Submitted:
25 April 2024
Posted:
25 April 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Bacterial Culture
2.2. Identification of RR120 Dye Decolorizing Bacteria
2.3. Decolourisation Studies
2.4. Aeration, Inoculum Size and Time for Dye Decolorization
2.4.1. Optimization by Taguchi Method
2.5. Development of Consortium
2.6. Immobilization of Dye-Degrading Bacteria
2.7. Decolourization of Various Azo Dyes and Mixture of Dyes
2.8. Qualitative Estimation of EPS Production by Dye-Degrading Bacteria
2.9. Dye Decolorization in the Presence of Metal Ions
2.10. Decolorization of Textile Fabric
2.11. Determination of Plant Growth-PromotingTraits
2.11.1. Qualitative Assessment of IAA Synthesis
2.11.2. Qualitative Assessment of Ammonium Production
2.12. Enzyme Assay
2.13. Decolorization and Biodegradation Analysis
2.13.1. UV-Vis Spectroscopic Study
2.13.2. Analysis of Dye Metabolites Using FTIR and GC-MS
3. Result
3.1. Identification of RR120 Dye Decolorizing Bacteria
3.2. Decolourisation Studies
3.3. Optimization of Various Parameters for Decolourizationo of Dye RR120
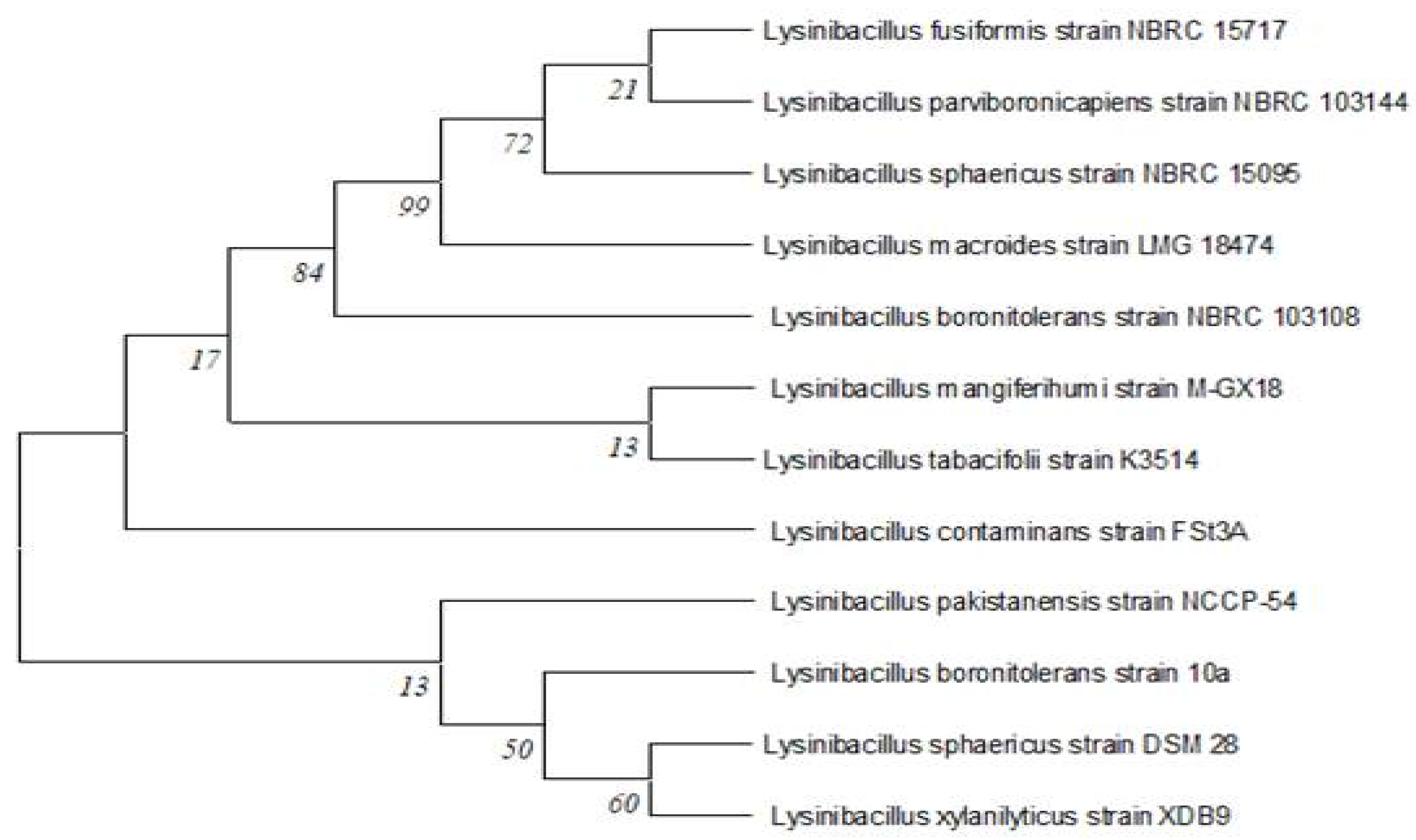
3.3.1. Effects of Aeration, Incubation Time and Inoculum Size
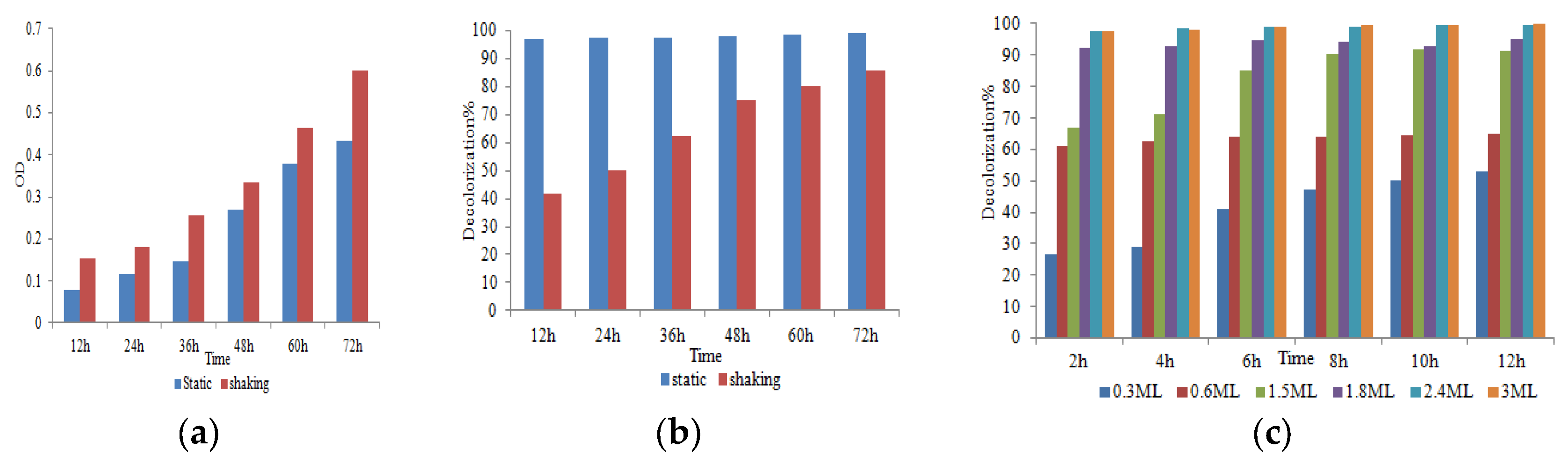
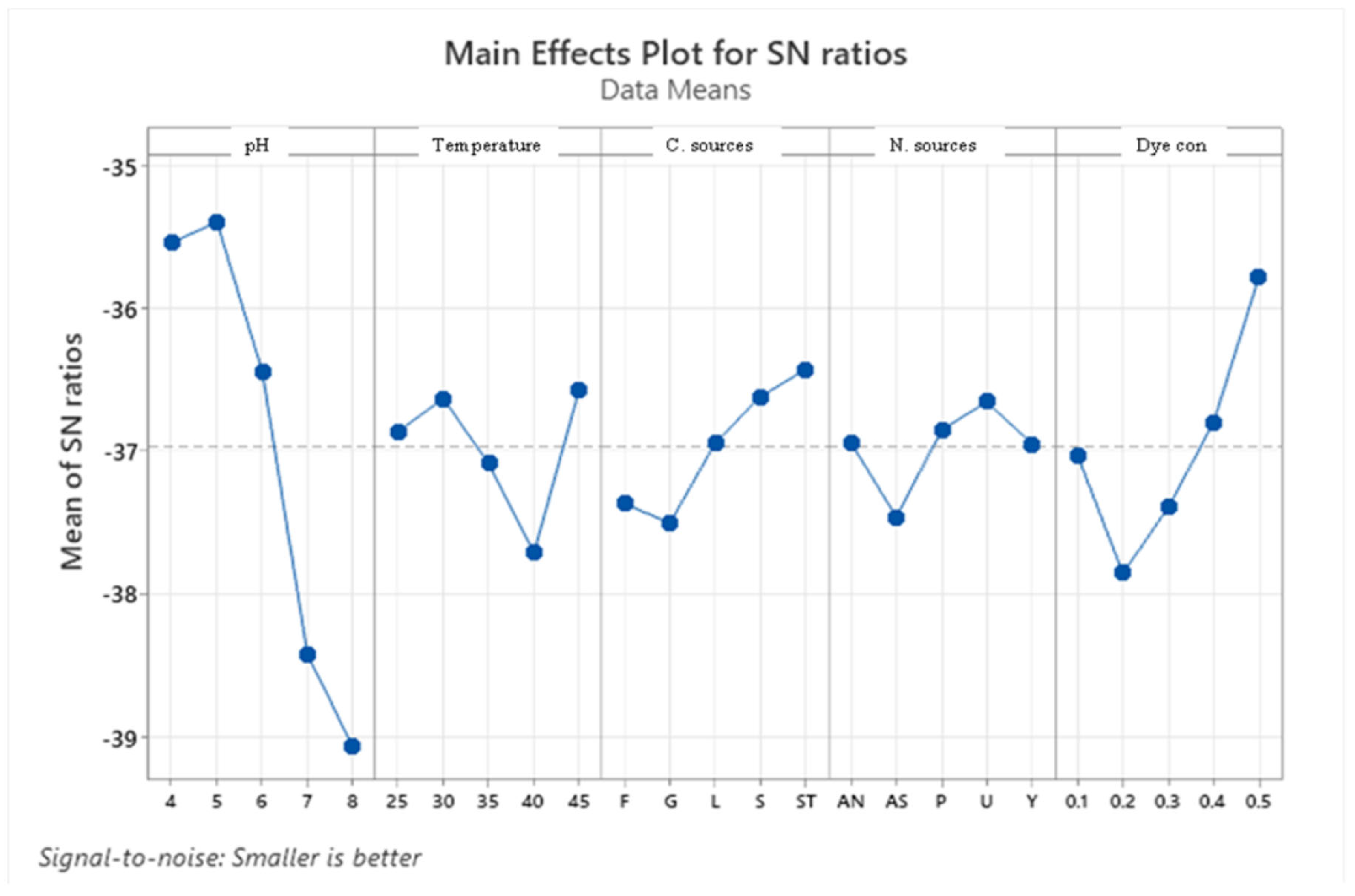
3.3.4. Taguchi Statistical Analysis
3.4. Dye Decolourization by Consortium
3.5. Immobilization of Dye-Degrading Lysinibacillus Capsici PB300(T) Bacteria
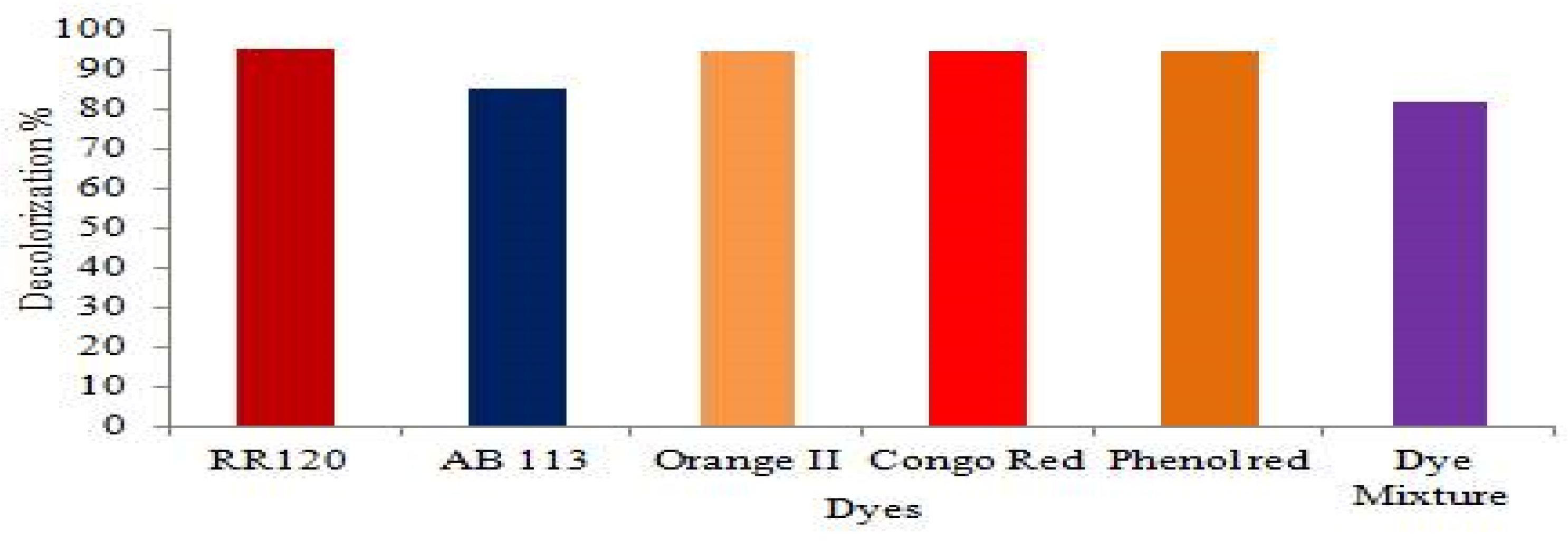
3.6. Decolourization of Reactive Dye and Dye Mixtures
3.7. EPS Production
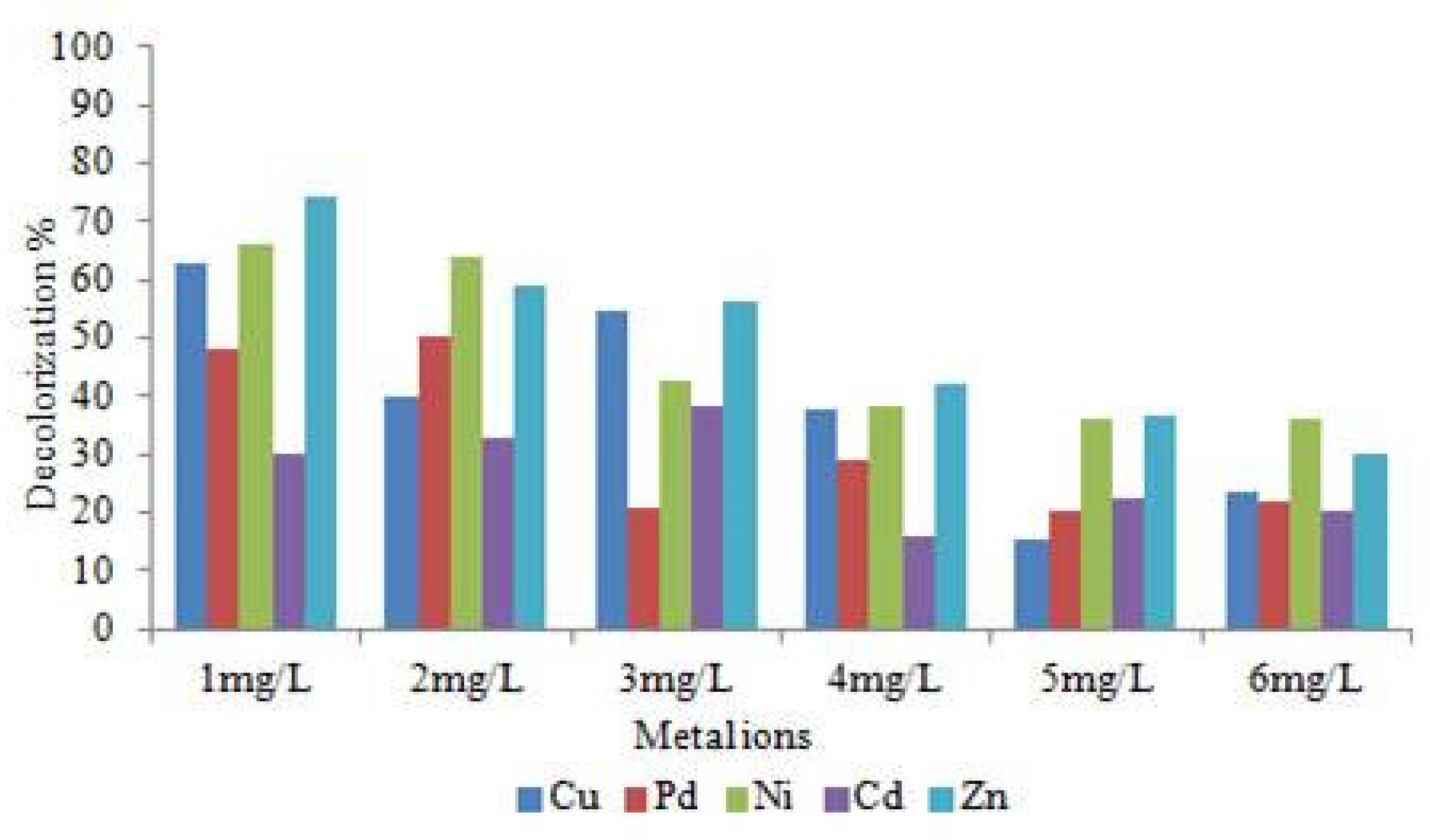
3.8. Effect of Heavy Mmetals Ions andNaCl on Decolourizationof RR120
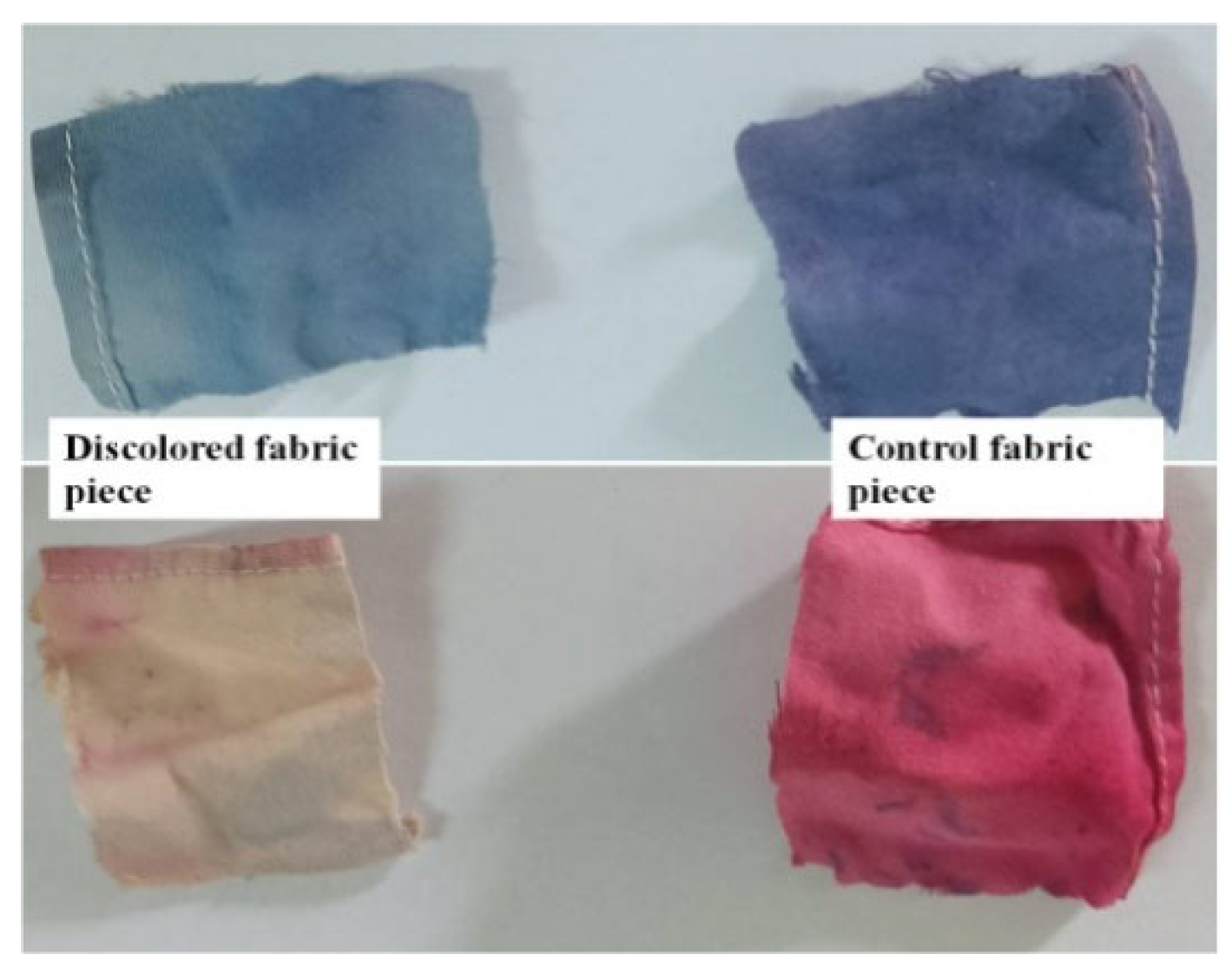
3.9. Decolourization of Textile Fabric
3.10. Qualitative Estimation of Plant Growth-Promoting IAA and Ammonium Production
3.12. Enzyme Assay
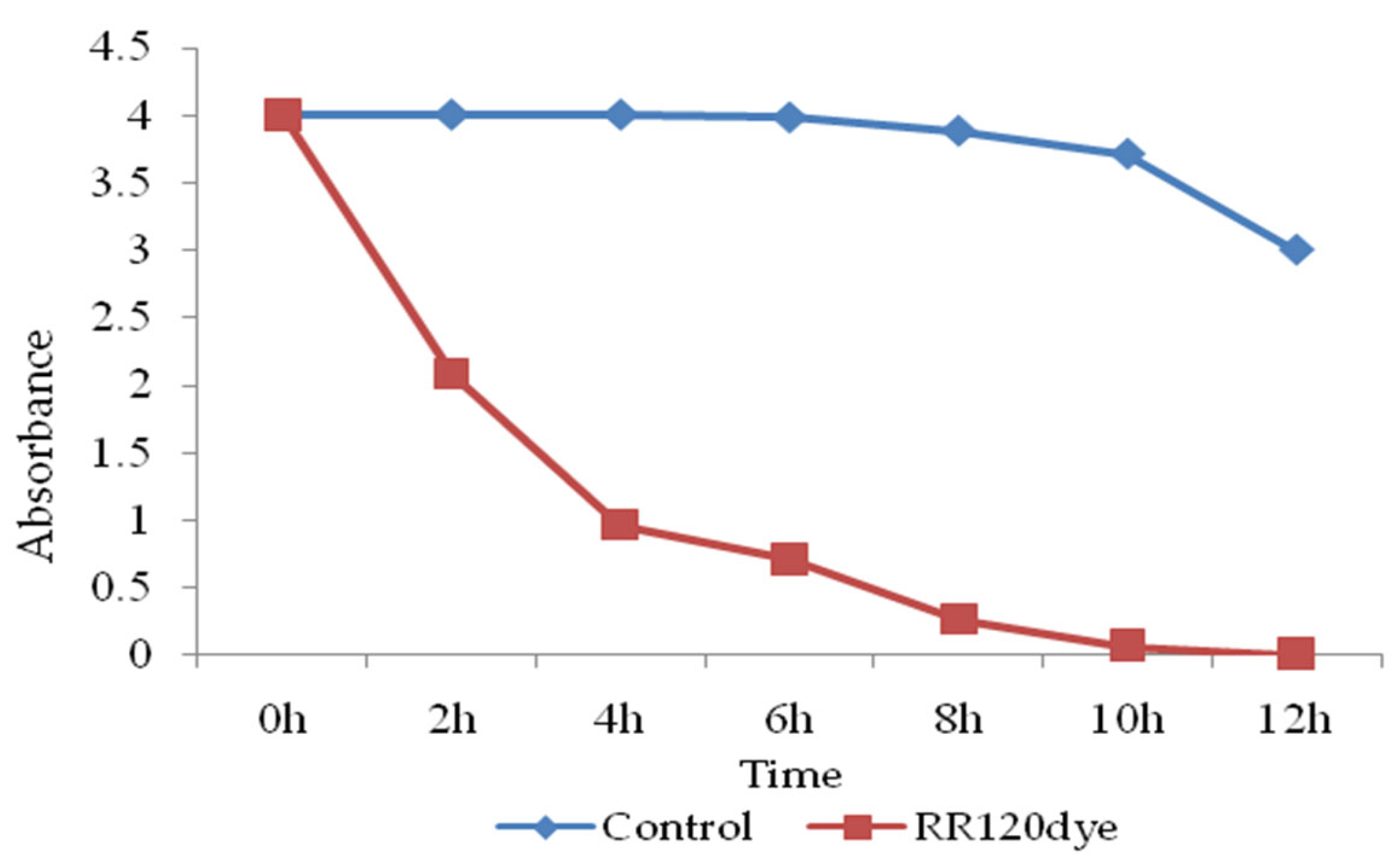
3.13. Decolorization and Biodegradation Analysis
3.13.1. UV-Vis Spectroscopic Analysis
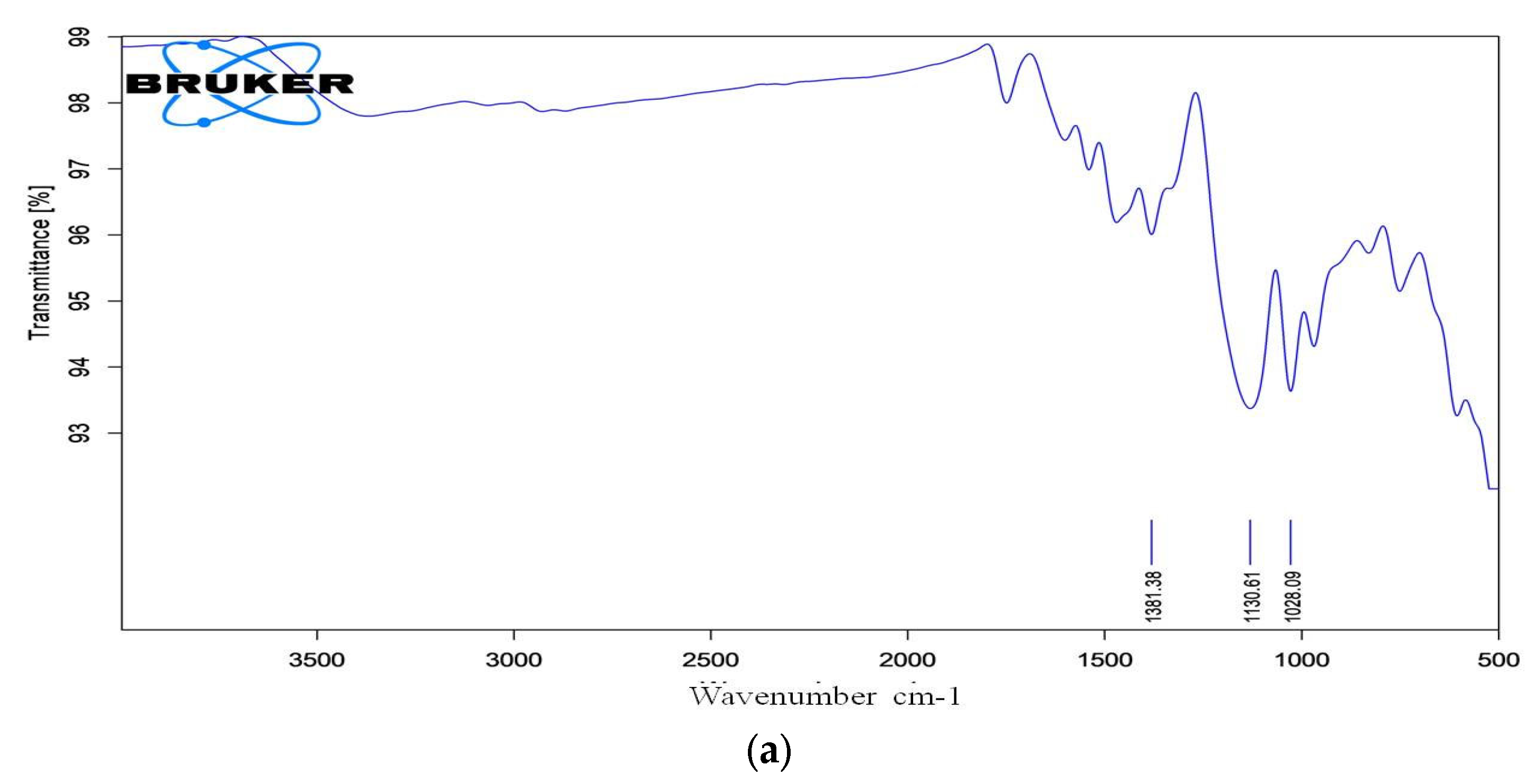
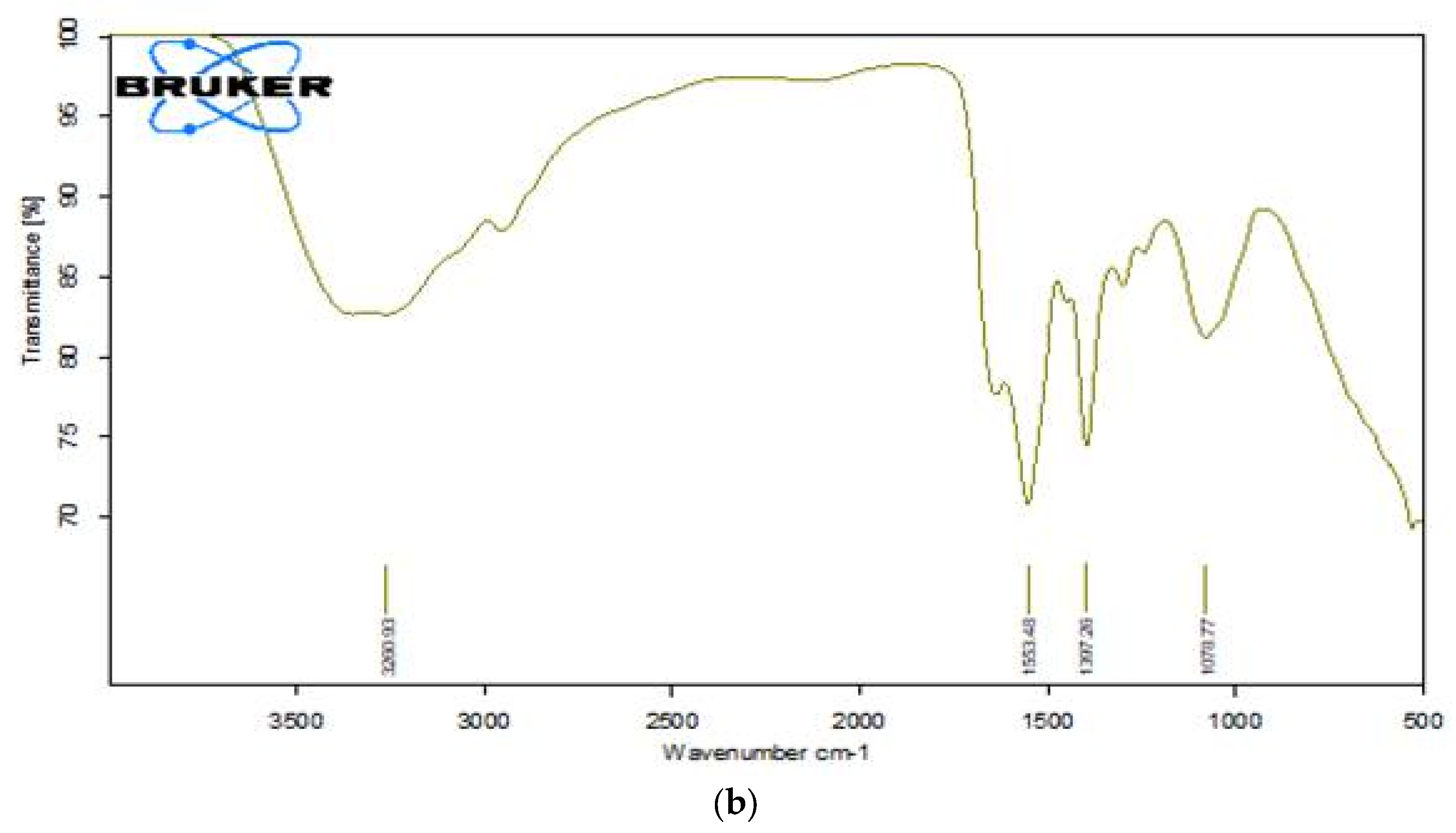
3.13.2. FTIR Analysis of Dye RR120 Metabolites
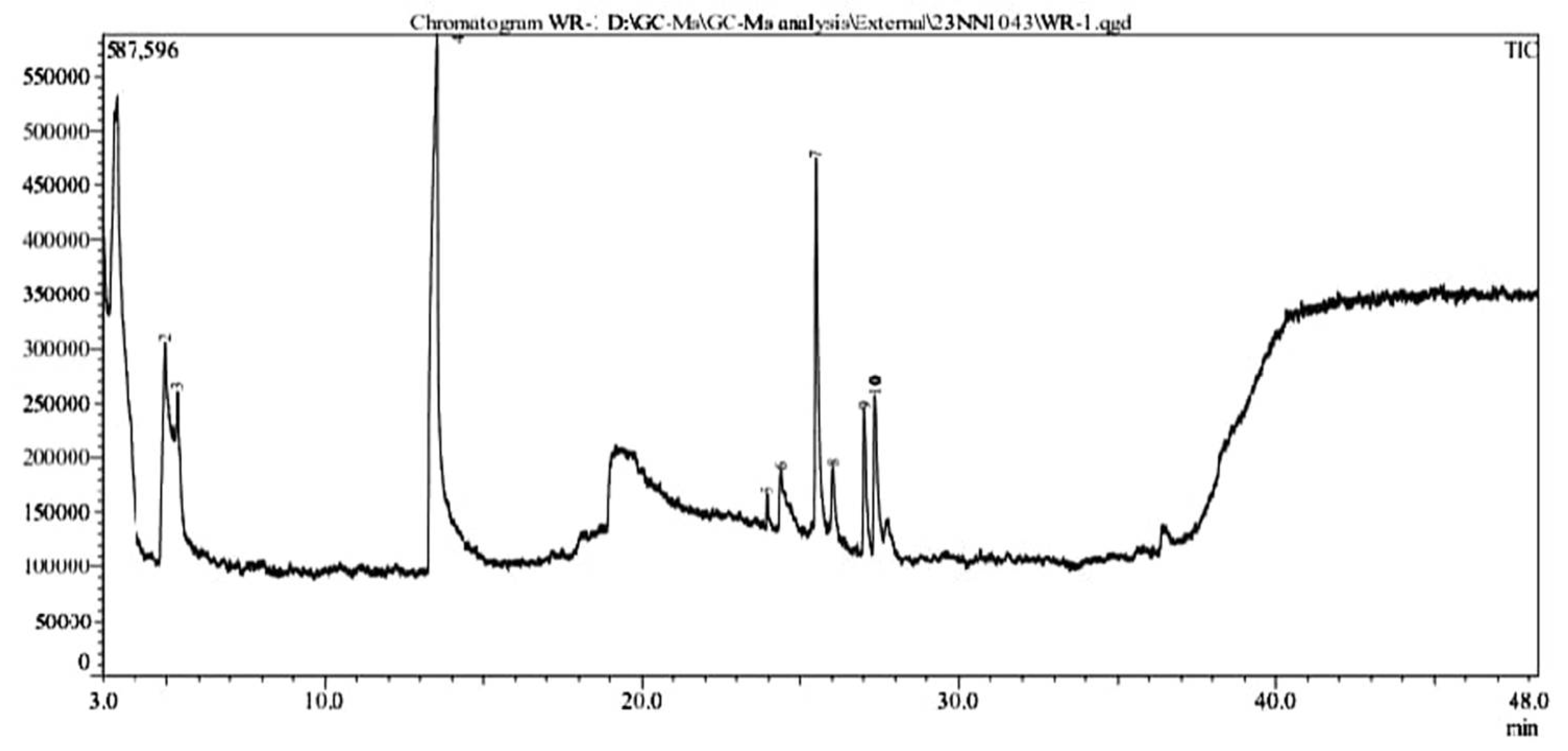
3.13.3. GC-MS Analysis of Metabolites Produced by Lysinibacillus capsici PB300(T)
4. Discussion
4.1. Isolation of Dye Decolorizing BacterialStrains
4.2. Effects of Various Parameters
4.3. RR120 Decolorization by Immobilised L. capsici PB300 (T)
4.4. RR120 Decolorization by Consortium Immobilised L. capsici PB300 (T)
4.5. Decolorization of Many Azo Dyes and Dye Mixtures
4.6. Heavy Metal Ions Tolerance Activity
4.7. Plant Growth Promotion and EPS Production
4.8. Enzyme Assay
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ayele, A.; Getachew, D.; Kamaraj, M.; Suresh, A. Phytoremediation of synthetic dyes: an effective and eco-friendly algal technology for the dye abatement. Journal of Chemistry 2021, 1–14. [Google Scholar]
- Rehaman, S.; Aravindan, G.; Karthick, G. Spectral studies of Azo Dye degradation using selected Biofertilizer of Pseudomonas fluorescens. Int. J. Life Sci. Pharma Res, 2021, 11, L73–79. [Google Scholar]
- Florencio, T.D.M.; de Godoi, L.A.; Rocha, V.C.; Oliveira, J.M.S.; Motteran, F.; Gavazza, S.; Damianovic, M.H.R.Z. Anaerobic structured-bed reactor for azo dye decolourization in the presence of sulfate ions. Journal of Chemical Technology & Biotechnology, 2021, 96, 1700–1708. [Google Scholar]
- Carolin, C.F.; Kumar, P.S.; Joshiba, G.J. A sustainable approach to decolourize methyl orange dye from aqueous solution using novel bacterial strain and its metabolites characterization. Clean Technologies and Environmental Policy 2021, 23, 173–181. [Google Scholar] [CrossRef]
- Kour, D.; Khan, S.S.; Kour, H.; Kaur, T.; Devi, R.; Rai, P.K.; Yadav, A.N. Microbe-mediated bioremediation: Current research and future challenges. Journal of Applied Biology and Biotechnology, 2023, 10, 6–24. [Google Scholar] [CrossRef]
- Kusumlata; Ambade, B.; Kumar, A.; Gautam, S. Sustainable Solutions: Reviewing the Future of Textile Dye Contaminant Removal with Emerging Biological Treatments. Limnological Review. 2024, 24, 126–49. [Google Scholar]
- Sreedharan, V.; Saha, P.; Rao, K.V.B. Dye degradation potential of Acinetobacter baumannii strain VITVB against commercial azo dyes. Bioremediation Journal 2021, 25, 347–368. [Google Scholar] [CrossRef]
- Lalnunhlimi, S.; Krishnaswamy, V. Decolorization of azo dyes (Direct Blue 151 and Direct Red 31) by moderately alkaliphilic bacterial consortium. brazilian journal of microbiology, 2016, 47, 39–46. [Google Scholar] [CrossRef]
- Lade, H.; Govindwar, S.; Paul, D. Low-cost biodegradation and detoxification of textile azo dye CI reactive blue 172 by Providencia rettgeri strain HSL1. Journal of Chemistry 2015.
- Chaturvedi, A.; Rai, B.N.; Singh, R.S.; Jaiswal, R.P. A computational approach to incorporate metabolite inhibition in the growth kinetics of indigenous bacterial strain Bacillus subtilis MN372379 in the treatment of wastewater containing Congo red dye. Applied Biochemistry and Biotechnology, 2021, 193, 2128–2144. [Google Scholar] [CrossRef]
- Chen, G.; An, X.; Li, H.; Lai, F.; Yuan, E.; Xia, X.; Zhang, Q. Detoxification of azo dye Direct Black G by thermophilic Anoxybacillus sp. PDR2 and its application potential in bioremediation. Ecotoxicology and Environmental Safety, 2021, 214, 112084. [Google Scholar] [CrossRef] [PubMed]
- Dixit, S.; Garg, S. Enzymatic degradation of sulphonated azo dye using purified azoreductase from facultative Klebsiella pneumoniae. Folia Microbiologica 2021, 66, 79–85. [Google Scholar] [CrossRef]
- Guembri, M.; Neifar, M.; Saidi, M.; Ferjani, R.; Chouchane, H.; Mosbah, A.; Ouzari, H.I. Decolorization of textile azo dye Novacron Red using bacterial monoculture and consortium: Response surface methodology optimization. Water Environment Research 2021, 93, 1346–1360. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Maqbool, Z.; Shahid, M.; Shahzad, T.; Muzammil, S.; Zubair, M.; Mahmood, F. Simultaneous removal of reactive dyes and hexavalent chromium by a metal-tolerant Pseudomonas sp. WS-D/183 harbouring plant growth-promoting traits. Int J Agric Biol 2020, 23, 241–252. [Google Scholar] [CrossRef]
- Kamal, I.M.; Abdeltawab, N.F.; Ragab, Y.M.; Farag, M.A.; Ramadan, M.A. Biodegradation, decolorization, and detoxification of di-azo dye direct Red 81 by halotolerant, alkali-thermo-tolerant bacterial mixed cultures. Microorganisms 2022, 10, 994. [Google Scholar] [CrossRef]
- Ayed, L.; Bekir, K.; Jabeur, C. Modelling and optimization of biodegradation of methylene blue by Staphylococcus aureus through a statistical optimization process: a sustainable approach for waste management. Water Science and Technology 2022, 86, 380–394. [Google Scholar] [CrossRef] [PubMed]
- Telke, A.; Kalyani, D.; Jadhav, J.; Govindwar, S. Kinetics and Mechanism of Reactive Red 141 Degradation by a Bacterial Isolate Rhizobium radiobacter MTCC 8161. Acta Chimica Slovenica 2008, 55. [Google Scholar]
- Pourbabaee, A.A.; Ramezani, S.; Javaheri Daneshmand, H. Biodegradation of malachite green by Klebsiella Terrigenaptcc 1650: the critical parameters were optimized using Taguchi optimization method. J. Bioremed. Biodeg, 2013, 4, 175. [Google Scholar]
- Marvi-Mashhadi, A.; Sharifmoghadam, M.R.; Goharimanesh, M.; Marvi-Mashhadi, M.; Dehghan, H.; Bahreini, M. Methyl red biodegradation based on Taguchi method by two novel bacteria. International Journal of Environmental Science and Technology 2021, 1–12. [Google Scholar] [CrossRef]
- Thiruppathi, K.; Rangasamy, K.; Ramasamy, M.; Muthu, D. Evaluation of textile dye degrading potential of ligninolytic bacterial consortia. Environmental Challenges, 2021, 4, 100078. [Google Scholar] [CrossRef]
- Hameed, B.B.; Ismail, Z.Z. Decolorization, biodegradation and detoxification of reactive red azo dye using non-adapted immobilized mixed cells. Biochemical Engineering Journal, 2018, 137, 71–77. [Google Scholar] [CrossRef]
- Kathiravan, M.N.; Praveen, S.A.; Gim, G.H.; Han, G.H.; Kim, S.W. Biodegradation of Methyl Orange by alginate-immobilized Aeromonas sp. in a packed bed reactor: external mass transfer modelling. Bioprocess and biosystems engineering, 2014, 37, 2149–2162. [Google Scholar] [CrossRef]
- Kaur, J.; Mudgal, G.; Negi, A.; Tamang, J.; Singh, S.; Singh, G.B.; Kesari, K.K. Reactive Black-5, Congo Red and Methyl Orange: Chemical Degradation of Azo-Dyes by Agrobacterium. Water, 2023, 15, 1664. [Google Scholar] [CrossRef]
- Islam, S.; Akanda, A.M.; Prova, A.; Islam, M.T.; Hossain, M.M. Isolation and identification of plant growth promoting rhizobacteria from cucumber rhizosphere and their effect on plant growth promotion and disease suppression. Frontiers in microbiology, 2016, 6, 1360. [Google Scholar] [CrossRef]
- Passari, A.K.; Mishra, V.K.; Leo, V.V.; Gupta, V.K.; Singh, B.P. Phytohormone production endowed with antagonistic potential and plant growth promoting abilities of culturable endophytic bacteria isolated from Clerodendrum colebrookianum Walp. Microbiological research, 2016, 193, 57–73. [Google Scholar] [CrossRef]
- Thanavel, M.; Bankole, P.O.; Selvam, R.; Govindwar, S.P.; Sadasivam, S.K. Synergistic effect of biological and advanced oxidation process treatment in the biodegradation of Remazol yellow RR dye. Scientific reports, 2020, 10, 20234. [Google Scholar] [CrossRef]
- Patel, V.R.; Bhatt, N. Isolation, development and identification of salt-tolerant bacterial consortium from crude-oil-contaminated soil for degradation of di-azo dye Reactive Blue 220. Water Science and Technology, 2015, 72, 311–321. [Google Scholar] [CrossRef]
- Vinayak, A.; Singh, G.B. Synthetic azo dye bio-decolorization by Priestia sp. RA1: process optimization and phytotoxicity assessment. Archives of Microbiology, 2022, 204, 318. [Google Scholar] [CrossRef]
- Singh, G.; Dwivedi, S.K. Decolorization and degradation of Direct Blue-1 (Azo dye) by newly isolated fungus Aspergillus terreus GS28, from sludge of carpet industry. Environmental technology & innovation 2020, 18, 100751. [Google Scholar]
- Abd El-Rahim, W.M.; Moawad, H.; Azeiz, A.Z.; Sadowsky, M.J. Biodegradation of azo dyes by bacterial or fungal consortium and identification of the biodegradation products. The Egyptian Journal of Aquatic Research. 2021, 47, 269–76. [Google Scholar] [CrossRef]
- Henagamage, A.P.; Peries, C.M. Degradation and decolorization of textile azo dyes by effective fungal-bacterial consortium. Molecular Biology Reports, 2023, 50, 8901–8914. [Google Scholar] [CrossRef]
- Thanavel, M.; Bankole, P.O.; Selvam, R.; Govindwar, S.P.; Sadasivam, S.K. Synergistic effect of biological and advanced oxidation process treatment in the biodegradation of Remazol yellow RR dye. Scientific reports, 2020, 10, 20234. [Google Scholar] [CrossRef]
- Si, J.; Peng, F.; Cui, B. Purification, biochemical characterization and dye decolorization capacity of an alkali-resistant and metal-tolerant laccase from Trametes pubescent. Bioresource Technology, 2013, 128, 49–57. [Google Scholar] [CrossRef]
- Haque, MM.; Hossen, MN.; Rahman, A.; Roy, J.; Talukder, MR.; Ahmed, M.; Ahiduzzaman, M.; Haque, MA. Decolorization, degradation and detoxification of mutagenic dye Methyl orange by novel biofilm-producing plant growth-promoting rhizobacteria. Chemosphere 2024, 346, 140568. [Google Scholar] [CrossRef]
- Shahid, M.; Mahmood, F.; Hussain, S.; Shahzad, T.; Haider, M.Z.; Noman, M.; Mustafa, G. Enzymatic detoxification of azo dyes by a multifarious Bacillus sp. strain MR-1/2-bearing plant growth-promoting characteristics. 3 Biotech 2018, 8, 1–12. [Google Scholar] [CrossRef]
- Mahmood, F.; Shahid, M.; Hussain, S.; Shahzad, T.; Tahir, M.; Ijaz, M.; Babar, S.A.K. Potential plant growth-promoting strain Bacillus sp. SR-2-1/1 decolorized azo dyes through NADH-ubiquinone: oxidoreductase activity. Bioresource Technology 2017, 235, 176–184. [Google Scholar] [CrossRef]
- Pham, VH.; Kim, J.; Chang, S.; Bang, D. Investigating bio-inspired degradation of toxic dyes using potential multi-enzyme producing extremophiles. Microorganisms 2023, 11, 1273. [Google Scholar] [CrossRef]
- Parshetti, G.K.; Telke, A.A.; Kalyani, D.C.; Govindwar, S.P. ; Decolorization and detoxification of sulfonated azo dye methyl orange by Kocuria rosea MTCC 1532. J. Hazard Mater. 2010, 176, 503–509. [Google Scholar] [CrossRef]
- Guo, G.; Hao, J.; Tian, F.; Liu, C.; Ding, K.; Xu, J.; Zhou, W.; Guan, Z. Decolorization and detoxification of azo day by halo-alkaliphilic bacterial consortium: systematic investigations of performance, pathway and metagenome. Ecotoxicol. Environ. Saf. 2020, 204, 111073. [Google Scholar] [CrossRef]
- Sharma, S.C.D.; Sun, Q.; Li, J.; Wang, Y.; Suanon, F.; Yang, J.; Yu, C.P. Decolorization of azo dye methyl red by suspended and co-immobilized bacterial cells with mediators anthraquinone-2, 6-disulfonate and Fe3O4 nanoparticles. International Biodeterioration & Biodegradation 2016, 112, 88–97. [Google Scholar]
- Singh, R.P.; Singh, P.K.; Singh, R.L. Bacterial decolorization of textile azo dye acid orange by Staphylococcus hominis RMLRT03. Toxicology international 2014, 21, 160. [Google Scholar] [CrossRef]
- Ameenudeen, S.; Unnikrishnan, S.; Ramalingam, K. Statistical optimization for the efficacious degradation of reactive azo dyes using Acinetobacter baumannii JC359. Journal of Environmental Management 2021, 279, 111512. [Google Scholar] [CrossRef]
- Radhika, B.; Aruna, K. Elucidating a pathway for degradation of azo dye reactive red 120 BY bacterial consortium. Journal of Applied Biological Sciences, 2022, 16, 396–417. [Google Scholar]
- Anwar, F.; Hussain, S.; Ramzan, S.; Hafeez, F.; Arshad, M.; Imran, M.; Abbas, N. Characterization of reactive red-120 decolorizing bacterial strain Acinetobacter junii FA10 capable of simultaneous removal of azo dyes and hexavalent chromium. Water, Air, & Soil Pollution 2014, 225, 1–16. [Google Scholar]
- Agrawal, S.; Tipre, D.; Dave, S.R. Biotreatment of azo dye containing textile industry effluent by a developed bacterial consortium immobilised on brick pieces in an indigenously designed packed bed biofilm reactor. World Journal of Microbiology and Biotechnology 2023, 39, 83. [Google Scholar] [CrossRef]
- Maheswari, N.U.; Sivagami, S. Biological degradation of textile dyes using marine Bacillus species. Int J Pure Appl Biosci 2016, 4, 123–128. [Google Scholar] [CrossRef]
- Mishra, S.; Maiti, A. Optimization of process parameters to enhance the bio-decolorization of Reactive Red 21 by Pseudomonas aeruginosa 23N1. International Journal of Environmental Science and Technology 2019, 16, 6685–6698. [Google Scholar] [CrossRef]
- Joe, M.H.; Lim, S.Y.; Kim, D.H.; Lee, I.S. Decolorization of reactive dyes by Clostridium bifermentans SL186 isolated from contaminated soil. World Journal of Microbiology and Biotechnology 2008, 24, 2221–2226. [Google Scholar] [CrossRef]
- Kurade, M.B.; Waghmode, T.R.; Govindwar, S.P. Preferential biodegradation of structurally dissimilar dyes from a mixture by Brevibacillus laterosporus. Journal of hazardous materials 2011, 192, 1746–1755. [Google Scholar] [CrossRef]
- Zhuang, M.; Sanganyado, E.; Xu, L.; Zhu, J.; Li, P.; Liu, W. High throughput sediment DNA sequencing reveals azo dye degrading bacteria inhabit nearshore sediments. Microorganisms 2020, 8, 233. [Google Scholar] [CrossRef]
- Kalpana, R.; Vignesh, N.S.; Vinothini, K.; Rajan, M.; Ashokkumar, B.; Brindhadevi, K.; Varalakshmi, P. Carbon quantum dots (CQD) fabricated from Exiguobacterium sp. VK2 exopolysaccharide (EPS) using hydrothermal reaction and its biodiesel applications. Fuel 2023, 333, 126426. [Google Scholar] [CrossRef]
- Kishor, R.; Purchase, D.; Saratale, G.D.; Ferreira, L.F.; Bilal, M.; Iqbal, H.M.; Bharagava, R.N. Environment friendly degradation and detoxification of Congo red dye and textile industry wastewater by a newly isolated Bacillus cohnni (RKS9). Environmental Technology & Innovation 2021, 22, 101425. [Google Scholar]

| Run | Factors | RR120 Decolourization % |
Mean values |
S/N Ratio | ||||
|---|---|---|---|---|---|---|---|---|
| pH | Temp. | Carbon source |
Nitrogen Source |
Dye concentration |
||||
| L1 | 4 | 25 | G | Y | 0.1 | 59.305 | 59.305 | -35.4618 |
| L2 | 4 | 30 | F | P | 0.2 | 67.63 | 67.63 | -36.6028 |
| L3 | 4 | 35 | S | AS | 0.3 | 64.04 | 64.04 | -36.129 |
| L4 | 4 | 40 | L | AN | 0.4 | 66.62 | 66.62 | -36.4721 |
| L5 | 4 | 45 | ST | U | 0.5 | 44.8 | 44.8 | -33.0256 |
| L6 | 5 | 25 | F | AS | 0.4 | 62.74 | 62.74 | -35.9509 |
| L7 | 5 | 30 | S | AN | 0.5 | 44.32 | 44.32 | -32.932 |
| L8 | 5 | 35 | L | U | 0.1 | 59.335 | 59.335 | -35.4662 |
| L9 | 5 | 40 | ST | Y | 0.2 | 65.825 | 65.825 | -36.3678 |
| L10 | 5 | 45 | G | P | 0.3 | 64.91 | 64.91 | -36.2462 |
| L11 | 6 | 25 | S | U | 0.2 | 70.515 | 70.515 | -36.9656 |
| L12 | 6 | 30 | L | Y | 0.3 | 66.255 | 66.255 | -36.4244 |
| L13 | 6 | 35 | ST | P | 0.4 | 57.39 | 57.39 | -35.1767 |
| L14 | 6 | 40 | G | AS | 0.5 | 72.945 | 72.945 | -37.2599 |
| L15 | 6 | 45 | F | AN | 0.1 | 65.86 | 65.86 | -36.3724 |
| L16 | 7 | 25 | L | P | 0.5 | 69.87 | 69.87 | -36.8858 |
| L17 | 7 | 30 | ST | AS | 0.1 | 84.27 | 84.27 | -38.5135 |
| L18 | 7 | 35 | G | AN | 0.2 | 98.67 | 98.67 | -39.8837 |
| L19 | 7 | 40 | F | U | 0.3 | 90.495 | 90.495 | -39.1325 |
| L20 | 7 | 45 | S | Y | 0.4 | 76.91 | 76.91 | -37.7197 |
| L21 | 8 | 25 | ST | AN | 0.3 | 89.72 | 89.72 | -39.0578 |
| L22 | 8 | 30 | G | U | 0.4 | 85.945 | 85.945 | -38.6844 |
| L23 | 8 | 35 | F | Y | 0.5 | 87.095 | 87.095 | -38.7999 |
| L24 | 8 | 40 | S | P | 0.1 | 92.75 | 92.75 | -39.3463 |
| L25 | 8 | 45 | L | AS | 0.2 | 94.135 | 94.135 | -39.475 |
| Level | pH | Temperature | Carbon source | Nitrogen source | Dye concentration | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean | S/N | Mean | S/N | Mean | S/N | Mean | S/N | Mean | S/N | |
| 1 | 60.48 | -35.54 | 70.43 | -36.86 | 74.76 | -37.37 | 73.04 | -36.94 | 72.3 | -37.03 |
| 2 | 59.43 | -35.39 | 69.68 | -36.63 | 76.35 | -37.51 | 75.63 | -37.47 | 79.35 | -37.86 |
| 3 | 66.59 | -36.44 | 73.31 | -37.09 | 71.24 | -36.94 | 70.51 | -36.85 | 75.08 | -37.4 |
| 4 | 84.04 | -38.43 | 77.73 | -37.72 | 69.71 | -36.62 | 70.22 | -36.65 | 69.92 | -36.8 |
| 5 | 89.93 | -39.07 | 69.32 | -36.57 | 68.4 | -36.43 | 71.08 | -36.95 | 63.81 | -35.78 |
| Delta | 30.5 | 3.68 | 8.4 | 1.15 | 7.95 | 1.08 | 5.41 | 0.81 | 15.55 | 2.08 |
| Rank | 1 | 1 | 3 | 3 | 4 | 4 | 5 | 5 | 2 | 2 |
| Bacterial | Code | Combination No. | Combination | Dye degradation% | |||||
| Lysinibacilluscapsici PB300(T) | A | 2h | 4h | 6h | 8h | 10h | 12h | ||
| 1. | A+B | 30.25 | 45.47 | 74.32 | 80.16 | 99.59 | 96.76 | ||
| 2. | A+C | 39 | 51.37 | 69.85 | 87.54 | 89.68 | 99.44 | ||
| 3. | A+D | 47.17 | 65.87 | 78.82 | 79.21 | 89.27 | 99.81 | ||
| Alcaligenesfaecalissubsp. Phenolicus DSM 16503(T) | B | 4. | B+C | 39.62 | 60.37 | 73.95 | 88.95 | 99.15 | 99.94 |
| 5. | B+D | 29.65 | 62.95 | 80.27 | 89.38 | 99.21 | 98.76 | ||
| 6. | C+D | 45.47 | 65.67 | 70.68 | 89.12 | 99.59 | 99.11 | ||
| Acinetobacterbaumanni ATCC 19606(T) | C | 7. | A+B+C | 51.4 | 61.35 | 87.81 | 99.73 | 99.82 | 100 |
| 8. | A+B+D | 40.42 | 65.6 | 87.04 | 99.56 | 99.79 | 99.46 | ||
| Pseudomonasaeruginosa JCM 5962(T) | D | 9. | A+C+D | 33.45 | 65 | 88.96 | 99.76 | 99.14 | 99.48 |
| 10. | A+B+C+D | 49.87 | 72.01 | 87.76 | 99.91 | 100 | 100 | ||
| No. | Compound | Molecular formula | R.Time (min.) | Molecular Weight (g/mol) | Peak Area % | Peak Height % |
|---|---|---|---|---|---|---|
| 1. | Butanoic acid | C5H10O2 | 3.433 | 102 | 49.38 | 23.88 |
| 2. | Pentanoic acid | C6H12O2 | 4.953 | 116 | 3.22 | 5.54 |
| 3. | Pentanoic acid | C6H12O2 | 5.345 | 116 | 0.90 | 2.85 |
| 4. | 2-Piperidinone | C5H9NO | 13.527 | 99 | 26.72 | 25.43 |
| 5. | Tridecanoic acid | C15H30O2 | 23.945 | 242 | 0.37 | 1.64 |
| 6. | 1,4-diazabicyclo[4.3.0]nonan-2,5-dione | C8H12N2O2 | 24.362 | 168 | 2.27 | 3.13 |
| 7. | Cyclo(L-prolyl-L-valine) | C10H16N2O2 | 25.498 | 196 | 8.77 | 19.45 |
| 8. | Pyrrolo[1,2-a]pyrazine-1,4-dione | C11H18N2O2 | 26.020 | 210 | 1.18 | 2.91 |
| 9. | L-Leucine | C25H47NO3 | 27.036 | 409 | 3.18 | 7.31 |
| 10. | Pyrrolo[1,2-a]pyrazine-1,4-dione | C11H18N2O2 | 27.356 | 210 | 4.01 | 7.81 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





