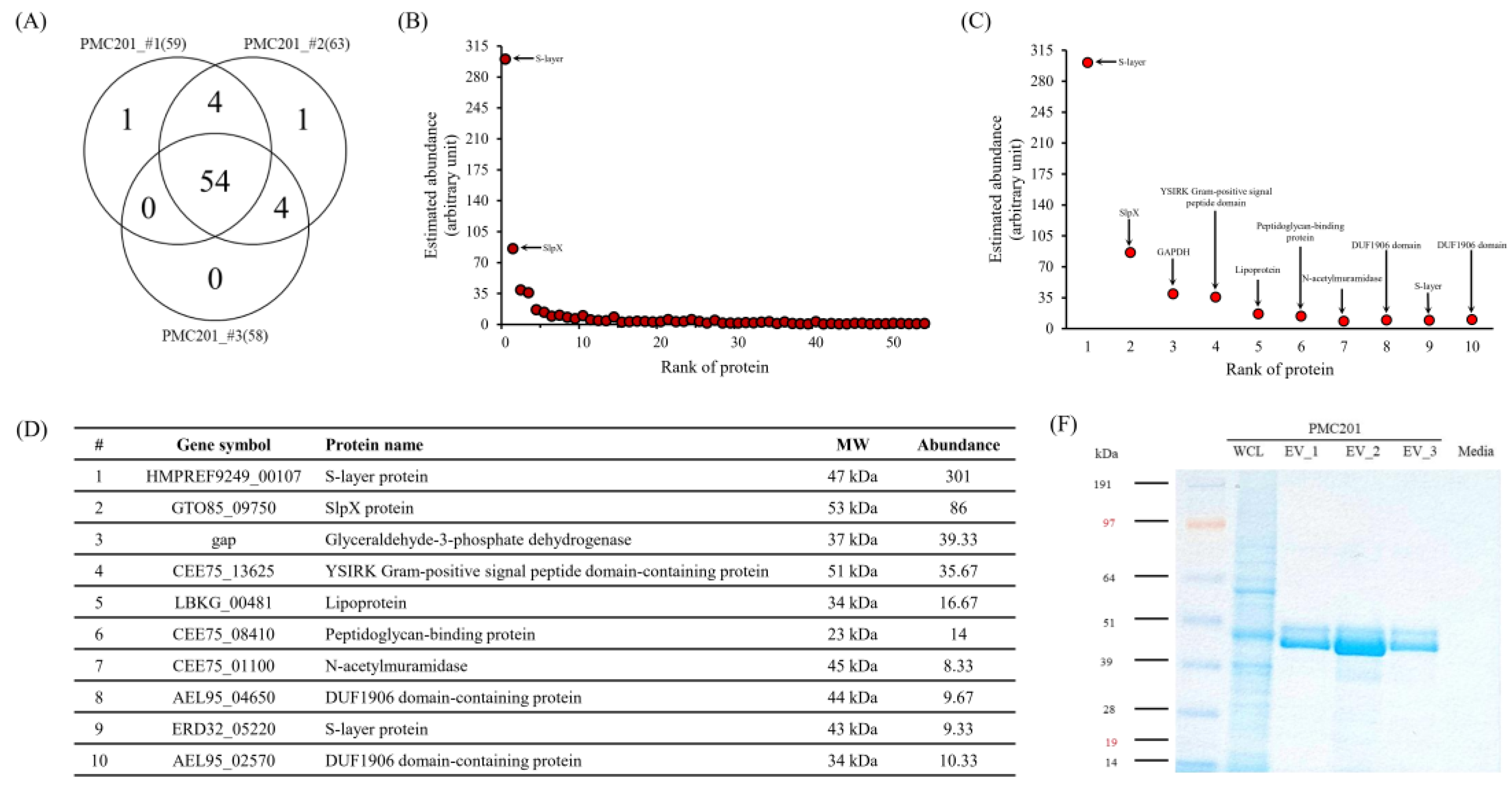
3.4. Proteome Profiling of EVs Derived from L. crispatus PMC201
3.4.1. LC-MS/MS Analysis
3.4.2. Confirmation of Protein Sizes through Coomassie Blue Staining
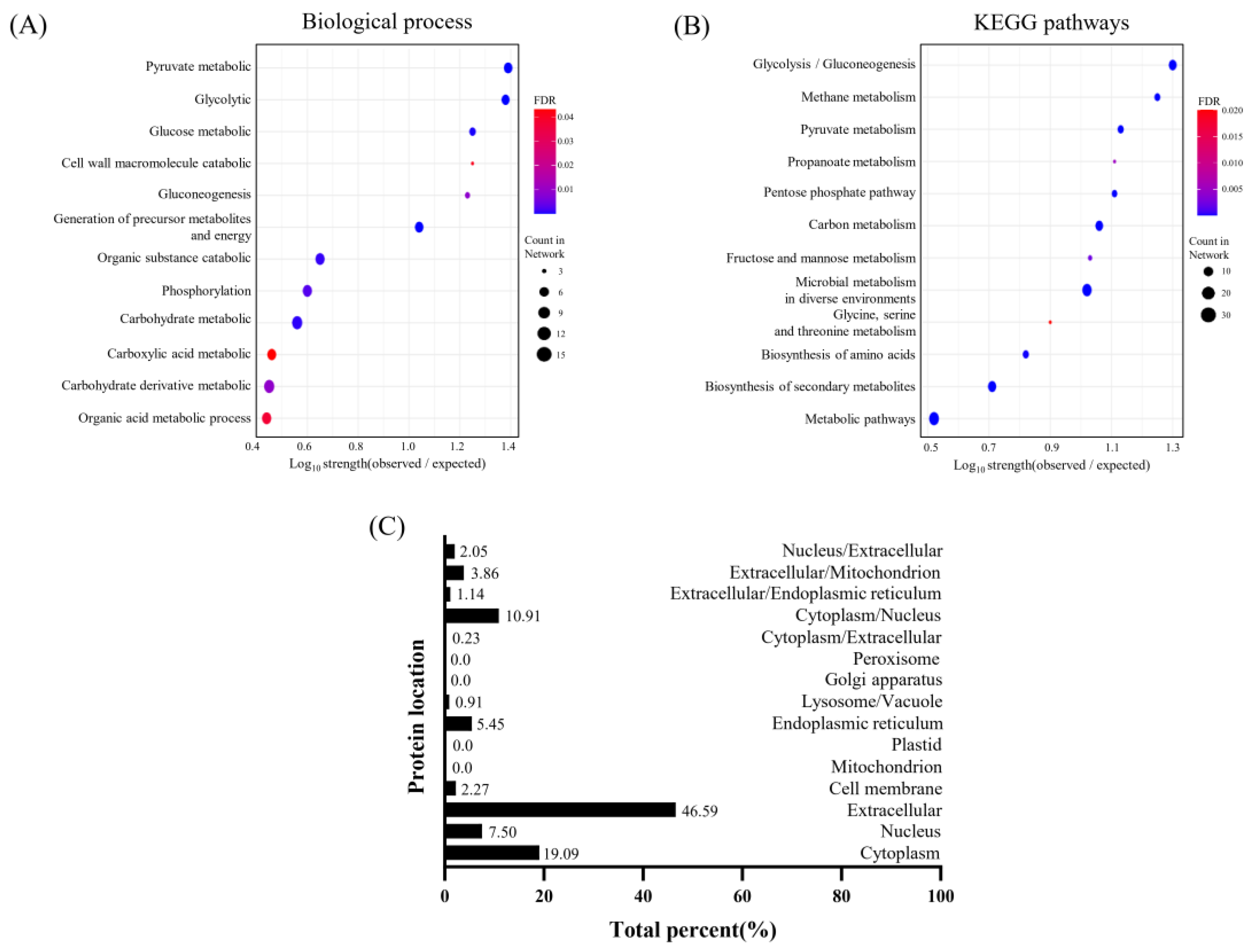
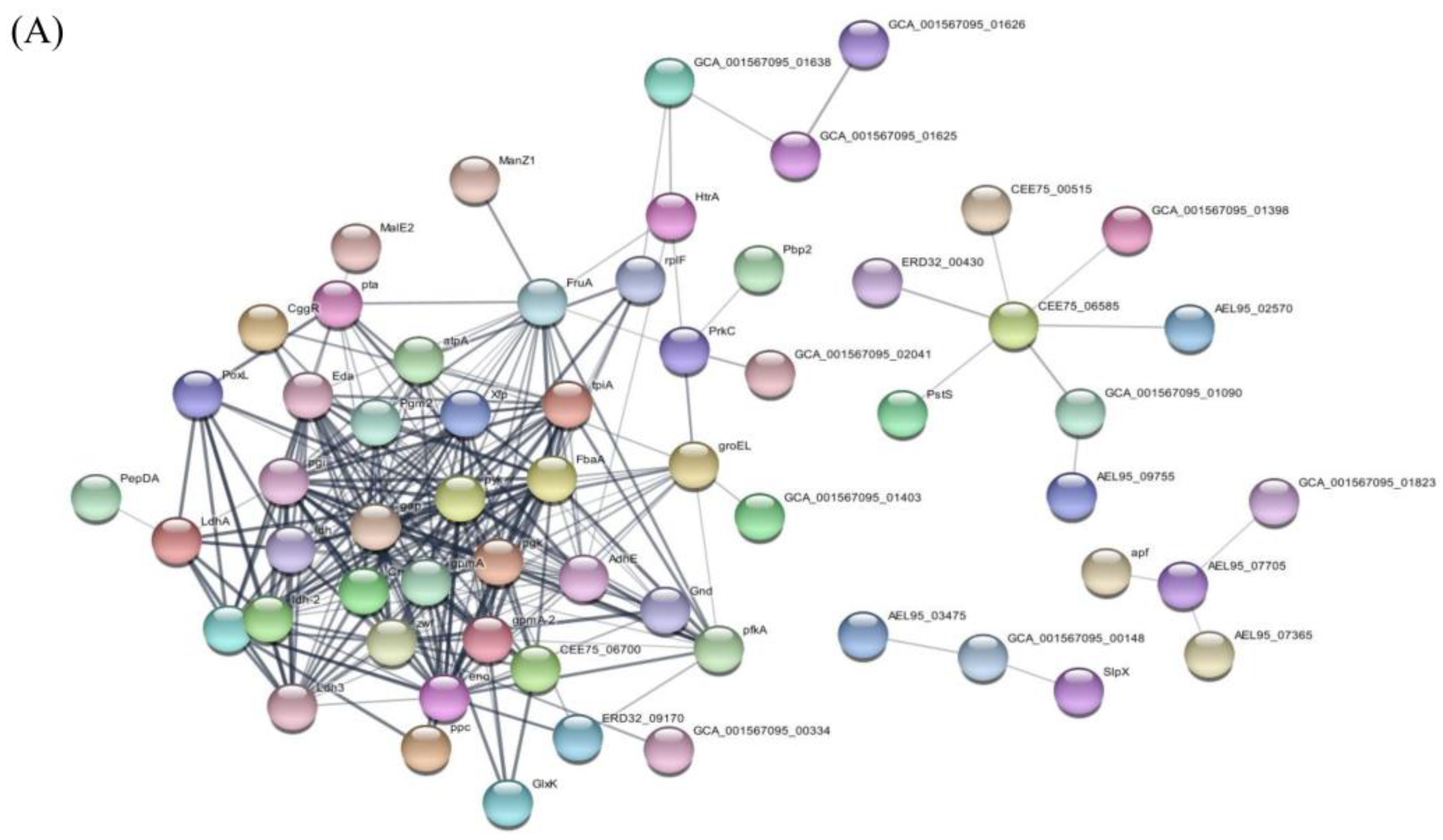
3.5. Prediction of Protein Pathways and Interaction of EVs Derived from L. crispatus PMC201
3.5.1. Cellular Components in Protein from EVs Derived from L. crispatus PMC201
3.5.2. Analysis of Biological Processes and KEGG Pathways in Proteins from EVs Derived from L. crispatus PMC201
3.5.3. Investigation into Subcellular Localization of Proteins EVs Derived from L. crispatus PMC201
3.5.4. Protein Interactions within EVs Derived from L. crispatus PMC201
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- FAO/WHO, “Health and Nutritional Properties of Probiotics in Food including Powder Milk with Live Lactic Acid Bacteria,” Oct. 2001.
- FDA, “Scientific Evaluation of the Evidence on the Beneficial Physiological Effects of Isolated or Synthetic Non-Digestible Carbohydrates Submitted as a Citizen Petition (21 CFR 10.30): Guidance for Industry.” [Online]. Available: http://www.regulations.gov.
- M. Kechagia et al., “Health Benefits of Probiotics: A Review,” ISRN Nutr, vol. 2013, pp. 1–7, Jan. 2013. [CrossRef]
- Z. Li et al., “Chemically and Biologically Engineered Bacteria-Based Delivery Systems for Emerging Diagnosis and Advanced Therapy,” Advanced Materials, vol. 33, no. 38. John Wiley and Sons Inc, Sep. 01, 2021. [CrossRef]
- B. Chugh and A. Kamal-Eldin, “Bioactive compounds produced by probiotics in food products,” Current Opinion in Food Science, vol. 32. Elsevier Ltd., pp. 76–82, Apr. 01, 2020. [CrossRef]
- L. G. Ruiz Rodríguez et al., “Diversity and functional properties of lactic acid bacteria isolated from wild fruits and flowers present in northern Argentina,” Front Microbiology, vol. 10, no. MAY, 2019. [CrossRef]
- A. Raheem, L. A. Raheem, L. Liang, G. Zhang, and S. Cui, “Modulatory Effects of Probiotics During Pathogenic Infections with Emphasis on Immune Regulation,” Frontiers in Immunology, vol. 12. Frontiers Media S.A., Apr. 08, 2021. [CrossRef]
- M. Bermudez-Brito, J. M. Bermudez-Brito, J. Plaza-Díaz, S. Muñoz-Quezada, C. Gómez-Llorente, and A. Gil, “Probiotic mechanisms of action,” Annals of Nutrition and Metabolism, vol. 61, no. 2. pp. 160–174, Oct. 2012. [CrossRef]
- M. Cunningham et al., “Shaping the Future of Probiotics and Prebiotics,” Trends in Microbiology, vol. 29, no. 8. Elsevier Ltd., pp. 667–685, Aug. 01, 2021. [CrossRef]
- K. S. Yoha, S. K. S. Yoha, S. Nida, S. Dutta, J. A. Moses, and C. Anandharamakrishnan, “Targeted Delivery of Probiotics: Perspectives on Research and Commercialization,” Probiotics and Antimicrobial Proteins, vol. 14, no. 1. Springer, pp. 15–48, Feb. 01, 2022. [CrossRef]
- S. Lin et al., “Surface-modified bacteria: synthesis, functionalization and biomedical applications,” Chemical Society Reviews, vol. 52, no. 19. Royal Society of Chemistry, pp. 6617–6643, Sep. 19, 2023. [CrossRef]
- M. Liu, J. M. Liu, J. Chen, I. P. W. Dharmasiddhi, S. Chen, Y. Liu, and H. Liu, “Review of the Potential of Probiotics in Disease Treatment: Mechanisms, Engineering, and Applications,” Processes, vol. 12, no. 2. Multidisciplinary Digital Publishing Institute (MDPI), Feb. 01, 2024. [CrossRef]
- P. Krzyżek, B. P. Krzyżek, B. Marinacci, I. Vitale, and R. Grande, “Extracellular Vesicles of Probiotics: Shedding Light on the Biological Activity and Future Applications,” Pharmaceutics, vol. 15, no. 2. MDPI, Feb. 01, 2023. [CrossRef]
- N. Díaz-Garrido, J. N. Díaz-Garrido, J. Badia, and L. Baldomà, “Microbiota-derived extracellular vesicles in interkingdom communication in the gut,” Journal of Extracellular Vesicles, vol. 10, no. 13. John Wiley and Sons Inc, Nov. 01, 2021. [CrossRef]
- J. A. Molina-Tijeras, J. J. A. Molina-Tijeras, J. Gálvez, and M. E. Rodríguez-Cabezas, “The immunomodulatory properties of extracellular vesicles derived from probiotics: A novel approach for the management of gastrointestinal diseases,” Nutrients, vol. 11, no. 5. MDPI AG, , 2019. 01 May. [CrossRef]
- R. M. Stubbendieck, C. 16. R. M. Stubbendieck, C. Vargas-Bautista, and P. D. Straight, “Bacterial communities: Interactions to scale,” Frontiers in Microbiology, vol. 7, no. AUG. Frontiers in Microbiology. [CrossRef]
- S. Domingues and K. M. Nielsen, “Membrane vesicles and horizontal gene transfer in prokaryotes,” Current Opinion in Microbiology, vol. 38, pp. 16–21, Aug. 2017. [CrossRef]
- N. Díaz-Garrido, J. N. Díaz-Garrido, J. Badia, and L. Baldomà, “Microbiota-derived extracellular vesicles in interkingdom communication in the gut,” Journal of Extracellular Vesicles, vol. 10, no. 13. John Wiley and Sons Inc, Nov. 01, 2021. [CrossRef]
- C. Mu, Y. C. Mu, Y. Yang, and W. Zhu, “Crosstalk between the Immune Receptors and Gut Microbiota,” Current Protein & Peptide Science, Volume 16, Issue 7, pp. 622-632, 2015. [CrossRef]
- B. Thay, A. B. Thay, A. Damm, T. A. Kufer, S. N. Wai, and J. Oscarsson, “Aggregatibacter actinomycetemcomitans outer membrane vesicles are internalized in human host cells and trigger NOD1- and NOD2-dependent NF-κB activation,” Infection and Immunity, vol. 82, no. 10, pp. 4034–4046, 2014. [CrossRef]
- M. Chatzidaki-Livanis, M. J. M. Chatzidaki-Livanis, M. J. Coyne, and L. E. Comstock, “An antimicrobial protein of the gut symbiont Bacteroides fragilis with a MACPF domain of host immune proteins,” Molecular Microbiology, vol. 94, no. 6, pp. 1361–1374, Dec. 2014. [CrossRef]
- R. A. ñahui Palomino, S. R. A. ñahui Palomino, S. Zicari, C. Vanpouille, B. Vitali, and L. Margolis, “Vaginal lactobacillus inhibits HIV-1 replication in human tissues ex vivo,” Frontiers in Microbiology, vol. 8, no. MAY, 17. 20 May. [CrossRef]
- Y. Lee et al., “Activity of Lactobacillus crispatus isolated from vaginal microbiota against Mycobacterium tuberculosis,” Journal of Microbiology, vol. 59, no. 11, pp. 1019–1030, 2021. [CrossRef]
- H. Tajdozian, Y. H. Tajdozian, Y. kyoung Lee, S. Kim, Y. Kyoung Jeong, and S. Kwak Kyung Chungnam Techno Park Jung-Hyun Ju, “Ecacy of lyophilized Lactobacillus sakei as a potential candidate for the prevention of carbapenem-resistant Klebsiella infection,” Research square, 2023. [CrossRef]
- J. Lee et al., “Proteomic analysis of extracellular vesicles derived from Mycobacterium tuberculosis,” Proteomics, vol. 15, no. 19, pp. 3331–3337, Oct. 2015. [CrossRef]
- D. Choi et al., “Quantitative proteomics and biological activity of extracellular vesicles engineered to express SARS-CoV-2 spike protein,” Journal of Extracellular Biology, vol. 1, no. 10, Oct. 2022. [CrossRef]
- C. Bäuerl, J. M. C. Bäuerl, J. M. Coll-Marqués, C. Tarazona-González, and G. Pérez-Martínez, “Lactobacillus casei extracellular vesicles stimulate EGFR pathway likely due to the presence of proteins P40 and P75 bound to their surface,” Scientific Reports, vol. 10, no. 1, Dec. 2020. [CrossRef]
- J. Kowal et al., “Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes,” Proceedings of the National Academy of Sciences, vol. 113, no. 8, pp. E968–E977, Feb. 2016. [CrossRef]
- W. Kim et al., “Lactobacillus plantarum-derived extracellular vesicles induce anti-inflammatory M2 macrophage polarization in vitro,” Journal of Extracell Vesicles, vol. 9, no. 1, Jan. 2020. [CrossRef]
- J. Ren, W. J. Ren, W. He, L. Zheng, and H. Duan, “From structures to functions: Insights into exosomes as promising drug delivery vehicles,” Biomaterials Science, vol. 4, no. 6. Royal Society of Chemistry, pp. 910–921, Jun. 01, 2016. [CrossRef]
- T. B. H. Geijtenbeek et al., “Mycobacteria target DC-SIGN to suppress dendritic cell function,” Journal of Experimental Medicine, vol. 197, no. 1, pp. 7–17, Jan. 2003. [CrossRef]
- L. Tallieux et al., “DC-SIGN is the major Mycobacterium tuberculosis receptor on human dendritic cells,” Journal of Experimental Medicine, vol. 197, no. 1, pp. 121–127, Jan. 2003. [CrossRef]
- M. G. Martinez, M. A. M. G. Martinez, M. A. Bialecki, S. Belouzard, S. M. Cordo, N. A. Candurra, and G. R. Whittaker, “Utilization of human DC-SIGN and L-SIGN for entry and infection of host cells by the New World arenavirus, Junín virus,” Biochemical and Biophysical Research Communications, vol. 441, no. 3, pp. 612–617, Nov. 2013. [CrossRef]
- E. A. Koppel, K. P. J. M. E. A. Koppel, K. P. J. M. van Gisbergen, T. B. H. Geijtenbeek, and Y. van Kooyk, “Distinct functions of DC-SIGN and its homologues L-SIGN (DC-SIGNR) and mSIGNR1 in pathogen recognition and immune regulation,” Cellular Microbiology, vol. 7, no. 2. pp. 157–165, Feb. 2005. [CrossRef]
- P. Konieczna et al., “Bifidobacterium infantis 35624 administration induces Foxp3 T regulatory cells in human peripheral blood: Potential role for myeloid and plasmacytoid dendritic cells,” Gut Microbes, vol. 61, no. 3, pp. 354–366, Mar. 2012. [CrossRef]
- P. Mastromarino, F. P. Mastromarino, F. Cacciotti, A. Masci, and L. Mosca, “Antiviral activity of Lactobacillus brevis towards herpes simplex virus type 2: Role of cell wall associated components,” Anaerobe, vol. 17, no. 6, pp. 334–336, Dec. 2011. [CrossRef]
- M. G. Martínez, M. M. G. Martínez, M. Prado Acosta, N. A. Candurra, and S. M. Ruzal, “S-layer proteins of Lactobacillus acidophilus inhibits JUNV infection,” Biochemical and Biophysical Research Communications, vol. 422, no. 4, pp. 590–595, Jun. 2012. [CrossRef]
- R. Hu et al., “Lactobacillus reuteri-derived extracellular vesicles maintain intestinal immune homeostasis against lipopolysaccharide-induced inflammatory responses in broilers,” Journal of Animal Science and Biotechnology, vol. 12, no. 1, Dec. 2021. [CrossRef]
- S. Wang et al., “Lipoteichoic acid from the cell wall of a heat killed Lactobacillus paracasei D3-5 ameliorates aging-related leaky gut, inflammation and improves physical and cognitive functions: from C. elegans to mice,” Geroscience, vol. 42, no. 1, pp. 333–352, Feb. 2020. [CrossRef]
- J. S. Ong et al., “Extracellular transglycosylase and glyceraldehyde-3-phosphate dehydrogenase attributed to the anti-staphylococcal activity of Lactobacillus plantarum USM8613,” Journal of Biotechnology, vol. 300, pp. 20–31, Jul. 2019. [CrossRef]
| Ingredient’s name | Commercial name of ingredients | Number of Hits |
|---|---|---|
| Soy peptone A2 SC | Soy peptone A2 SC | 10g |
| Yeast extract | Amberferm | 10g |
| Glucose | Dextrose | 20g |
| Tween 80 | Tween 80 | 1ml |
| Magnesium sulfate | Magnesium sulfate | 0.1g |
| Sodium chloride | Sodium chloride | 5g |
| Culture media / Isolation method |
MRS / SEC | FGM / SEC | FGM / UC |
|---|---|---|---|
| Mean | 136+/-7.1 nm | 119.8+/-5.3nm | 166.9+/-13.5 nm |
| Mode | 107.6+/-8.7 nm | 88.05+/-6.7 nm | 110.1+/-7.0 nm |
| SD | 62.3+/-12.6 nm | 58.6+/-12.1 nm | 89.9+/-4.1 nm |
| Concentration(particles/mL) | 1.06107+/-9.60106 | 2.31108+/-1.02107 | 3.50108+/-2.54107 |
| Accession | Identified Proteins | Alternate ID | Molecular Weight |
|---|---|---|---|
| K1NK16 | S-layer protein | HMPREF9249_00107 | 47 kDa |
| A0A6M1GK10 | S-layer protein (Fragment) | G5T20_09495 | 44 kDa |
| A0A5M9Z605 | S-layer protein | F1C07_06475 | 47 kDa |
| A0A854PP49 | Surface layer protein A domain-containing protein | AYP82_00340 | 47 kDa |
| A0A5M9Z4Y9 | S-layer protein OS=Lactobacillus crispatus | F1C07_06510 | 47 kDa |
| A0A854PLV2 | Surface layer protein A domain-containing protein | AYP82_00370 | 47 kDa |
| A0A109DEN3 | Surface layer protein A domain-containing protein | AEL95_05125 | 27 kDa |
| A0A109DCK1 | S-layer protein (Fragment) | AEL95_10035 | 49 kDa |
| A0A135YNS0 | Bacterial surface layer protein (Fragment) | HMPREF3209_02474 | 60 kDa |
| A0A7H9EBL4 | SlpX protein | GTO85_09750 | 53 kDa |
| A0A135YZ62 | Bacterial surface layer protein | ERD32_09040 | 53 kDa |
| C7XGC7 | Glyceraldehyde-3-phosphate dehydrogenase | gap | 37 kDa |
| A0A4R6CPL7 | YSIRK Gram-positive signal peptide domain-containing protein | CEE75_13625 | 51 kDa |
| E3R2H8 | Lipoprotein | LBKG_00481 | 34 kDa |
| A0A4R6CS48 | Peptidoglycan-binding protein | CEE75_08410 | 23 kDa |
| A0A4R6CWH5 | N-acetylmuramidase | CEE75_01100 | 45 kDa |
| A0A109DEL0 | DUF1906 domain-containing protein | AEL95_04650 | 44 kDa |
| A0A135Z2L1 | S-layer protein | ERD32_05220 | 43 kDa |
| A0A109DTG6 | DUF1906 domain-containing protein | AEL95_02570 | 34 kDa |
| A0A109DRM6 | PDZ domain-containing protein | AEL95_06810 | 43 kDa |
| A0A109DF87 | Cell division protein | AEL95_03480 | 63 kDa |
| A0A4R6CR16 | Gram-positive cocci surface proteins LPxTG domain-containing protein (Fragment) | CEE75_10850 | 64 kDa |
| A0A109DF34 | Surface layer protein A domain-containing protein | AEL95_03475 | 27 kDa |
| A0A120DIM9 | Enolase | eno | 47 kDa |
| A0A125P6L3 | Uncharacterized protein | AEL95_00975 | 24 kDa |
| A0A109DWU7 | Glucose-6-phosphate isomerase | pgi | 49 kDa |
| A0A135ZEW6 | HNH endonuclease | HMPREF3209_00721 | 35 kDa |
| A0A120DI73 | DUF1002 domain-containing protein | AEL95_06965 | 34 kDa |
| A0A4Q0LXU7 | Autotransporter | ERD32_01305 | 17 kDa |
| A0A4R6CRX5 | Membrane transport protein MMPL domain-containing protein | CEE75_08755 | 124 kDa |
| D0DGZ7 (+2) | Uncharacterized protein | HMPREF0508_00896 | 23 kDa |
| D0DH00 | Phage minor structural protein, N-terminal domain protein | HMPREF0508_00899 | 72 kDa |
| A0A135ZH00 | Penicillin-binding protein, transpeptidase domain protein | HMPREF3209_00031 | 74 kDa |
| A0A135ZG11 | Copper oxidase | ERD32_05870 | 59 kDa |
| A0A109DGH2 | Hydrolase | AEL95_00120 | 32 kDa |
| A0A109DGC6 | Glycosidase | AEL95_00130 | 20 kDa |
| A0A109DES4 | Surface layer protein A domain-containing protein | AEL95_04840 | 27 kDa |
| A0A109DE36 | Pyruvate oxidase | spxB | 67 kDa |
| K1MKS9 | DUF3383 family protein | HMPREF9249_02106 | 48 kDa |
| A0A109DCM5 | Lysin | AEL95_09750 | 42 kDa |
| A0A109DME9 | Large ribosomal subunit protein | rplF | 19 kDa |
| A0A109DVU6 | Phosphoglycerate kinase | pgk | 43 kDa |
| A0A135Z6Y8 | Dipeptidase | HMPREF3209_01335 | 53 kDa |
| A0A109DFB8 | Extracellular solute-binding protein | AEL95_02275 | 44 kDa |
| A0A4V6PQ54 | Phosphate-binding protein | CEE75_08510 | 31 kDa |
| A0A226V487 | non-specific serine/threonine protein kinase | pknB | 75 kDa |
| A0A120DIW3 | Surface layer protein A domain-containing protein | AEL95_02090 | 46 kDa |
| A0A4Q0LY00 | DUF2184 domain-containing protein | ERD32_01340 | 35 kDa |
| A0A109DIV5 | Aggregation promoting factor | AEL95_07365 | 25 kDa |
| A0A125P6A1 | Cell surface protein | AEL95_06085 | 42 kDa |
| A0A120DIL8 | D-2-hydroxyacid dehydrogenase | AEL95_03565 | 38 kDa |
| A0A109DF57 | Beta-lactamase family protein | AEL95_03595 | 39 kDa |
| A0A135ZB87 | Signal peptide protein, YSIRK family | CEE75_00270 | 33 kDa |
| A0A109DGV4 | Foldase protein PrsA | prsA | 33 kDa |
| A0A2I1WJR8 | Peptidase M13 | CYJ80_03380 | 73 kDa |
| A0A109DF15 | L-lactate dehydrogenase | ldh | 35 kDa |
| D0DH11 | DUF3277 domain-containing protein | HMPREF0508_00910 | 14 kDa |
| A0A120DIB4 | Bifunctional oligoribonuclease/PAP phosphatase NrnA | AEL95_05665 | 35 kDa |
| A0A109DJD1 | PTS mannose family transporter subunit IID | AEL95_07920 | 34 kDa |
| A0A2N5KZX0 | Bifunctional metallophosphatase/5'-nucleotidase | CYJ79_03285 | 91 kDa |
| A0A6M1GF03 | Phage holin | G5T20_00725 | 14 kDa |
| A0A109DVJ8 | Aggregation promoting factor surface protein | AEL95_07990 | 13 kDa |
| A0A135Z466 | pullulanase | pulA | 140 kDa |
| A0A109DPT6 | DUF5052 domain-containing protein | AEL95_09755 | 23 kDa |
| GO-term | description | count in network | strength | false discovery rate |
|---|---|---|---|---|
| GO:0005576 | Extracellular region | 8 of 29 | 0.91 | 0.0014 |
| GO:0009274 | Peptidoglycan-based cell wall | 4 of 12 | 0.99 | 0.0263 |
| GO:0005618 | Cell wall | 5 of 14 | 1.02 | 0.0122 |
| GO:0030115 | S-layer | 3 of 3 | 1.47 | 0.0145 |
| GO-term | description | count in network | strength | false discovery rate |
|---|---|---|---|---|
| GO:0006082 | Organic acid metabolic process | 12 of 128 | 0.44 | 0.0358 |
| GO:1901135 | Carbohydrate derivative metabolic process | 15 of 159 | 0.45 | 0.0088 |
| GO:0019752 | Carboxylic acid metabolic process | 11 of 113 | 0.46 | 0.0413 |
| GO:0005975 | Carbohydrate metabolic process | 15 of 121 | 0.56 | 0.00074 |
| GO:0016310 | Phosphorylation | 12 of 90 | 0.6 | 0.0027 |
| GO:1901575 | Organic substance catabolic process | 12 of 80 | 0.65 | 0.001 |
| GO:0006091 | Generation of precursor metabolites and energy | 10 of 27 | 1.04 | 8.09E-06 |
| GO:0006094 | Gluconeogenesis | 4 of 7 | 1.23 | 0.0085 |
| GO:0006006 | Glucose metabolic process | 6 of 10 | 1.25 | 0.00027 |
| GO:0016998 | Cell wall macromolecule catabolic process | 3 of 5 | 1.25 | 0.0393 |
| GO:0006096 | Glycolytic process | 9 of 11 | 1.38 | 1.33E-06 |
| GO:0006090 | Pyruvate metabolic process | 10 of 12 | 1.39 | 2.85E-07 |
| Pathway | description | count in network | strength | false discovery rate |
|---|---|---|---|---|
| lcr01100 | Metabolic pathways | 31 of 240 | 0.52 | 2.42E-08 |
| lcr01110 | Biosynthesis of secondary metabolites | 20 of 100 | 0.71 | 6.37E-08 |
| lcr01230 | Biosynthesis of amino acids | 10 of 39 | 0.82 | 6.75E-05 |
| lcr00260 | Glycine, serine and threonine metabolism | 4 of 13 | 0.9 | 0.0201 |
| lcr01120 | Microbial metabolism in diverse environments | 27 of 66 | 1.02 | 6.09E-17 |
| lcr00051 | Fructose and mannose metabolism | 5 of 12 | 1.03 | 0.0023 |
| lcr01200 | Carbon metabolism | 17 of 38 | 1.06 | 5.45E-11 |
| lcr00030 | Pentose phosphate pathway | 8 of 16 | 1.11 | 1.22E-05 |
| lcr00640 | Propanoate metabolism | 4 of 8 | 1.11 | 0.0052 |
| lcr00620 | Pyruvate metabolism | 10 of 19 | 1.13 | 4.05E-07 |
| lcr00680 | Methane metabolism | 9 of 13 | 1.25 | 4.05E-07 |
| lcr00010 | Glycolysis / Gluconeogenesis | 18 of 23 | 1.3 | 1.10E-14 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





